Abstract
We used the mitochondrial control region and nuclear microsatellites to assess the distribution patterns, population structure, demography and landscape genetics for the hedgehogs Erinaceus europaeus and Erinaceus roumanicus in a transect of the mid-European zone of sympatry. E. roumanicus was less frequent and restricted to regions with lower altitudes. Demographic analyses suggested recent population growth in this species. A comparison of patterns in the spatial variability of mitochondrial and nuclear DNA indicated less sex-biased dispersal and higher levels of gene flow in E. roumanicus. No evidence of recent hybridisation or introgression was detected. We interpreted these results by comparing with phylogeographic and palaeontological studies as well as with the occurrence of hybridisation in the Russian contact zone. We propose that Central Europe was colonised by E. roumanicus by the beginning of the Neolithic period and that there was a subsequent reinforcement stage as well as the formation of a zone of sympatry after the complete reproductive isolation of both species.
Keywords: Erinaceus, landscape genetics, population structure, mitochondrial DNA, microsatellites, reproductive isolation
Introduction
Proximate mechanisms of speciation can be illuminated by describing the spatial organisation of genetic variability. Different aspects of this process can be addressed within the field of phylogeography (Avise et al., 1987) and landscape genetics (Manel et al., 2003). Phylogeography is a well-established discipline that focuses on the historical causes of genetic variation patterns using phylogenetic methodology and sequence data. Landscape genetics is a new approach that focuses on the contemporary processes affecting the distribution of genetic variability. Population genetic approaches and microsatellite data are primarily used in landscape genetics. However, unlike population genetics, this field uses individuals as study units and a posteriori defined populations. Although there is some overlap between those disciplines (Wang, 2010), these fields enable researchers to address different biological questions (Storfer et al., 2010).
Over the past two decades, the phylogeographic approach has provided evidence with regard to the frequency of allopatric speciation in nature occurring in variety of patterns. In gradual allopatric models, the diversification of nascent lineages could be driven by both neutral evolution, which involves stochastic genetic processes, and adaptive evolution responding to selection effects of potentially different environmental conditions and ecological niche commitments (Coyne and Orr, 2004). If secondary contacts of an allopatrically evolved population arise, further adaptive processes occur that are associated with species interactions. Many examples of these patterns were found in Central Europe because this region is a crossroad of postglacial colonisation routes for many terrestrial species due to Pleistocene climatic oscillations in European biota (Hewitt, 2004). These changes caused recurrent latitudinal and altitudinal range shifts of warm temperate ecosystems. These ecosystems retracted and survived in southern refugia in connection with the topography of the Mediterranean sea during glaciations, and then spread north during interglacial periods.
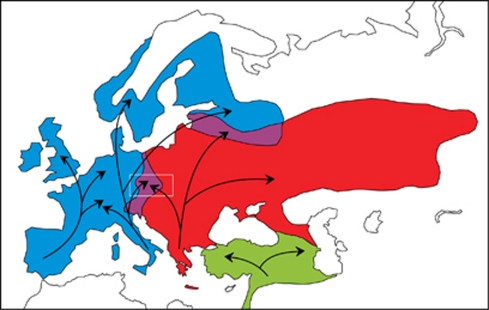
Hedgehogs of the genus Erinaceus played an especially important role as a model for revealing the Quaternary phylogeography of Europe. As insectivores, the hedgehogs were undoubtedly greatly influenced by climate-dependent changes in the availability of insect food during the Quaternary period and the dramatic range dynamic must have left clear imprints in the genetic architecture of this taxon (Santucci et al., 1998; Seddon et al., 2001, 2002; Berggren et al., 2005). Recently, three species of the genus Erinaceus were recognised in the western Palearctic (Aulagnier et al., 2009). The west European hedgehog (Erinaceus europaeus) inhabits Western Europe and has a range that spreads to Scandinavia, the Baltic republics and northern Russia. Three mitochondrial clades within this species correlate with origins in the Iberian, Apennine and Sicilian refugia, and these clades contrast the homogenous nuclear gene pool within the entire species. The range of northern white-breasted hedgehogs (Erinaceus roumanicus) covers Eastern Europe and Ponto-Mediterranean regions and likely originated in the Balkan refugium. The range of southern white-breasted hedgehogs (E. concolor) includes Asia Minor and Levant, but it is isolated from the range of E. roumanicus by the Bosporus Strait and Caucasus Mountains (Seddon et al., 2002). Although the imprints of glacial oscillations are obvious, the roots of the divergence are Pliocene (3.2–4.5 Myr; Seddon et al., 2001). The distribution of E. europaeus and E. roumanicus is parapatric on a macrogeographical scale, and contact zones are situated in Central Europe (Poland, the Czech Republic, Austria and Italy) and north-eastern Europe (Latvia, Estonia and Russia to the Ural mountains; Figure 1). There is a broad zone of sympatry, however, in the Czech Republic (Figure 2). It is not clear if, and to what extent, past episodes of secondary contacts occurred during particular interglacial range pulses.
