Abstract
T-cell intracellular antigen-1 (TIA-1) regulates developmental and stress-responsive pathways through distinct activities at the levels of alternative pre-mRNA splicing and mRNA translation. The TIA-1 polypeptide contains three RNA recognition motifs (RRMs). The central RRM2 and C-terminal RRM3 associate with cellular mRNAs. The N-terminal RRM1 enhances interactions of a C-terminal Q-rich domain of TIA-1 with the U1-C splicing factor, despite linear separation of the domains in the TIA-1 sequence. Given the expanded functional repertoire of the RRM family, it was unknown whether TIA-1 RRM1 contributes to RNA binding as well as documented protein interactions. To address this question, we used isothermal titration calorimetry and small-angle X-ray scattering (SAXS) to dissect the roles of the TIA-1 RRMs in RNA recognition. Notably, the fas RNA exhibited two binding sites with indistinguishable affinities for TIA-1. Analyses of TIA-1 variants established that RRM1 was dispensable for binding AU-rich fas sites, yet all three RRMs were required to bind a polyU RNA with high affinity. SAXS analyses demonstrated a `V' shape for a TIA-1 construct comprising the three RRMs, and revealed that its dimensions became more compact in the RNA-bound state. The sequence-selective involvement of TIA-1 RRM1 in RNA recognition suggests a possible role for RNA sequences in regulating the distinct functions of TIA-1. Further implications for U1-C recruitment by the adjacent TIA-1 binding sites of the fas pre-mRNA and the bent TIA-1 shape, which organizes the N- and C-termini on the same side of the protein, are discussed.
Keywords: RRM, protein-RNA interactions, small-angle X-ray scattering, isothermal titration calorimetry, pre-mRNA splicing, translational silencing
Introduction
The TIA-1 and the TIA-1-related (TIAR) proteins are essential, so that mice lacking TIA-1 or TIAR show high rates of embryonic lethality1; 2. TIA-1 and TIAR play dual roles in promoting premRNA splicing and repressing mRNA translation. In its capacity as a pre-mRNA splicing factor, TIA-1 activates the 5′ splice sites of transcripts including those encoding the fas apoptosis-promoting receptor3, cystic fibrosis transmembrane conductance regulator4, and Survival Motor Neuron 25 among others. To accomplish its nuclear function in pre-mRNA splicing, TIA-1 binds U-rich RNA sites located downstream of weak 5′ splice sites (Fig. 1a). There, TIA-1 engages the U1 small nuclear ribonucleoprotein (snRNP) component of the spliceosome via protein-protein interactions with its U1-C subunit6; 7. As an alternative to recruiting the U1 snRNP, TIAR has been found to enhance association of the U6 snRNP with pre-mRNA sites8; 9. In the cytoplasm, TIA-1 and TIAR regulate translation of various mRNAs including tnf-α and cox-2 mRNAs that are involved in apoptotic pathways and can be defective in inflammation and arthritis1; 10; 11. In this capacity, TIA-1 typically represses translation by binding 3′ untranslated regions marked by class II AU-rich elements (AREs), a series of partially overlapping AUUUA pentamers within a uridine-rich context (Fig. 1a)12. The mechanism of TIA-1-induced translational repression has been characterized for conditions of environmental stress, such as oxidation, heat, or starvation (reviewed in13; 14). Under these conditions, eIF2α phosphorylation inhibits translation initiation, and TIA-1 or TIAR escort the unproductive translation initiation complexes with the mRNA to stress granules15; 16; 17.
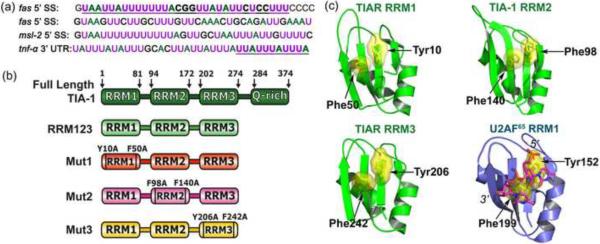
Fig. 1.
(a) Nucleotide sequence comparisons of RNA sites regulated by TIA-1, either adjacent the 5′ splice sites (SS) of fas intron 5, fas intron 6, or msl-2 intron 1, or an AU-rich region within the tnf-α 3′ UTR. Adenosines are colored green, and uridines are magenta. Oligonucleotides used in this study (underlined and in bold) were derived from the 5′ splice site of fas intron 5 for calorimetry or the minimal AU-element of tnf-α RNA for SAXS. (b) Schematic diagram of full length TIA-1 and the RRM123, Mut1, Mut2, and Mut3 constructs used in this study. (c) TIA-1 or TIAR RRM structures compared with the second U2AF65 RRM bound to a polyU-tract (TIA-1 RRM2 PDB ID 3BS9; TIAR RRM1 PDB ID 2CQI, TIAR PDB ID RRM3 1×4G; U2AF65 RRM2 PDB ID 2G4B). For clarity, the TIAR residues are numbered according to human TIA-1 sequence. The RNP1 and RNP2 consensus residues that were altered by site-directed mutagenesis are labeled in (b) and (c).
Although TIA-1 and TIAR functions are becoming better understood, much remains to be learned concerning their underlying molecular actions. The TIA-1 and TIAR proteins share 80% sequence identity and similar domain organizations of three RNA recognition motifs (RRMs) and a C-terminal, Q-rich domain (Fig. 1b)18; 19. The Q-rich domain acts as a protein-protein interaction motif to promote U1-C interactions or TIA-1 aggregation and does not contribute to the RNA affinity of TIA-17; 20. Instead, attention on the region responsible for recognizing the U-rich elements of splice sites and 3′ UTRs has focused on the RRMs of the TIA-1 or TIAR. Structures of individual TIA-1 or TIAR RRMs (RIKEN Structural Genomics Initiative and21; 22) establish that all three RRMs possess a canonical αβ fold prevalent among this class of RNA binding domain23. The TIA-1 or TIAR RRMs display consensus ribonucleoprotein (RNP1 and RNP2) residues at the expected positions for RNA binding (Fig. 1c). Indeed, the importance of the central RRM2 of TIA-1 for RNA recognition is well-established, since the isolated RRM2 domain binds the msl-2 5′ splice site region7, viral RNAs24, and SELEX RNAs25 in gel shift experiments. The C-terminal TIA-1 RRM3 also binds RNA, since the isolated RRM3 domain of TIA-1 or TIAR affinity-precipitates cellular RNAs25. In contrast, the role of the N-terminal RRM1 of TIA-1 or TIAR is less clear. In the absence of the other domains, the isolated TIA-1 or TIAR RRM1 lacks detectable RNA binding7; 25; 26, although the isolated TIAR RRM1 binds DNA26. Instead, some reports suggest that TIA-1 RRM1 primarily assists a C-terminal Q-rich domain of TIA-1 to interact with the U1 snRNP-C (U1-C) subunit, and hence stabilizes U1 snRNP association with regulated splice sites6; 7.
Here we used isothermal titration calorimetry (ITC) to investigate the role of the TIA-1 RRMs in RNA recognition, and complementary small-angle X-ray scattering (SAXS) to determine the solution shape of the TIA-1 RRM-containing domain in the presence and absence of RNA. ITC characterization of site-directed mutant proteins demonstrates that all three RRMs are required for high affinity recognition of polyU RNAs, but that RRM1 is dispensable for lower affinity binding to a natural fas pre-mRNA site. The SAXS analyses indicate that TIA-1 undergoes a conformational change in the RNA-bound state. Ab initio models of TIA-1 reveal an obtuse `V' shape positioning the amino- and carboxy-termini on the same face of the molecule; a result that has important implications for U1 snRNP recruitment to the 5′ splice site.
