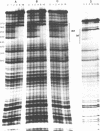
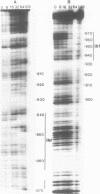
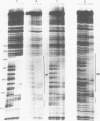
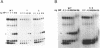Abstract
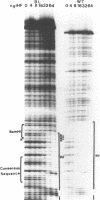
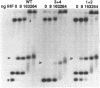
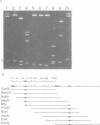
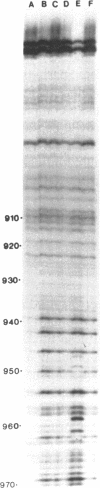
In bacteriophage Mu the converging early and repressor transcriptions are both stimulated by binding of IHF to the same region, which is located just upstream of the early promoter (Pe) and 100 base pairs downstream of the repressor promoter (Pc). Within this region two sequences are present (ihfa and ihfb) that match the consensus sequence for IHF binding. These sequences are partially overlapping and in inverted orientation. In this paper we describe the effect of mutations in the non-overlapping part of ihfa and ihfb on the binding of IHF. We show that IHF has a very strong preference to bind to ihfb even when a mutated ihfa has a better match with the consensus. A stretch of A residues located nine base pairs from the ihfb sequence appears to play an important role in the stability of the DNA-IHF complex, but not in the discrimination between the two putative binding sites. In addition we describe the effect of the mutations on the stimulation of early and repressor transcription. We show that for activation of the Pc promoter a stable complex between IHF and the DNA is required, whereas for normal Pe stimulation a much weaker DNA-IHF interaction is sufficient.
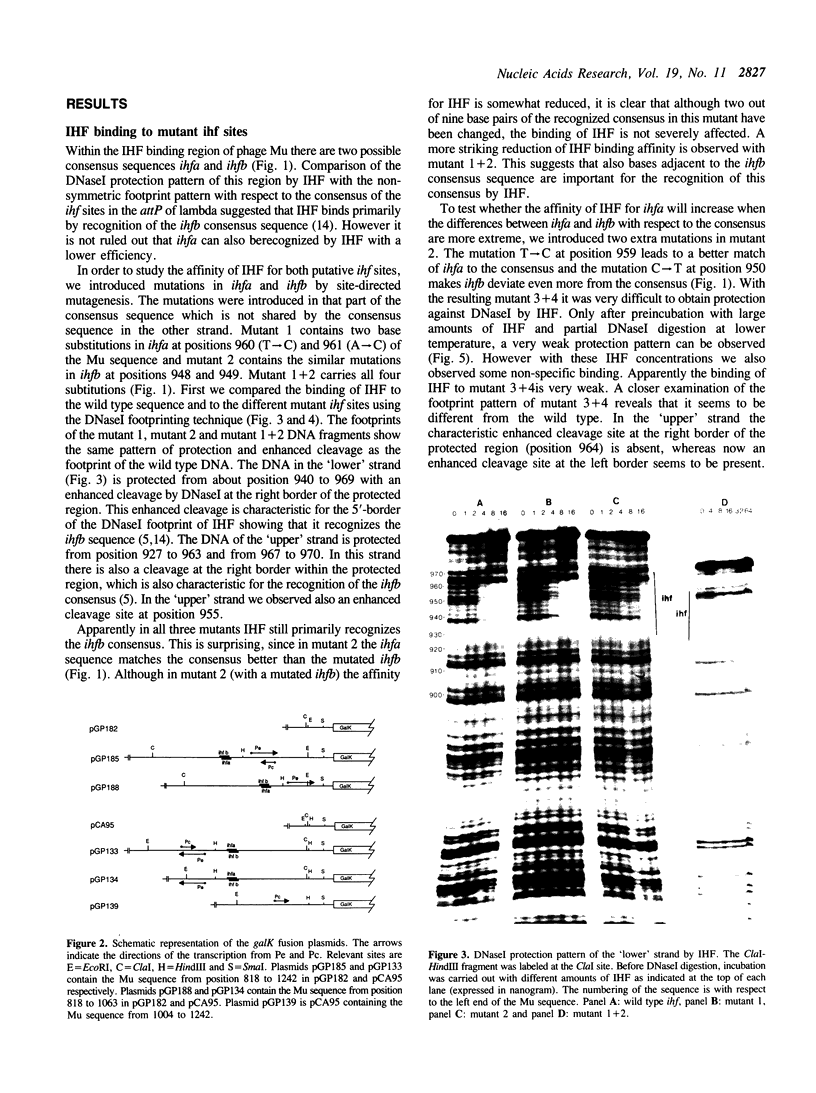
Full text
PDF





Images in this article
Selected References
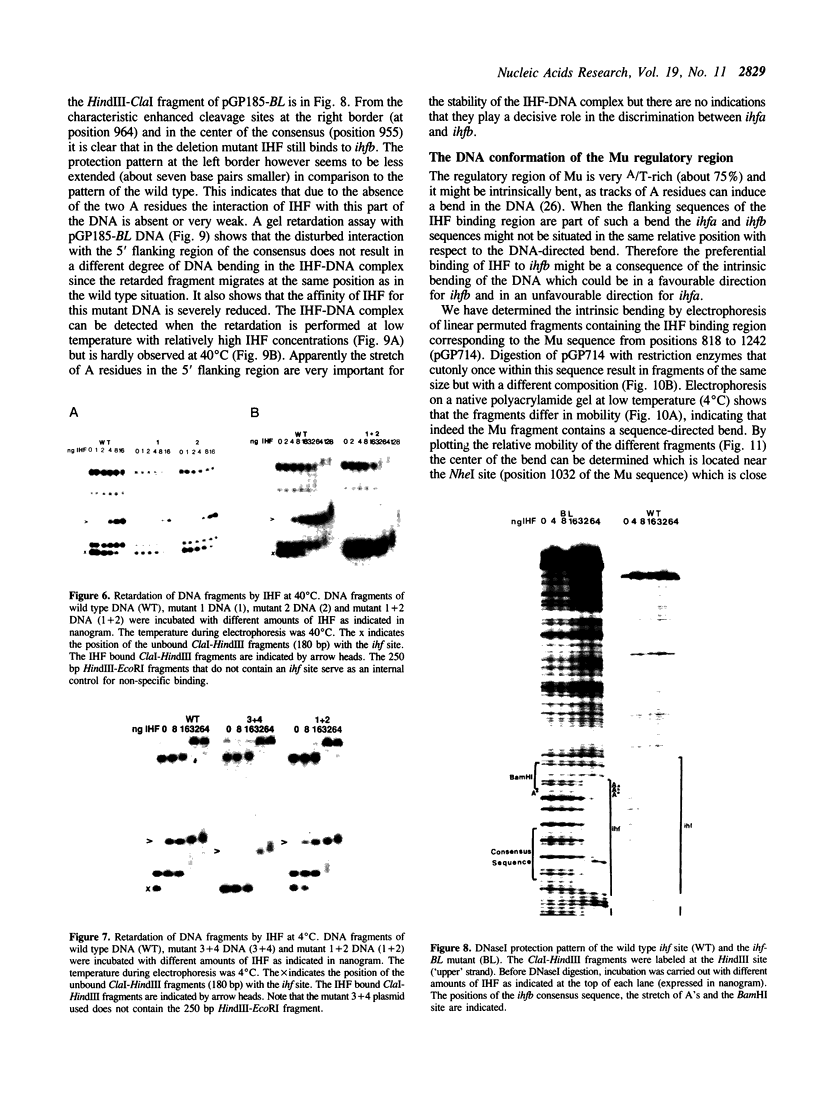
These references are in PubMed. This may not be the complete list of references from this article.