Figure 1.
Distribution range map of Erinaceus europaeus (blue), E. roumanicus (red) and E. concolor (green) in the western Palearctic (modified according to Reeve (1994) and Aulagnier et al. (2009)). The sympatry zones are violet. The arrows show colonisation routes from refuges after the last ice age (according to Hewitt (2000)). The white rectangle indicates the study area within the sympatric zone.
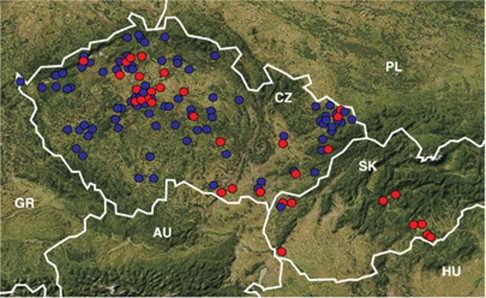
Figure 2.
Sampling for the study in the sympatry zone of Erinaceus europaeus (blue) and E. roumanicus (red). In areas with dense sampling (Prague, Ostrava), not all individuals are displayed owing to space limitations. Country codes: AU, Austria; CZ, Czech Republic; GR, Germany; HU, Hungary; PL, Poland; SK, Slovak Republic. The map was created using the website http://www.mapy.cz.
In contrast to their phylogeography, which is well studied, there is a lack of knowledge about genetic structure at intermediate geographic scales. In addition, information about the population genetic structure of E. europaeus outside Great Britain and of E. roumanicus is missing completely. This information can be obtained within a landscape genetics framework. We have focused our sampling on a transect of the zone of sympatry in Central Europe, which was useful for several reasons. The differences in biology between E. europaeus and E. roumanicus have seldom been studied through comparative approaches, and there is a relative lack of knowledge about the eastern species. Comparison in the sympatric zone is especially useful as it can be performed with a single geographic background, and potential diversification might be intensified by character displacement. Finally, the sympatric zone is suitable for studying species interactions after secondary contact. Assessing the level of species cohesion in this zone is an important factor to understand speciation process in hedgehogs. To address these topics, we performed detailed individual-based sampling from 152 localities that transect the mid-European suture zone and the adjacent allopatric populations from west to east across approximately 800 km. The aims of this study were (1) to provide a basic picture of the distribution and hypsometric data of both species in the transect of the sympatric zone; (2) to assess and compare the population structure at intermediate geographic scales using both population and landscape genetics approaches to make inferences for patterns of gene flow and other aspects of biology. For that purpose, we used fast-evolving markers (nine nuclear microsatellites and mitochondrial control region). The combination of mitochondrial and nuclear markers enabled us to employ the specific features of particular genetic pools and to study both female- and male-mediated gene flow; (3) to reconstruct the demographic history of both species in the Central Europe by using both traditional and coalescent methods; and (4) to compare the genetic integrity of populations living in allopatry and sympatry. By assessing the occurrence of cytonuclear conflicts and performing admixture analyses, we aimed to find traces of potential hybridisation and introgression between both taxa and to evaluate the degree of reproductive isolation.