Results
Two identical sites for TIA-1 binding a fas RNA target
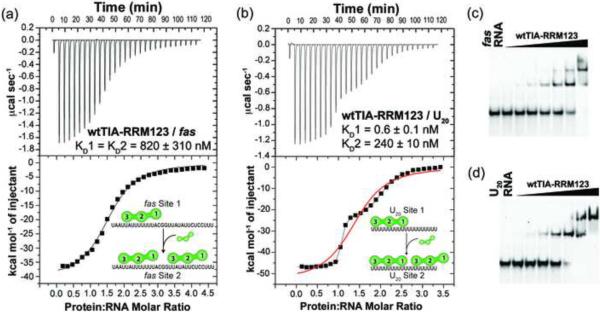
We used ITC to investigate the affinity, enthalpy and entropy changes for TIA-1 binding a target site from the fas pre-mRNA (Fig. 2, Table 1). A nearly full-length TIA-1 construct containing three RRMs (wtTIA-RRM123, residues 1–274) (Fig. 1b) was titrated into a 31-nucleotide AU-rich RNA derived from the 5′ splice site of fas exon 5 (fas RNA, Fig. 1a). Two identical binding sites for wtTIA-RRM123 were observed in the fas RNA isotherm (<χ>2 5.7E5 for identical site vs. <χ>2 2.5E5 for nonidentical site fits) (Fig. 2a), consistent with the fas RNA sequence composition of two 14-nucleotide AU-tracts separated by a `CGG' triplet (Fig. 1a). An electrophoretic mobility shift assay (EMSA) of wtTIA-RRM123 binding fluorescently-labeled fas RNA revealed two shifted bands, consistent with a 2:1 stoichiometry of proteins to RNA (Fig. 2c). The apparent equilibrium dissociation constants for wtTIA-RRM123 / fas RNA were on the weaker end of the spectrum for RRM-containing proteins (KD 0.8 ± 0.3 μM) (Table 1) – for example, the tandem RRMs of Sex-lethal or poly(A) binding protein exhibit KDs of ~100 nM for their preferred binding sites27. The TIA-1 / fas RNA binding events were driven by large, exothermic enthalpy changes and accompanied by large unfavorable entropy changes (Table 1), which under similar conditions, appears to be a thermodynamic signature of RRM-containing proteins recognizing specific RNA sites27.
Fig. 2.
Comparison of wild-type TIA-1 (wtTIA-RRM123) binding RNA sites. Representative isotherms for (a) wtTIA-RRM123 (140 μM) titrated into fas RNA (7 μM) and fit with an identical binding sites model. A nonidentical sites model produced similar quality fit. (b) wtTIA-RRM123 (70 μM) into U20 RNA (7 μM). The curve resulting from the model of nonidentical sites is shown in black, whereas that of identical sites is overlaid in red. The KD values and proposed models for each binding event are inset. The RRM domains of each TIA-1 molecule are numbered from N- to C-terminus here and throughout. The enthalpy and entropy changes of binding are given in Table 1. (c–d) Qualitative electrophoretic mobility shift assays demonstrate the formation of two complexes during titration of wtTIA-RRM123 into fluorescein-labeled (c) fas or (d) U20 RNA sites (2.5 μM). The final protein concentrations were 0.7, 1.1, 1.8, 2.9, 4.6, 11.5 and 18.5 μM for the fas RNA titration, and 0.6, 1.0, 1.6, 2.6, 6.1, 9.7 and 15.6 μM for the U20 RNA titration.
Table 1.
Affinities and thermodynamic values for RNA binding by TIA-1 variantsa
| Interaction | KD (nM) | Protein:RNA Stoichiometry | ΔH (kcal mol−1) | −TΔSb(kcal mol−1) |
|---|---|---|---|---|
| wtTIA-RRM123 → fas RNA | 820 ± 310 | 1.9 ± 0.2 | −38.9 ± 4.0 | 30.4 ± 4.2 |
| TIA-Mut1 → fas RNA | 420 ±10 | 2.0 ± 0.2 | −36.7 ± 0.4 | 27.9 ± 0.8 |
| wtTIA-RRM123 → U20 RNA Site 1 | 0.5 ± 0.1 | 1.0 ± 0.1 | −47.0 ± 0.3 | 34.2 ± 0.2 |
| Site 2 | 240 ± 10 | 1.0 ± 0.1 | −24.7 ± 0.3 | 15.5 ± 0.3 |
| U20 RNA → wtTIA-RRM123 Site 1 | 2.7 ± 1.2 | 1.2 ± 0.1 | −43.7 ± 6.8 | 31.8 ± 6.5 |
| Site 2 | 620 ± 70 | 1.2 ± 0.1 | −24.4 ± 6.7 | 15.8 ± 6.6 |
| U20 RNA → TIA-RRM23 c | 120 ± 10 | 2.2 ± 0.2 | −29.6 ± 0.5 | 20.0 ± 0.5 |
| U20 RNA → TIA-Mut1 | 180 ± 10 | 1.8 ± 0.1 | −32.6 ± 3.3 | 23.3 ± 3.3 |
| U20 RNA → TIA-Mut2 Site 1 | 130 ± 20 | 1.2 ± 0.1 | −33.3 ± 0.8 | 23.7 ± 0.8 |
| Site 2 d | 27000 ± 9000 | 0.4 ± 0.1 | −72.5 ± 0.7 | 66.2 ± 7.2 |
| U20 RNA → TIA-Mut3 Site 1 | 14.4 ± 6.1 | 1.0 ± 0.1 | −34.7 ± 1.2 | 23.8 ± 1.0 |
| Site 2 | 1270 ± 60 | 1.2 ± 0.1 | −28.0 ± 1.8 | 19.8 ± 1.8 |
Average values and standard deviations of three independent experiments.
Calculated using the equation −TΔS = ΔG − ΔH at T = 303 K, where ΔG = −RT ln (KD−1).
Values from ref. 27
Apparent stoichiometries for weak binding sites depend on the Kd, macromolecule and ligand concentrations as well as the number of binding sites (ref. 49).
Distinct high- and low-affinity sites for TIA-1 RRM123 binding a 20-uridine RNA
PolyU RNAs are the preferred binding sites of TIA-1 and TIAR25. For comparison with our previous studies of RRM-containing proteins27, we measured the enthalpy changes and affinity of wtTIA-RRM123 binding a 20-nucleotide polyU RNA (U20) (Fig. 2, Table 1). A model of two nonidentical sites with dramatically different affinities (KD1 0.5 nM and KD2 240 nM, <χ>2 8.0E4 for nonidentical site fit) fit the wtTIA-RRM123 / U20 RNA isotherm significantly better than an identical sites model (<χ>2 6.1E6 for identical site fit) (Fig. 2b). An EMSA of wtTIA-RRM123 with fluorescently-labeled U20 exhibited two shifted bands with different mobilities, consistent with the observation of two nonidentical binding sites by ITC (Fig. 2d). The affinity of wtTIA-RRM123 for the stronger U20 site was 1600-fold greater than for the fas RNA sites.