- Bonnefoy E., Rouvière-Yaniv J. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 1991 Mar;10(3):687–696. doi: 10.1002/j.1460-2075.1991.tb07998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. E. coli integration host factor binds to specific sites in DNA. Cell. 1984 Dec;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
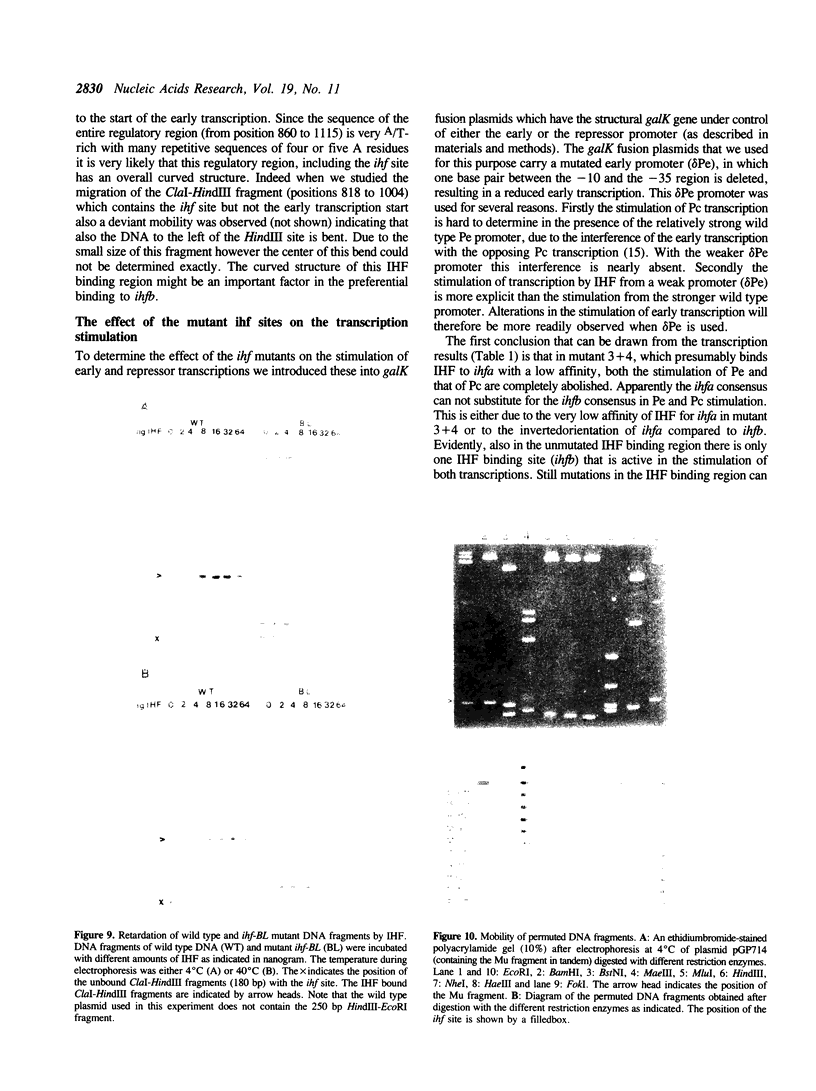
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
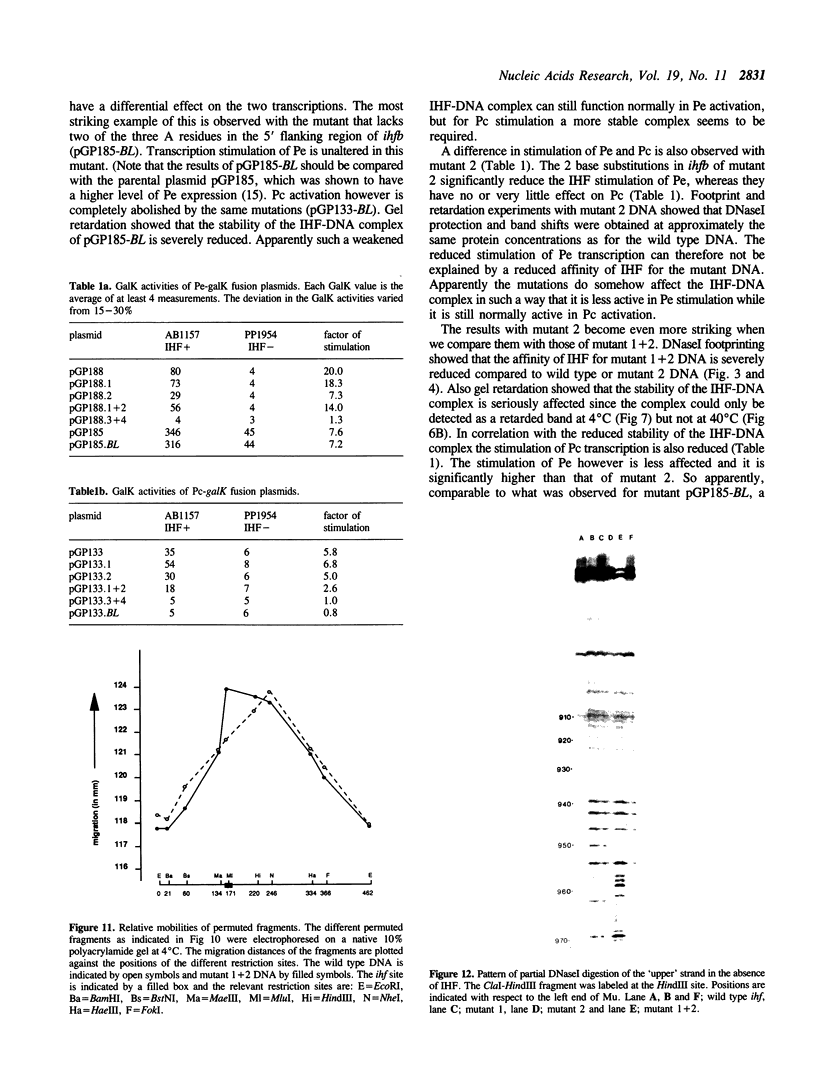
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Gamas P., Chandler M. G., Prentki P., Galas D. J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987 May 20;195(2):261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., van Heuvel M., Moolenaar G. F., van de Putte P. Regulation of Mu transposition. II. The escherichia coli HimD protein positively controls two repressor promoters and the early promoter of bacteriophage Mu. Gene. 1984 Dec;32(3):419–426. doi: 10.1016/0378-1119(84)90017-9. [DOI] [PubMed] [Google Scholar]
- Groenen M. A., Kokke M., van de Putte P. Transposition of mini-Mu containing only one of the ends of bacteriophage Mu. EMBO J. 1986 Dec 20;5(13):3687–3690. doi: 10.1002/j.1460-2075.1986.tb04700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Collier D. A., Kilpatrick M. W., Krause H. M. Supercoiling and integration host factor change the DNA conformation and alter the flow of convergent transcription in phage Mu. J Biol Chem. 1989 Feb 15;264(5):3035–3042. [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. C., MacWilliams M. P., Gumport R. I., Gardner J. F. Genetic analysis of Escherichia coli integration host factor interactions with its bacteriophage lambda H' recognition site. J Bacteriol. 