From all of these comparisons of sister species, we can gain valuable insights into the microevolutionary processes connected with speciation, such as ecological adaptation, niche differentiation, life-history trade-offs, different responses to climate changes and social system differentiation.
Materials and methods
Sampling and DNA isolation
We collected muscle or ear tissues from individuals who died in rescue centres or from road-killed animals as well as hair follicles from animals captured during the night transects in the study area (see Supplementary Appendix A). The tissues were stored in absolute ethanol at −20 °C. We collected samples from E. europaeus in the Czech Republic (n=152), Slovak Republic (n=1), Germany (n=22), France (n=1), Sicily (n=1) and Portugal (n=1). The E. roumanicus samples came from the Czech Republic (n=50), Slovak Republic (n=11), Poland (n=1), Romania (n=2) and Bulgaria (n=1). The E. concolor samples came from Turkey (n=2) and Lebanon (n=1). All samples were collected during the 2007–2009 seasons and they were georeferenced. Animals outside the sympatric zone were used as purebred outgroups for admixture analysis in NewHybrids and for haplotype network. For detailed analyses of demography and landscape genetics, the continuously sampled part of the range was used (Figure 2). We isolated total genomic DNA using the DNA Blood and Tissue Kit (Qiagen, Prague, Czech Republic). The number of specimens included in the mitochondrial/microsatellite data sets varied depending on the sequencing and genotyping success (see Supplementary Appendix A).
Mitochondrial DNA sequencing
For amplification of the 5′ segment of the mitochondrial control region (410 bp), including the left hypervariable domain, we used the primers ProL-He, which was located in the tRNAPro gene region, and DLH-He, which was located in the conserved sequence block of the E region (Seddon et al., 2001). Polymerase chain reaction (PCR) conditions were set according to Seddon et al. (2001). PCR products were subsequently purified using the QIAquick PCR Purification Kit (Qiagen) and then sequenced using the BigDye Terminator v.3.1 Cycle Sequencing Kit and analysed on a 3130 Genetic Analyzer (Applied Biosystems, Prague, Czech Republic). The sequenced parts of the mitochondrial genome were edited using SeqMan 5.05 (Swindell and Plasterer, 1996) and aligned using BioEdit 7.0.9.0 (Hall, 1999) encompassing Clustal W (Thompson et al., 1994). The haplotype data were submitted to GenBank (Accession Numbers HM462024–HM462052).
Microsatellite genotyping
For microsatellite genotyping, we used primers developed for E. europaeus (Becher and Griffiths, 1997; Henderson et al., 2000), and after preliminary analyses, we chose nine loci with sufficient amplification efficiencies and polymorphisms (EEU2, EEU3, EEU4, EEU5, EEU6, EEU12H, EEU37H, EEU43H and EEU54H). Forward primers were fluorescently labelled and divided into three multiplexes according to annealing temperature (see Supplementary Appendix B). PCR was performed as described in Becher and Griffiths (1997) and Henderson et al. (2000). The fragmentation analysis mixture contained PCR products, formamide and size standard (Gene ScanTM 500 LIZ Size Standard; Applied Biosystems). Fragmentation analyses were run on an ABI Prism 3100 Avant Genetic Analyzer (Applied Biosystems) with polymer POP4 and standard DS-33. The sizes of the microsatellite loci were assigned by the program GeneMarker v.1.85 (www.softgenetics.com).