`Reverse' titrations of U20 RNA into wtTIA-RRM123 confirmed the presence of two distinct binding sites (<χ>2 1.0E5 for nonidentical site vs. 8.1E6 for identical fits) (Fig. 3a). Control titrations of wtTIA-RRM123 (Supplementary Fig. S1) or U20 RNA27 into buffer under identical conditions assured that the distinct binding sites were not produced by heats of protein or RNA dilution. The U20 RNA-into-wtTIA-RRM123 reverse titration cannot be directly compared with the forward titrations of wtTIA-RRM123-into-U20 RNA, since distinct association and/or dissociation events may contribute to the apparent thermodynamic changes for each case. Nevertheless, the signs and magnitudes of the enthalpy and entropy changes remained similar between the forward and reverse titrations, and the affinities were similar in magnitude (two- to three-fold changes, Table 1).
Fig. 3.
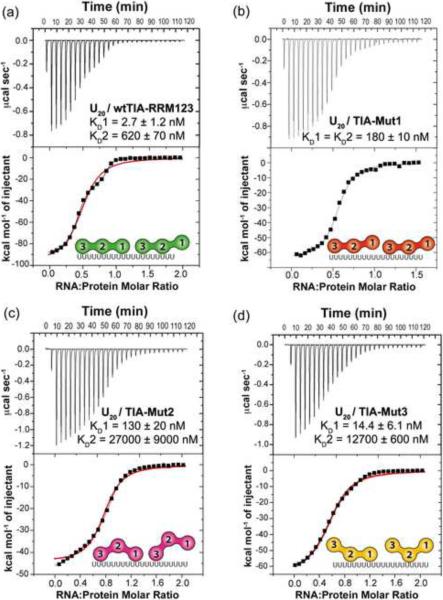
Comparison of U20 RNA binding to TIA-1 variants. Representative isotherms for the titration of (a) U20 RNA (70 μM) into wtTIA-RRM123 (7 μM), (b) U20 RNA (42 μM) into TIA-Mut1 (6 μM), (c) U20 RNA (80 μM) into TIA-Mut2 (9 μM), (d) U20 RNA (50 μM) into TIA-Mut3 (5 μM). TIA-Mut1 was fit with an identical sites model, since the fit of a nonidentical sites model was similar. The wtTIARRM123, TIA-Mut2 and TIA-Mut3 were fit with a nonidentical sites model (black line). For comparison, the fits of the identical sites models are overlaid (red line). The KD values and proposed models for the saturated RNA complexes are inset. The enthalpy and entropy changes of the titrations are given in Table 1.
The shape of the wtTIA-RRM123 / U20 isotherm indicated that TIA-1 bound the higher affinity site before the lower affinity site during the titration of protein into the RNA. This apparent negative cooperativity between TIA-1 molecules binding the U20 RNA site unexpected given that identical binding sites had been observed previously for other RRM-containing proteins binding U20 RNA27. The uniform sequence and largely unstructured nature of the U20 homopolymer28 does not offer distinct binding sites. Instead, we hypothesized that the majority of the consecutive uridines provided a high affinity binding site for association of the first TIA-1 molecule, which left a subset of uridines available for weaker association with the second TIA-1 molecule (inset model of Fig. 2b). This model is consistent with the lower affinity of wtTIA-RRM123 for the 31-nucleotide fas RNA, based on the fas RNA sequence composition of two short, uridine-rich tracts interrupted by GC-nucleotides (Fig. 1a and inset model of Fig. 2a).
TIA-1 RRM1 is required for high affinity U20 RNA binding
Among known structures of RRM/RNA complexes, the core RRM fold typically binds 3–5 nucleotides in a consistent 5′-to-3′ orientation23. The possibility that higher and lower affinity TIA-1 sites were composed of different length uridine tracts further predicted that different TIA-1 RRMs were involved in each binding event. For example, the uridines remaining exposed after association of the first TIA-1 molecule could be sufficient to bind only a few RRMs of a second molecule (Fig. 2, inset models). Previously, we had determined the enthalpy and entropy changes for U20 RNA binding by a minimal TIA-1 construct composed of the central and C-terminal RRMs (TIA-RRM23, residues 94–274), under the identical conditions to those used here (100 mM NaCl, 25 mM BES pH 7.4 at 30°C) (Table 1)27. Already, this study offered the opportunity to compare U20 RNA binding by the wtTIA-RRM123 and hence evaluate the contribution of TIA-1 RRM1. In the absence of RRM1, a model of two identical sites adequately fit the TIA-RRM23 / U20 isotherm (<χ>2 1.1E6 for identical vs. 3.6E5 for nonidentical site fits). The two TIA-RRM23 sites in the U20 RNA displayed similar affinities, enthalpy, and entropy changes to the weaker wtTIA-RRM123 / U20 RNA site (Table 1). These observations supported the idea that all three RRMs were required for the high affinity U20 RNA site, and that the nucleotides remaining exposed after association of one wtTIA-RRM123 molecule served as a weaker binding site for a subset of RRMs in a second molecule.
To further test the hypothesis that TIA-1 RRM1 selectively contributes to the higher affinity U20 site, within the context of the wtTIA-RRM123 construct, we mutated two key RRM1 residues to alanine (Y10A/F50A, TIA-1 Mut1) (Fig. 1b). These ribonucleoprotein consensus residues (RNP1 and RNP2) typically stack with the RNA bases and are important for the RNA affinities of canonical RRMs23; 29; 30; 31 (Fig. 1c). As such, the Y10A/F50A double point mutations were expected to inhibit the RNA interactions of RRM1 without affecting those of RRM2 or RRM3. For direct comparison with the previous TIA-RRM23 experiment, the U20 RNA was titrated into the mutant protein in the sample cell. The U20 RNA / TIA-Mut1 isotherm fit an identical site model (<χ>2 6.0E5 for identical vs. 4.9E5 for nonidentical site fits) (Fig. 3b). The affinity of TIA-Mut1 for the U20 RNA was comparable to that of TIA-RRM23, as well as to the weaker wtTIA-RRM123 site (two-fold affinity changes, Table 1). The observation of identical, low affinity sites for TIA-Mut1 in the U20 RNA supported a model in which RRM1 is required for high affinity RNA binding by TIA-1.
TIA-1 RRM2 and RRM3 contribute to both U20 RNA binding sites
We then proceeded to test the contributions of the central RRM2 and C-terminal RRM3 of TIA-1 to U20 RNA binding in an analogous manner as for RRM1. The consensus aromatic residues of RRM2 (F98A/F140A) or RRM3 (Y206A/F242A) RNP motifs were mutated to alanine, respectively generating TIA-Mut2 and TIA-Mut3 variants of wtTIA-RRM123 (Fig. 1b). The U20 RNA was titrated into TIA-Mut2 or TIA-Mut3 to evaluate the respective contributions of RRM2 or RRM3 to the distinct sites. Two nonidentical binding sites remained evident for U20 RNA binding by either mutant (TIA-Mut2: <χ>2 1.2E6 for identical vs. 6.6E4 for nonidentical site fits; TIA-Mut3: <χ>2 1.3E6 for identical vs. 5.7E4 for nonidentical site fits) (Fig. 3c,d). The RRM2 mutations significantly reduced the TIA-Mut2 affinities for both types of U20 RNA sites, respectively by 48- or 44-fold for the higher or lower affinity sites (Table 1). The importance of RRM2 for RNA recognition by TIA-1 was expected, since this isolated domain binds the msl-2 5′ splice site region7, viral RNAs24, and SELEX RNAs25 in gel shift experiments. Consistent with the previously established ability of TIA-1 RRM3 to affinity-precipitate cellular RNAs25, the affinities of TIA-Mut3 for the U20 RNA sites decreased by five- and two-fold respectively for the high and low affinity sites. Altogether, these results demonstrated that TIA-1 RRM2 and RRM3 both contributed to the distinct U20 RNA binding sites, although RRM2 dominated the affinity as previously suggested7. In contrast, RRM1 contributed solely to the higher affinity binding site of the wtTIA-RRM123 / U20 RNA complex.