1991 Jan;173(2):609–617. doi: 10.1128/jb.173.2.609-617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Miller H. I. Primary structure of the himA gene of Escherichia coli: homology with DNA-binding protein HU and association with the phenylalanyl-tRNA synthetase operon. Cold Spring Harb Symp Quant Biol. 1984;49:691–698. doi: 10.1101/sqb.1984.049.01.078. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet. 1981;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Galas D. J. Escherichia coli integration host factor bends the DNA at the ends of IS1 and in an insertion hotspot with multiple IHF binding sites. EMBO J. 1987 Aug;6(8):2479–2487. doi: 10.1002/j.1460-2075.1987.tb02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- van Rijn P. A., Goosen N., Turk S. C., van de Putte P. Regulation of phage Mu repressor transcription by IHF depends on the level of the early transcription. Nucleic Acids Res. 1989 Dec 25;17(24):10203–10212. doi: 10.1093/nar/17.24.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn P. A., Goosen N., van de Putte P. Integration host factor of Escherichia coli regulates early- and repressor transcription of bacteriophage Mu by two different mechanisms. Nucleic Acids Res. 1988 May 25;16(10):4595–4605. doi: 10.1093/nar/16.10.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sluis C. A., Moolenaar G. F., Backendorf C. Regulation of the uvrC gene of Escherichia coli K12: localization and characterization of a damage-inducible promoter. EMBO J. 1983;2(12):2313–2318. doi: 10.1002/j.1460-2075.1983.tb01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]