Analyses of genetic variability
For visualisation of the relationships within mitochondrial haplotypes, we calculated the median-joining network (Bandelt et al., 1999) included in Network 4.5.1.2 (www.fluxus-engineering.com). This method enables illustration of alternative genealogical hypotheses and is useful for shallow divergence data sets. For this analysis, we used data of E. europaeus from Germany and Czech Republic, E. roumanicus from Czech Republic, Poland and Bulgaria, and E. concolor from Lebanon and Turkey. We charted the haplotypes into the map and indicated the overlap with previously published mitotypes (Seddon et al., 2001) to interpret the phylogeographic pattern in wider geographic context (see Supplementary Appendix C). The historical demographical processes were described by several methods. First, we used summary statistics of genetic variability, including tests for haplotype diversity (h), nucleotide diversity (π) and neutrality tests (Fu and Li's F*, Fu and Li's D*, Fu's F's and Tajima's D) were computed in DnaSp v.5 (Rozas et al., 2003). The raggedness index (rg) and expansion parameter tau (τ) were computed in Arlequin 3.11 (Excoffier et al., 2005). The parameter τ estimates the time from expansion in mutation units, and a higher τ value represents an older expansion. Estimations of population size in the recent past were computed using the mismatch distribution test (Schneider and Excoffier, 1999) in DnaSp v.5 (Rozas et al., 2003), which compared the observed frequencies of pairwise differences with the frequencies expected under demographic models. For a more detailed estimation of population size, coalescent-based Bayesian skyline plots (Drummond et al., 2005) were used. This method uses the Markov chain Monte Carlo (MCMC) procedure to estimate the posterior distribution of effective population size. We inferred the model of sequence evolution in Modeltest 3.7 (Posada and Crandall, 1998) using Akaike criterion. We found K81uf+I variant for E. europaeus and HKY for E. roumanicus. To standardise the model for both species and to avoid bias due to under-parametrisation, we have chosen more complex general time reversible model for both species. The Bayesian skyline plot analysis was conducted using the program BEAST 1.4.8 (Drummond and Rambaut, 2007) with strict molecular clock enforced. The MCMC procedure was run three times for each species with 30 000 000 iterations, and the genealogy and parameters of the model were stored every 1000 iterations. The convergence of chains was assessed in Tracer v.1.4, and burn-in was set to 10 000 000 iterations. The results were combined in LogCombiner.
Within the microsatellite data set, genotyping errors due to PCR artefacts (for example, stuttering, −A allele occurrence, short allele dominance and null alleles) were tested using a Monte Carlo simulation of expected allele–size differences using Micro-Checker (Van Oosterhout et al., 2004). The distribution of allele frequencies, number of private alleles, estimation of null alleles and inbreeding coefficient (FIS) were evaluated by GenePop v.4 (Rousset, 2008). The number of alleles, expected (He) and observed (Ho) heterozygosity, and deviations from Hardy–Weinberg equilibrium (HWE) for each locus were determined by Arlequin 3.11 (Excoffier et al., 2005). The significance of HWE was tested by 100 000 MCMC steps. Allelic richness (AR) was estimated using FSTAT 2.9.3 (Goudet, 2001). This rarefaction-derived statistics enables to compare nuclear diversity between population samples of different N.
The basic assumption of the relationships between individual nuclear genotypes was assessed using factorial correspondence analysis in Genetix (Belkhir et al., 2004). Each individual was classified as E. roumanicus or E. europaeus according to mitochondrial DNA (mtDNA) determination before the analysis. This approach enables the detection of potential discrepancies between mitochondrial and nuclear determination and possible cytonuclear conflicts caused by mtDNA introgression events between species. To test for population substructure and assign all individuals to potential subpopulations as well as identify possible hybrids, we used Bayesian clustering with the admixture model of correlated allele frequencies (Falush et al., 2003) in Structure 2.2 (Pritchard et al., 2000). The length of the MCMC was 1 000 000 steps after 100 000 steps of a burn-in period. The likeliest number of populations (K) was estimated during independent runs starting with K=1 and ending with K=10. To identify intraspecific substructures, the analyses were also performed for each species separately. For direct estimates of recent hybridisation and introgression in nuclear loci, we performed admixture analyses via Bayesian clustering based on a model estimating the posterior probability of classifying individuals to a priori defined hybrid categories, which was implemented by the program NewHybrids 1.1 (Anderson and Thompson, 2002). We set the program to test for the presence of six genotype frequency classes that theoretically might have occurred after two generations of interbreeding between the species (pure E. europaeus, pure E. roumanicus, F1 hybrid, F2 hybrid, backcross F1 with E. europaeus and backcross F1 with E. roumanicus). The purebred origin of populations living in allopatry was set before the run. The burn-in period had 10 000 steps, and the analysis had 100 000 steps. The convergence of chains was tested visually during independent runs of the program.