TIA-1 RRM1 is dispensable for fas RNA binding
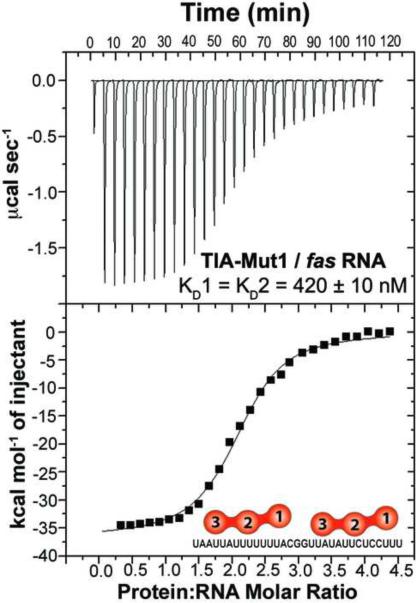
Given the comparable affinities of wtTIA-RRM123 / fas RNA, TIA-RRM23 / U20 RNA and TIA-Mut1 / U20 RNA, it appeared plausible that RRM2 and RRM3 were primarily responsible for recognizing the fas splice site. To evaluate this possibility, we used ITC to characterize TIA-Mut1 binding to the fas RNA (Fig. 4). The affinities, enthalpy, and entropy changes for fas RNA recognition were nearly unchanged by the RRM1 point mutations (<2-fold difference within experimental error). The comparable affinities of wtTIA-RRM123 and TIA-Mut1 for the fas RNA were consistent with the notion that only RRM2 and RRM3 recognize this pre-mRNA sequence. Instead, RRM1 selectively enhanced the affinity TIA-1 for long polyU tracts.
Fig. 4.
Representative isotherm for TIA-Mut1 (140 μM) titrated into fas RNA (7 μM) fit with an identical binding sites model.
TIA-1 compresses when bound to an AU-rich RNA site
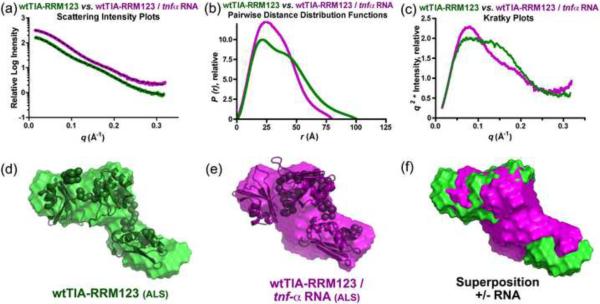
The involvement of all three RRMs in RNA recognition by TIA-1 raised the question of the arrangement of these RRMs in the three-dimensional structure of the protein and its RNA complex. To address this question, we used small-angle X-ray scattering (SAXS) to characterize the low resolution, solution shapes of wtTIA-RRM123, either in isolation or bound to a prototypical, `AU-rich' (ARE) site from the 3′ untranslated region of the tnf-α mRNA (Fig. 5). Gel shift assays confirmed that an 11-nucleotide tnf-α RNA site formed a homogeneous complex with wtTIA-RRM123 (Supplementary Fig. S3). Since the tnf-α RNA oligonucleotide contributed only 11% of the total scattering mass, the SAXS analyses of the wtTIA-RRM123 and its tnf-α RNA complex primarily reflected the conformations of the protein components. SAXS data for the wtTIA-RRM123 or the wtTIA-RRM123 / tnfα RNA complex were collected at the SIBYLS beamline of the Advanced Light Source. Three different concentrations of each sample (ranging from 1.6 to 6.7 mg / mL) were scaled and merged as described in the Experimental Methods. The samples were monodisperse by dynamic light scattering (data not shown), and the Guinier plots of the different concentrations were linear and resulted in radii of gyration (RG) that were identical within error (Supplementary Fig. S4).
Fig. 5.
Small-angle X-ray scattering data and ab initio models for wtTIA-RRM123 in the absence (green) and presence (purple) of tnf-α RNA site (5′-UUAUUUAUUUA-3′). (a) The experimental X-ray scattering profiles displaced by arbitrary units along the y-axis for clarity. The fits of theoretical scattering from the most typical ab initio models generated using DAMMIN are overlaid (dashed lines). (b) The P(r) functions were scaled to equalize the areas under the integrated functions. (c) Kratky plots were calculated after scaling the scattering intensities. The averaged and filtered DAMMIN envelopes of (d) wtTIA-RRM123 and (e) wtTIA-RRM123 / tnf-α RNA were overlaid with the most typical BUNCH rigid body models of the individual RRMs (PDB codes given in Fig. 1c), and the envelopes were superimposed in (f). The radii of gyration (Rg), maximum intraparticle sizes (Dmax), χ2 and NSD values are given in Table 2. Scattering data were collected at ALS.
In the absence of RNA, the molecular dimensions of wtTIA-RRM123 were approximately 100 Å in the maximum dimension (Dmax) and 30 Å in particle size, RG (Table 2, Fig. 5b). The asymmetric shape and distinct shoulder of the protein's pairwise distance distribution [P(r)] function suggested an elongated arrangement of the distinct RRM domains. Accordingly, the ab initio envelope of the apo-protein calculated using the program DAMMIN32 assumed an obtuse `V' shape with three distinct lobes (Fig. 5d). Although the detailed orientations of the RRMs could not be distinguished at the resolution of the SAXS data, a rigid body fit determined using the program BUNCH32 placed each of the three RRMs within a distinct lobe of the ab initio molecular envelope (NSD 1.7 Å) (Fig. 5d). Following binding to the tnf-α RNA site, the dimensions of the wtTIA-RRM123 / RNA complex appeared to decrease substantially by 20 Å in Dmax and 5 Å in RG, respectively to 80 Å and 25 Å (Table 2, Fig. 5b). Consistent with closely integrated RRMs when bound to RNA, the shoulder of the P(r) function became less prominent and shifted to a shorter distance. This could be visualized in the three dimensional, ab initio envelope of the wtTIA-RRM123 / RNA, which compressed in length but continued to assume three lobes in a subtle `V' shape. Since the contribution of RNA to the overall scattering mass was minimal, a rigid body model docked the apo-RRMs in a close configuration and gave reasonable agreement with the SAXS data (Fig. 5d, Table 2). The compaction of the wtTIA-RRM123/RNA complex also was observed in the low resolution regions of the Kratky plots, where a gradual plateau in the absence of RNA changed to a slimmer, inverted bell-shaped curve in the RNA-bound state. The absence of extended, rising tails in the high resolution regions of the Kratky plots provided evidence that wtTIA-RRM123 remained folded to a similar extent both in the absence and presence of RNA (Fig. 5c); i.e. the apparent changes in the molecular dimensions were not simply an outcome of a global order-to-disorder transition in the structure.
Table 2.