To analyse the spatial architecture of mitochondrial and nuclear variability, we performed individual-based landscape genetics analyses with the R package Geneland (Guillot et al., 2005) using Bayesian clustering. For mitochondrial (haploid) haplotype data, a multinomial distribution of genotypes conditionally based on allele frequencies, population memberships and linkage equilibrium was assumed. For the microsatellite data, we chose a model to determine the likeliest number of clusters (K) in HWE and performed MCMC iterations in five independent runs (the MCMC model had 1 000 000 steps stored every 100 steps, minimum K=1, maximum K=10). After comparing the results of the analyses, we selected a run with the highest posterior probability and post-processed it for graphical display. This approach enables visualisation of the two-dimensional shape of possible subpopulations and therefore might provide a more realistic picture than summary statistics. The difference in altitudinal distribution between both species was tested by comparison of elevation above mean sea level of particular records using a non-parametric Mann–Whitney U-test in Statistica v.6.0.
Results
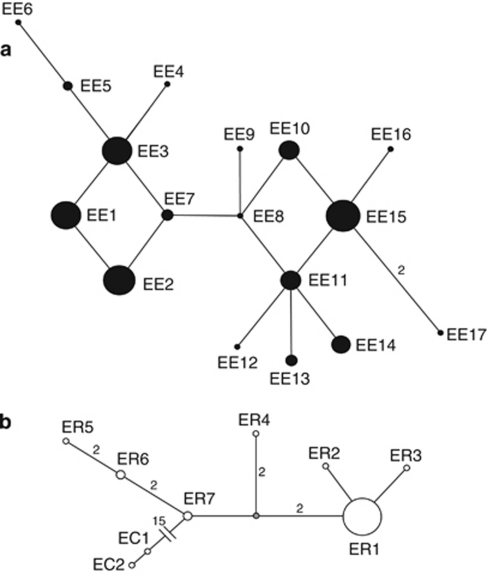
Based on samples from a sympatric zone in the Czech Republic, E. europaeus was approximately three times more frequent (N=152) than E. roumanicus (N=50) in our data set. MtDNA haplotype networks (Figure 3) showed complex and moderately diversified topology with 17 haplotypes for E. europaeus, whereas the E. roumanicus population structure with seven haplotypes was dominated by the ER1 variant (possessed by 45 of 55 sequenced individuals). The rest of the haplotypes differed from ER1 by a few mutations and were represented by a small number of individuals. Both widespread and geographically private haplotypes were found in both species (see Supplementary Appendix C). The haplotype composition of the investigated region showed similarities with refugial as well as northern areas in both species. The dominant ER1 haplotype of E. roumanicus was detected only in northern Italy outside the study area, indicating possible source role of this region.
Figure 3.
Median-joining network of mitochondrial control region haplotypes for: (a) Erinaceus europaeus (E1–E17; N=154) from Czech Republic, Germany and Slovak republic; (b) E. roumanicus (ER1–ER7; N=55) from Czech Republic, Slovak Republic and Poland; and E. concolor (EC1–EC2; N=2) from Turkey and Lebanon. Haplotypes are denoted as circles with a size proportional to haplotype frequency. Numbers nearby the connecting lines represent how many mutation steps are between two connected haplotypes and are shown only if the number of steps was more than one. The hypothesised haplotype is represented by untitled dot.
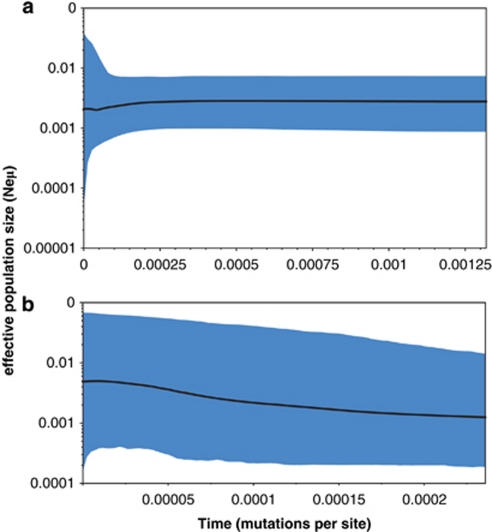
Demographic parameters showed contrasting patterns in both species (see Table 1 for descriptive parameters of mtDNA variability). Nucleotide and haplotype diversity values were higher for E. europaeus than for E. roumanicus. Neutrality tests for E. roumanicus yielded negative values, but none of these results were significant. On the other hand, neutrality tests for E. europaeus yielded positive values. The raggedness statistic was not significant. In addition, the parameter τ was much higher for E. europaeus (41.8) than for E. roumanicus (4.1). The mismatch distribution of E. roumanicus seemed to be unimodal compared with the distribution of E. europaeus. Bayesian skyline plots (Figure 4) indicated that the population size was nearly constant for E. europaeus in the recent past, whereas the population size of E. roumanicus increased slightly (the time to coalescence of all haplotypes was shorter in this case owing to less diversity among sequences).
Table 1. Genetic variability and neutrality tests of the mitochondrial control region for Erinaceus europaeus and Erinaceus roumanicus from the Czech and Slovak Republic.
| N | Nh | π±s.d. | h±s.d. | F | D | F's | TD | rg | τ | |
|---|---|---|---|---|---|---|---|---|---|---|
| E. roumanicus | 54 | 6 | 0.00182±0.00050 | 0.289±0.077 | −0.9804 | −0.85999 | −0.882 | −0.78708 | 0.4627 | 3.0 |
| E. europaeus | 134 | 10 | 0.00611±0.00013 | 0.861±0.009 | 1.7662* | 1.17076 | 0.499 | 2.09875* | 0.0687 | 41.8 |
Abbreviations: D, Fu and Li's D*; F, Fu and Li's F*; Fs, Fu's Fs; h, haplotype diversity; N, number of individuals; Nh, number of haplotypes; π, nucleotide diversity; rg, raggedness index; s.d., standard deviation; τ, time from expansion in mutation units TD, Tajima's D.
*P<0.05.
Figure 4.
Bayesian skyline plots based on partial sequences of the mitochondrial control region. Time is measured in mutation units per nucleotide position. The y axis represents a correlate of population size (Neμ). Black lines illustrate median Ne estimation, and grey areas show the 95% confidence interval. (a) Erinaceus europaeus (N=134) and (b) E. roumanicus (N=54).
Each of the nine microsatellites was polymorphic and had between 2–17 and 4–22 alleles for E. roumanicus and E. europaeus, respectively (see Table 2 for descriptive parameters of microsatellite variability). Analysis of the microsatellite data set showed no evidence of genotyping error owing to stuttering or large allele dropout, but the analysis also indicated the presence of null alleles in loci EEU43H and EEU3, which may have led to deviations from HWE. Estimation of the frequency of non-amplified alleles varied by species and by locus, but all values of the null allele estimated frequencies were smaller than 0.1. Both species showed a significant absence of heterozygotes and deviated from HWE (Table 2). Nuclear diversity was comparable for both species. The expected heterozygosity was higher for E. roumanicus, but the observed heterozygosity was higher for E. europaeus. Both species differed in the distribution of allele frequencies and in the number of species-specific alleles with frequencies higher than 0.05 (21 for E. europaeus and 15 for E. roumanicus).