Molecular dimensions and quality indicators derived from small-angle X-ray scattering data.a
| RGg (Å) | RGP (Å) | Dmax (Å) | <χab> | <NSDab> | |
|---|---|---|---|---|---|
| Comparison +/− RNA:b | |||||
| wtTIA-RRM123 (ALS) | 27.2 ± 0.3 | 31.4 ± 0.1 | 100 ± 10 | 0.98 ± 0.02 (1.24) | 0.66 ± 0.03 (1.02) |
| wtTIA-RRM123 / tnf-α RNA | 24.7 ± 0.1 | 25.4 ± 0.1 | 80 ± 8 | 1.04 ± 0.01 (1.95) | 0.71 ± 0.04 (0.87) |
| Comparison +/− RRM1 mutations:c | |||||
| wtTIA-RRM123 (CHESS) | 30.3 ± 0.3 | 30.6 ± 0.1 | 100 ± 10 | 0.99 ± 0.01 (1.29) | 0.72 ± 0.02 (1.12) |
| TIA-Mut1 | 29.6 ± 0.3 | 30.4 ± 0.1 | 100 ± 10 | 1.04 ± 0.01 (1.00) | 0.72 ± 0.03 (1.04) |
RGg, radius of gyration value from Guinier analysis of the final merged data; RGP, radius of gyration value from P(r) analysis; Dmax, maximum size; <χab> values are the average discrepancies between the experimental data and the computed scattering calculated from ab initio models resulting from ten independent DAMMIN trials. <NSDab> is the average normalized spatial discrepancy among ten ab initio models resulting from DAMMIN. Values in parentheses are for the `most typical' (reference) BUNCH model shown in Fig. 5d–e or Fig. 7d–e.
Data collected at Advanced Light Source (ALS) beamline 12.3.1.
Data collected at Cornell High Energy Synchrotron Source (CHESS) beamline F2.
Structural response of three `RRMs' to RNA binding differs between TIA-1 and U2AF65
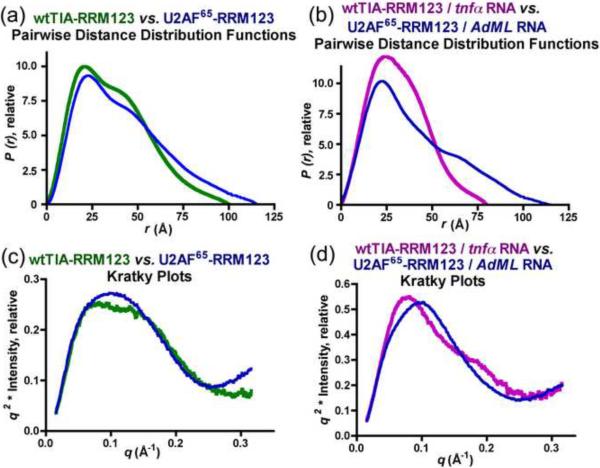
Previously, we used SAXS to characterize the structure of the RRM-containing domain of an essential pre-mRNA splicing factor, U2AF65 in the presence and absence of a 12-nucleotide RNA site33 (Fig. 6). This U2AF65 fragment (U2AF65-RRM123) bears striking parallels to wtTIA-RRM123. Both TIA-1 and U2AF65 recognize uridine-rich RNAs and contain three tandem RRM-like domains, and in both cases, one of these RRM-like domains has a major function in protein-protein interactions. The RRM-like, protein-protein interaction domain of U2AF65 is denoted a `U2AF homology motif' (UHM) to reflect its interactions with a protein partner SF1 in lieu of RNA. Although RRM1 of TIA-1 lacks sequence features of a UHM, a parallel role in U1-C rather than RNA interactions had been proposed7.
Fig. 6.
Small-angle X-ray scattering data for wtTIA-RRM123 in the absence (green) or presence (purple) of the tnf-α RNA site respectively compared with U2AF65 RRM123 in the absence or presence of its RNA site (AdMl Py tract, 5′-UCCCUUUUUUUUCC-3′)32. (a, b) Pairwise distance distribution functions and (c, d) Kratky plots.
The availability of SAXS data under identical conditions for TIA-1, U2AF65, and their RNA complexes allowed us to compare their solution shapes at low resolution (Fig. 6). The act of RNA-binding provoked little or no apparent changes in the P(r) functions and Kratky plots of U2AF65-RRM123, unlike the altered shape and dimensions of the RNA-bound compared with the apo-wtTIA-RRM123. Since the `UHM' of U2AF65-RRM123 lacks detectable RNA interactions in isolation, the absence of RNA binding by this domain could mask subtle changes in the U2AF65-RRM123 / RNA structure. In contrast with the minimal changes following association of U2AF65-RRM123 with RNA, the act of RNA binding reduces the dimensions of a ternary U2AF65-RRM123 / SF1 / RNA complex by approximately 2 Å in RG and 15 Å in Dmax33. Since U2AF65 and SF1 recognize adjoining sequences of the target RNA, this outcome is expected from the context of the U2AF65-RRM123 / SF1 / RNA complex locking the U2AF65 UHM adjacent to the SF1-bound RNA site. By analogy with the U2AF65-RRM123 / SF1 / RNA complex, the structural changes observed in the wtTIA-RRM123 / tnf-α RNA complex compared with apo-wtTIA-RRM123 support the conclusion that all three TIA-1 RRMs are involved in high affinity RNA recognition.
Similar TIA-1 conformation following mutation of RRM1 RNP motifs
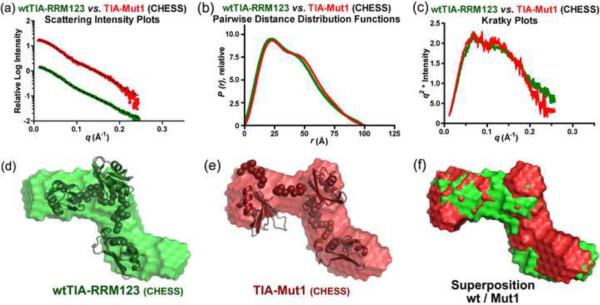
It remained possible that RRM1 influenced the U20 RNA affinity of TIA-1 indirectly by positioning RRM2 and RRM3 rather than directly by contacting the RNA. To address this question, we compared SAXS analyses of TIA-Mut1 with wtTIA-RRM123 (Fig. 7). The TIA-Mut1 SAXS data was collected at the F2 beamline of the Cornell High Energy Synchrotron Source (CHESS). To avoid possible differences due to the experimental setup (buffer corrections and/or resolution), the wtTIA-RRM123 data also was recollected at the F2 beamline. The CHESS data was collected, scaled and merged in an analogous manner as for the wtTIA-RRM123 and wtTIA-RRM123 / tnf-α RNA samples at ALS, with the exception of beamline-specific hardware, exposure times, and software for buffer corrections (as described in the Methods). As for the ALS data, similar RG values were derived from the Guinier plots at three different concentrations, confirming that both the wild-type and mutant data were free of interparticle effects (Supplementary Fig. S4). The wtTIA-RRM123 data collected at CHESS was a reasonable match with the wtTIA-RRM123 data collected at ALS (Supplementary Fig. S5) apart from a decrease in the maximum resolution (q 0.25 Å−1 vs. 0.32 Å−1, respectively) (Fig. 7a).
Fig. 7.
Small-angle X-ray scattering data for wtTIA-RRM123 (green) compared TIA-Mut1 (red). (a) X-ray scattering plots, (b) pairwise distance distribution functions, and (c) Kratky plots are shown. The averaged and filtered DAMMIN envelopes of (d) wtTIA-RRM123 and (e) TIA-Mut1 are overlaid with the most typical BUNCH rigid body models of the individual RRMs (PDB codes given for Fig. 1c), or (f) superimposed. The radii of gyration (Rg), maximum intraparticle sizes (Dmax), χ2 g and NSD values are given in Table 2. Scattering data were collected at CHESS.