Table 2. Descriptive characteristics of microsatellite genetic variability for Erinaceus europaeus (N=131) and Erinaceus roumanicus (N=61) in the Czech and Slovak Republic.
| Locus | E. roumanicus |
E. europaeus |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size range | Na | AR | HE | HO | Fis | Size range | Na | AR | HE | HO | Fis | |
| EEU2 | 269–284 | 13 | 12.98 | 0.8821 | 0.7966 | 0.0976 | 262–283 | 17 | 15.73 | 0.8676* | 0.8203* | 0.0547 |
| EEU3 | 134–179 | 17 | 16.70 | 0.6835* | 0.5574* | 0.1858 | 147–165 | 12 | 9.82 | 0.7468* | 0.6692* | 0.1042 |
| EEU4 | 142–169 | 10 | 9.91 | 0.7969* | 0.6229* | 0.2197 | 142–168 | 14 | 12.81 | 0.8261* | 0.7939* | 0.0392 |
| EEU5 | 117–145 | 10 | 8.95 | 0.6391* | 0.5082* | 0.2203 | 105–138 | 9 | 6.86 | 0.5159 | 0.4923 | 0.0458 |
| EEU6 | 152–174 | 10 | 9.90 | 0.8126* | 0.6393* | 0.2146 | 144–165 | 14 | 10.41 | 0.5301* | 0.5038* | 0.0497 |
| EEU12H | 91–93 | 2 | 1.95 | 0.0325 | 0.0328 | — | 89–95 | 4 | 3.28 | 0.1105 | 0.0923 | — |
| EEU37H | 239–248 | 4 | 3.95 | 0.6578 | 0.6066 | 0.0785 | 231–278 | 22 | 16.63 | 0.7981 | 0.7538 | 0.0557 |
| EEU43H | 160–198 | 16 | 16.00 | 0.8744* | 0.7414* | 0.1532 | 144–174 | 13 | 9.87 | 0.6067* | 0.4662* | 0.1827 |
| EEU54H | 281–298 | 9 | 8.87 | 0.7193* | 0.5333* | 0.2602 | 281–296 | 12 | 9.38 | 0.7376 | 0.7239 | 0.017 |
| Mean | 10.33 | 9.84 | 0.73175 | 0.6673 | 0.1768 | 13.44 | 10.48 | 0.6872 | 0.695 | 0.0686 | ||
For each locus, the following are given: size range; Na, number of alleles; AR, allelic richness; HE, expected heterozygosity; HO, observed heterozygosity; and FIS, inbreeding coefficient. The mean values for all loci are given at the bottom of the table.
*Significant departure from HWE (P<0.05).
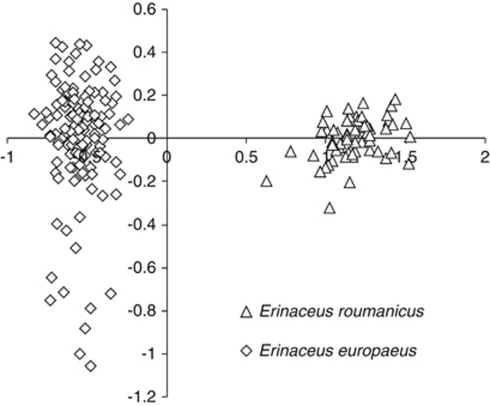
Factorial correspondence analysis clearly sorted E. europaeus and E. roumanicus into separate clusters with no evidence of overlap (Figure 5). There was no discrepancy between the mitochondrial and nuclear species determination, which could be interpreted as an absence of mitochondrial introgression. The result of the analysis in Structure agreed with this interpretation. All of the individuals were ranked to the cluster with a very high probability (the average value was 0.995 for E. europaeus and 0.996 for E. roumanicus). Similarly, there were no genotypes of mixed origin in NewHybrids analysis. All of the individuals from the contact zone were assigned to the pure E. roumanicus category or the pure E. europaeus category with posterior probabilities greater than 0.9 (see Supplementary Appendix D).
Figure 5.
Factorial correspondence analysis based on nine microsatellite loci, showing a multivariate relationship between individuals of Erinaceus europaeus (N=130) and E. roumanicus (N=49) in the Czech and Slovak Republics.