The RG and Dmax dimensions of TIA-Mut1 remained indistinguishable from those of wtTIA-RRM123 within the error of measurement (Table 2). The features of the P(r) functions and Kratky plots appeared unchanged between the TIA-Mut1 and wtTIA-RRM123 samples (Fig. 7b,c). Likewise, the ab initio molecular envelopes and rigid body placement of the RRMs closely matched between the wild-type and mutant proteins (NSD 0.57 between DAMFILT models) (Fig. 7d–f). Thus, the RNP mutations of RRM1 had no detectable effect on the relative positioning of the three RRM domains, and instead were likely to contribute to RNA binding directly as typical among canonical RRMs.
Discussion
RRMs are among the most prevalent RNA interaction motifs of eukaryotic proteins (reviewed in23). The structures of canonical RRM-containing proteins share salient features, and conserved aromatic RNP residues of the β-strands generally stack with the RNA bases in a consistent 5′-to-3′ direction of the RNA strand relative to the RRM fold. We resolved distinct binding sites using ITC, a technique with sufficient sampling, and investigated the contributions of TIA-1 RRMs to RNA recognition using point mutations of well-characterized RNP residues (for example29; 34; 35). We confirmed that as expected for canonical RRMs, the RNP mutations of TIA-1 RRM2 or RRM3 reduced the affinity of the intact RRM-containing domain for both types of nonidentical binding sites within the U20 RNA. Since isolated TIA-1 RRM2 or RRM3 domains have been shown to associate with cellular RNAs25, these RNP mutations of RRM2 or RRM3 presumably interfere with RNA contacts at both types of U20 RNA site. Using this approach, we further demonstrated that TIA-1 RRM1 significantly contributes to a high-affinity U20 RNA site. Mutation or deletion of TIA-1 RRM1 decreased association with the high affinity U20 site to a similar level as a second, weak binding site. In agreement with this finding, it has been previously noted that the TIAR RRM123 domain (which shares 88% sequence identity with the TIA-1 RRM123) binds a population of uridine-rich SELEX RNAs with higher affinity than TIAR fragments composed of only RRM2 and RRM3 (RRM23)25.
As the numbers of known structures increase, RRM subclasses have emerged with similar folds but specialized features for protein-protein rather than protein-RNA interactions. In one subclass, which is exemplified by the Y14 / mago nashi and Upf2 / Upf3 complexes36; 37; 38, the aromatic RNP residues are conserved but engaged in contacts with a neighboring protein rather than RNA. In the `UHM' subclass, which is exemplified by U2AF subunits39; 40, the RNP residues are degenerate and secondary sites have evolved for protein contacts. In addition, a role analogous to that of the UHM in protein-protein interactions had been previously proposed for TIA-1 RRM17 based on the complementary observations that TIA-1 RRM1 lacks detectable RNA binding in isolation7; 25; 26, yet can enhance U1-C recruitment when flanked by a C-terminal Q-rich domain7. The results presented here support a direct role for TIA-1 RRM1 in U20 RNA interactions. Given that TIA-1 RRM1 lacks detectable RNA affinity when in isolation7; 25, we conclude that RRM1 must recognize RNA synergistically with RRM2 and RRM3. Crystal structures have shown that RNP motifs can mediate protein-protein interactions between distinct RRMs of the same polypeptide41; 42, although it is not clear if these conformations represent the major population in solution43; 44. SAXS analyses of the TIA-Mut1 protein demonstrated that the average distances separating the TIA-1 RRMs remain unchanged with RRM1 mutations. Therefore, the RNP residues of RRM1 are likely to contribute to RNA affinity through direct contacts, rather than by indirectly perturbing the positions of the RRMs relative to the RNA. In support of the hypothesis of direct RNA interactions, the RNP residues of TIA-1 are similar to those of canonical, RNA-binding RRMs (for example, Fig. 1c).
The similar association of TIA-1 with RRM1 mutations to the lower affinity U20 RNA site argues that TIA-1 RRM1 is not involved in binding this weak site, but rather selectively contributes to the high affinity U20 RNA site. A model for this interaction based on the known footprint of ~4 nucleotides and directional RNA binding for canonical RRMs is shown in the inset of Figure 2b and in Figure 8a. In this model, all three RRMs of TIA-1 contribute to the high affinity U20 site. In addition, the affinity of TIA-1 for the strong site is substantially reduced by RNP mutations in any of the RRMs. Consistent with a typical binding site of approximately four nucleotides per RRM, the uridines that remain exposed after the first TIA-1 molecule has bound would be sufficient to accommodate only a subset of the TIA-1 RRMs, and as such would constitute the lower affinity binding site. Given the consistent 5′-to-3′ direction of an RNA strand relative to the RRM fold and the steric restrictions of linked RRMs, either terminal TIA-1 RRM (RRM1 or RRM3) could be formally excluded from the second binding site. Accordingly, the impact of RRM3 mutations on the weak U20 site is less than that on the strong site (a decrease in affinity by five- and two-fold, respectively), suggesting that RRM2-RRM3 binding to the weak U20 site is exchanged with RRM1-RRM2 some of the time. However, TIA-Mut1 or RRM23 affinities for this weak U20 site are not inhibited, and are enhanced three-fold following RRM1 mutation or deletion, which is consistent with the predominant occurrence of RRM2-RRM3 interactions at this site. In agreement with these findings, TIA-1 RRM1 has the weakest RNA affinity of the three RRMs when studied in isolation7; 25; 26. Taken together, we propose a model whereby RRM2 and RRM3 of TIA-1 contact the second U20 (Figures 2b and 8a).
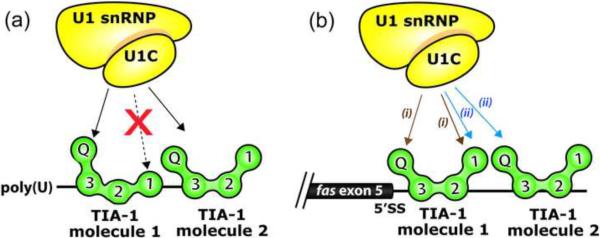
Fig. 8.
Models for U1-C interactions with TIA-1. The TIA-1 RRM1 contributes to TIA-1 binding to long uridine tracts (a), but is dispensable for binding tandem AU-tracts such as those following fas exon 5 (b). The latter case may free RRM1 to interact with other molecules such as U1-C. The `V' shape of TIA-1 aligns the N- and C-terminal domains along one side of the molecule, consistent with the intramolecular participation of both domains in U1-C interactions7 as schematically diagrammed with the (i) brown arrows. The association of two TIA-1 molecules with the AU-rich sequences following fas exon 5 leads to an alternative model for U1-C recruitment to the fas 5′ splice site via intermolecular interactions with TIA-1, as indicated by (ii) blue arrows.
In a distinct manner from the higher affinity U20 RNA site, the RRM1 mutations had little effect on the affinities and thermodynamic characteristics for TIA-1 binding the fas RNA. The affinity of TIA-1 for the natural fas pre-mRNA sequence was comparable to that for the lower affinity U20 site without mutation or deletion of RRM1. Differences between the fas and high-affinity U20 RNA sites are unlikely to simply result from avidity effects due to higher local concentrations of RNA sites, since the fas RNA is longer than the U20 RNA. Instead, based on the binding data presented here, we propose that RNA sequences with distinct uridine content would engage TIA-1 RRM1 to different extents, and as such have the potential to regulate TIA-1 interactions during translation or pre-mRNA splicing. Extensive polyU tracts would engage RRM1 in the RNA-bound state, whereas G/C interruptions would leave RRM1 available to interact with other proteins and/or RNA cofactors. Accordingly, the AU-tracts of well-characterized TIA-1 target sites, such as the tnfα 3′ UTR, fas, and msl-2 splice sites shown in Figure 1a, are interrupted by different numbers and locations of G/C base pairs.
The C-terminal Q-rich domain of TIA-1 interacts with U1-C, and these interactions are enhanced by the presence of the N-terminal RRM16; 7. Proximity in the three-dimensional structure of TIA-1 would enable both of these domains to interact concurrently with U1-C, despite linear separation in the TIA-1 polypeptide. We observe a bent `V' shape by SAXS that positions the N- and C-termini towards the same face of the TIA-1 molecule (Fig. 5). Moreover, the consistent 5′-to-3′ direction of canonical RRMs bound single-stranded RNAs23 would place the N-terminal domain of one TIA-1 molecule adjacent the C-terminal domain of a second TIA-1 molecule bound to the fas splice site. As such, U1-C could interact either with a single TIA-1 molecule (left arrows labeled (i) in Fig. 8b) or simultaneously with two TIA-1 molecules bound to adjacent sites within the 31-nucleotide regulated fas segment (right arrows labeled (ii) in Fig. 8b). In summary, we have demonstrated the selective involvement of RRM1 in recognizing polyU sites, identified a `V' shape of the TIA-1 RRM-containing domain, and described the association of two TIA-1 molecules with the fas RNA site. Taken together, these findings suggest compelling means for the synergistic action of the RRM1 and Q-rich domains of TIA-1 during regulation of RNA sites.
Materials and Methods
Protein expression and purification
Human wtTIA-RRM123 (residues 1–274) was subcloned from full length TIA-1 (NCBI Refseq NP_071320) into pGEX-6p-2. Double mutations Y10A/F50A, F98A/F140A or Y206A/F242A respectively generated the TIA-Mut1, Mut2, and Mut3 constructs. The glutathione-S-transferase (GST) fusion proteins were expressed in Escherichia coli BL21 Rosetta-2 cells (Merck Novagen), purified by glutathione affinity chromatography, and the GST tag was cleaved overnight. A second round of affinity chromatography removed the cleaved GST and the TIA-1 fragment was purified further by cation-exchange chromatography using an SP HiTrap column (GE Healthcare). Proteins used for SAXS data collection were purified further by size exclusion chromatography as described below. Synthetic RNAs were purchased from Dharmacon, Inc. and deprotected according to the manufacturer's instructions.
Electrophoretic mobility shift assay
TIA-1 proteins and fluorescently labeled RNA (final concentrations indicated in Figure 2 and Supplementary Figure S2 and S3 legends) were incubated in binding buffer containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.4, 50 mM KCl, 8 mM MgCl2, 5% (v/v) glycerol, and 1U of SUPERase-In (Applied Biosystems) on ice for 15–30 minutes. Binding reactions (10 μl final volume) were then electrophoresed in 0.5× TBE (45 mM Tris-borate, 1 mM ethylenediaminetetraacetic acid) through an 8% polyacrylamide gel (acrylamide to bis-acrylamide 37.5:1, w/w) at 10 V/cm for approximately 5 hours at 4°C. Gels were visualized using a STORM 860 (Molecular Dynamics) to detect blue fluorescence (excitation wavelength, 490 nm) and processed using ImageQuant (Molecular Dynamics).
Isothermal titration calorimetry
The wtTIA-RRM123 and RNP variants were dialyzed overnight into 100 mM NaCl, 25 mM N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES) pH 7.4, 0.2 mM tris(2-carboxyethyl)phosphine (TCEP) prior to ITC experiments. The heats of binding were measured using a VP-ITC calorimeter (GE Healthcare, Inc.) with a stirring rate of 310 rpm at 30°C. Except for wtTIA-RRM123 titration into U20 RNA, the isotherms were corrected for the heats of protein or RNA dilution by subtracting the average of 3–5 data points from the saturated tail of the titration. Because a limited number of data points were available at saturation, the titration of TIA-1 RRM123 into U20 RNA was corrected by subtracting the y-intercept from the linear regression of a protein-into-buffer titration (Supplementary Fig. S1). The isotherms were fit using the least-squares method of the Origin v7.0 software (GE Healthcare, Inc.). Protein and RNA concentrations are given in the figure legends.
SAXS data collection and analyses
Samples for SAXS data collection were purified by size exclusion chromatography (Superdex-75, GE Healthcare) in 25 mM HEPES pH 7.4, 3% glycerol, 0.2 mM TCEP and either 300 mM NaCl in protein-only samples or 200 mM NaCl in samples containing proteins and RNA. To prepare the wtTIA-RRM123 / RNA complex for SAXS, an equimolar amount of the 11-nucleotide tnf-α RNA (sequence 5′-UUAUUUAUUUA-3′) was added to wtTIA-RRM123 and the complex was concentrated using a 2 kDa molecular weight cut-off filter (Sartorius). The final concentrations and the presence of RNA in the final complexes were respectively estimated and verified by the ratios of absorbance at 260 nm and 280 nm compared with standards of known wtTIA-RRM123 / tnf-α RNA composition (A260:A280 ratios of 1.8:1.0 for a one-to-one complex, compared with 0.6:1.0 for the protein only and 2.8:1.0 for the RNA only, data not shown). The filtrate was recovered as a blank for SAXS data correction. All samples were monodisperse by dynamic light scattering in the indicated buffers (data not shown).
SAXS data for wtTIA-RRM123 and wtTIA-RRM123 / tnf-α RNA were collected at the SIBYLS beamline 12.3.1 of the Advanced Light Source (Lawrence Berkeley National Laboratory) using a MARCCD X-ray detector located 1.6 m from a sample chamber to collect data in the q-spacing 0.01−0.32 Å−1, where q =4π sinθ /λ (2θ is the scattering angle and λ = 1.03 Å is the wavelength). SAXS data for wtTIA-RRM123 and TIA-Mut1 were collected at the F2 beamline of the Cornell High Energy Synchrotron Source using ADSC Quantum-210 X-ray detector located 0.8 m from a sample chamber to collect data in the q-spacing 0.01−0.25 Å−1 (λ = 1.25 Å). All samples were cooled at 10° C during data collection. Short and long exposures of each sample at a given concentration (6 s and 60 s for ALS data; 3 min and 9 min for CHESS data) were scaled and merged using the program PRIMUS 45. Three different concentrations were merged following comparison to rule out possible interparticle effects based on Guinier approximations of the radii of gyration (RgG from q < 1.3/Rg) 46 (Supplementary Fig. S4). All SAXS data were analyzed using programs from the ATSAS suite (http://www.emblhamburg.de/ExternalInfo/Research/Sax/software.html). Since the single-chain model assumed by GASBOR would be inappropriate for a protein / RNA complex, DAMMIN was used for ab initio modeling 32. Rigid body models for the individual RRMs (PDB IDs 3BS9, 2CQI, 1X4G), connected by ab initio models of the interdomain linkers, were positioned in a manner consistent with the scattering data using the program BUNCH 47. Superpositions shown in the figures used the program SUPCOMB 48.
Supplementary Material
Three tandem RRMs are required for TIA-1 to bind a polyU RNA with high affinity.
The N-terminal RRM is not required for TIA-1 to bind a fas splice site RNA.
The average solution conformation of three TIA-1 RRMs is an obtuse `V' shape.
The tandem TIA-1 RRMs become more compact when bound to RNA.
Acknowledgements
J. Valcárcel graciously shared the TIA-1 cDNA as the basis for expression plasmid construction. We thank M. Swenson and A.O. Kumar for preliminary ITC experiments, Drs. G.L. Hura and R. Gillilan for assistance with SAXS data collection, and Drs. J.E. Wedekind, J.F. Kielkopf, and M. Sattler for insightful discussions. This work was funded by a grant from the National Institutes of Health (NIH R01 GM070503) to C.L.K. SAXS data collected at the SIBYLS beamline of the Advanced Light Source, Lawrence Berkeley National Laboratory, were supported in part by the U.S. Department of Energy (DOE) program Integrated Diffraction Analysis Technologies and the DOE program Molecular Assemblies Genes and Genomics Integrated Efficiently under contract number DE-AC02-05CH11231. SAXS data collected at the F2 beamline of the Cornell High Energy Synchrotron source were supported by the National Science Foundation award DMR-0936384 and the NIH/NCRR award RR-01646.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers SAXS data and models will be made accessible through BIOISIS† using the following IDs: 1TIAKP, wtTIA-RRM123 (ALS data); 1TIAKX, wtTIA-RRM123/tnf-α RNA (ALS data); 2TIAKP, wtTIA-RRM123 (CHESS data); 1TM1KP, TIA-Mut1 (CHESS data). † http://www.bioisis.net
Supplementary Data Supplementary data to this article can be found online at …
References
- 1.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck AR, Miller IJ, Anderson P, Streuli M. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci U S A. 1998;95:2331–2336. doi: 10.1073/pnas.95.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Zuccato E, Buratti E, Stuani C, Baralle FE, Pagani F. An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J Biol Chem. 2004;279:16980–16988. doi: 10.1074/jbc.M313439200. [DOI] [PubMed] [Google Scholar]
- 5.Singh NN, Seo J, Ottesen EW, Shishimorova M, Bhattacharya D, Singh RN. TIA-1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol Cell Biol. 2010 doi: 10.1128/MCB.00945-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gesnel MC, Theoleyre S, Del Gatto-Konczak F, Breathnach R. Cooperative binding of TIA-1 and U1 snRNP in K-SAM exon splicing activation. Biochem Biophys Res Commun. 2007;358:1065–70. doi: 10.1016/j.bbrc.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 7.Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol Cell Biol. 2003;23:5959–5971. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol Cell Biol. 2008;28:1240–51. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD, Prescott SM. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J Exp Med. 2003;198:475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes R, Alcalde J, Izquierdo JM. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009;10:R87. doi: 10.1186/gb-2009-10-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–70. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 13.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–21. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–6. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 15.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–68. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschald OR, Malec V, Krasteva G, Hasan D, Kamlah F, Herold S, Rose F, Seeger W, Hanze J. TIAR and TIA-1 mRNA-binding proteins co-aggregate under conditions of rapid oxygen decline and extreme hypoxia and suppress the HIF-1alpha pathway. J Mol Cell Biol. 2010;2:345–56. doi: 10.1093/jmcb/mjq032. [DOI] [PubMed] [Google Scholar]
- 18.Beck AR, Medley QG, O'Brien S, Anderson P, Streuli M. Structure, tissue distribution and genomic organization of the murine RRM-type RNA binding proteins TIA-1 and TIAR. Nucleic Acids Res. 1996;24:3829–3825. doi: 10.1093/nar/24.19.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami A, Tian Q, Streuli M, Poe M, Edelhoff S, Disteche CM, Anderson P. Intron-exon organization and chromosomal localization of the human TIA-1 gene. J Immunol. 1994;152:4937–4945. [PubMed] [Google Scholar]
- 20.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–98. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwasako K, Takahashi M, Tochio N, Abe C, Tsuda K, Inoue M, Terada T, Shirouzu M, Kobayashi N, Kigawa T, Taguchi S, Tanaka A, Hayashizaki Y, Guntert P, Muto Y, Yokoyama S. Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode. Biochemistry. 2008;47:6437–50. doi: 10.1021/bi7024723. [DOI] [PubMed] [Google Scholar]
- 22.Kumar AO, Swenson MC, Benning MM, Kielkopf CL. Structure of the central RNA recognition motif of human TIA-1 at 1.95A resolution. Biochem Biophys Res Commun. 2008;367:813–9. doi: 10.1016/j.bbrc.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Li Y, Kedersha N, Anderson P, Emara M, Swiderek KM, Moreno GT, Brinton MA. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J Virol. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dember LM, Kim ND, Liu KQ, Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 26.Suswam EA, Li YY, Mahtani H, King PH. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005;33:4507–4518. doi: 10.1093/nar/gki763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaughlin KJ, Jenkins JL, Kielkopf CL. Large favorable enthalpy changes drive specific RNA recognition by RNA recognition motif proteins. Biochemistry. 2011;50:1429–1431. doi: 10.1021/bi102057m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inners LD, Felsenfeld G. Conformation of polyribouridylic acid in solution. J Mol Biol. 1970;50:373–89. doi: 10.1016/0022-2836(70)90199-3. [DOI] [PubMed] [Google Scholar]
- 29.Chi SW, Muto Y, Inoue M, Kim I, Sakamoto H, Shimura Y, Yokoyama S, Choi BS, Kim H. Chemical shift perturbation studies of the interactions of the second RNA-binding domain of the Drosophila sex-lethal protein with the transformer pre-mRNA polyuridine tract and 3′ splice-site sequences. Eur J Biochem. 1999;260:649–60. doi: 10.1046/j.1432-1327.1999.00157.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee AL, Volkman BF, Robertson SA, Rudner DZ, Barbash DA, Cline TW, Kanaar R, Rio DC, Wemmer DE. Chemical shift mapping of the RNA-binding interface of the multiple-RBD protein sex-lethal. Biochemistry. 1997;36:14306–17. doi: 10.1021/bi970830y. [DOI] [PubMed] [Google Scholar]
- 31.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–86. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta A, Jenkins JL, Kielkopf CL. RNA induces conformational changes in the SF1-U2AF65 splicing factor complex. J Mol Biol. 2010;405:1128–1138. doi: 10.1016/j.jmb.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 35.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis of polypyrimidine tract recognition by the essential pre-mRNA splicing factor, U2AF65. Mol Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–9. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 37.Shi H, Xu RM. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev. 2003;17:971–6. doi: 10.1101/gad.260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–7. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 39.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–26. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 41.Crichlow GV, Zhou H, Hsiao HH, Frederick KB, Debrosse M, Yang Y, Folta-Stogniew EJ, Chung HJ, Fan C, De la Cruz EM, Levens D, Lolis E, Braddock D. Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition. EMBO J. 2008;27:277–89. doi: 10.1038/sj.emboj.7601936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae E, Reiter NJ, Bingman CA, Kwan SS, Lee D, Phillips GN, Jr., Butcher SE, Brow DA. Structure and interactions of the first three RNA recognition motifs of splicing factor prp24. J Mol Biol. 2007;367:1447–58. doi: 10.1016/j.jmb.2007.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin-Tumasz S, Reiter NJ, Brow DA, Butcher SE. Structure and functional implications of a complex containing a segment of U6 RNA bound by a domain of Prp24. RNA. 2010;16:792–804. doi: 10.1261/rna.1913310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins JL, Shen H, Green MR, Kielkopf CL. Solution conformation and thermodynamic characteristics of RNA binding by the splicing factor U2AF65. J Biol Chem. 2008 doi: 10.1074/jbc.M806297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crys. 2003;36:1277–1282. [Google Scholar]
- 46.Guinier A, Fournet G. Small-angle scattering of X-rays. Wiley Interscience; New York: 1955. [Google Scholar]
- 47.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–50. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Cryst. 2000;34:33–41. [Google Scholar]
- 48.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–7. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.