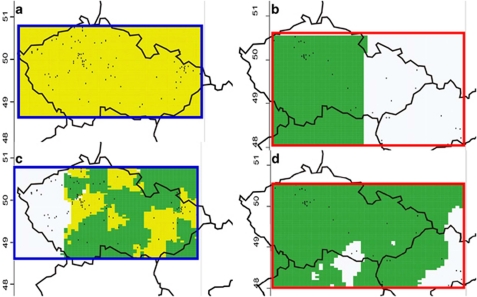
The landscape analyses of both nuclear and mtDNA data in E. roumanicus revealed two subpopulations with an abrupt transition zone (Figure 6). E. europaeus did not show a pronounced population structure from microsatellites, but mtDNA showed a mosaic structure of three subpopulations (Figure 6). Analyses of population structure were repeated in Structure, which confirmed that there were two subpopulations of E. roumanicus, but no structure of E. europaeus in the nuclear data (see Supplementary Appendix D). Mann–Whitney U-test showed that there is a significant difference (P=0.000001, U=1932) in altitudinal distribution between both species. Although E. europaeus was widespread (the altitudinal distribution was continuous up to 500 m above mean sea level and there were 12 records between 500 and 700 m above mean sea level), the distribution of E. roumanicus was correlated with lower altitudes (only four records exceeded 300 m above mean sea level; Figure 2), reflecting influxes of the Pannonic system to the Hercynian and Carpathian provinces.
Figure 6.
Tessellation maps illustrating the spatial distribution of subpopulations within Erinaceus europaeus and E. roumanicus, inferred by the Bayesian clustering method implemented in GeneLand. Black dots represent sample sites. (a) Microsatellites of E. europaeus, (b) microsatellites of E. roumanicus, (c) mtDNA of E. europaeus and (d) mtDNA of E. roumanicus.
Discussion
Distributional patterns in the zone of sympatry
Comparing our results with faunal data was difficult given that systematic research on hedgehogs has not been conducted in the Czech Republic since the 1970s. Moreover, discrimination based solely on external characteristics (for example, fur coloration and spine striping) could be biased owing to considerable individual variability and the changes during ontogeny and moulting. The altitudinal differences in distribution of both species could be the result of spatially specific adaptations (because a large part of the E. roumanicus range is located in steppe and forest–steppe biomes) and may be further strengthened by character displacement. Further research on ecology is needed to test this hypothesis. The distribution in Central Europe, however, cannot be considered microallopatry because we found localities with syntopic occurrence of both species (for example, Prague). Differences in temperature sensitivity among hedgehogs have also been discussed in the literature. However, the conclusions of particular authors differed. Kratochvíl (1975) showed a shorter hibernation period and thus a larger resistance to low temperatures in E. roumanicus in the Czech Republic compared with what Holz and Niethammer (1990a, 1990b) showed for E. europaeus in Austria. It is obvious that these characteristics may show clinal variability in both species owing to ecological adaptations dependent on the geographical origins of particular populations.
Population structure and demography
The shallow mitochondrial genetic structure of both species could be interpreted as the result of low or modest contemporary matrilineal gene flow. The more complex structure of E. europaeus, however, required more time to evolve, or the matrilineal gene flow was restricted in this species. The outcomes of demographic analyses suggested relatively stable population sizes in E. europaeus. Analyses conducted in E. roumanicus demonstrated contrasting patterns compared to E. europaeus and suggested a moderate population growth.
In parameters describing variability of nuclear markers, both species demonstrated more similarity than in mtDNA measures. Several factors could have contributed to this finding. First, given that mtDNA is haploid and has an effective population size four times smaller than that of nuclear loci, mtDNA is more sensitive to bottlenecks. If the spread of E. roumanicus was more rapid, connected with leptokurtic dispersal and successive bottlenecking, the resulting loss of genetic diversity would be more obvious in the mtDNA. Second, this pattern may indicate that dispersal was more sex-biased towards female philopatry in E. europaeus than in E. roumanicus.
Several loci with excess homozygosity in both species were probably affected by the occurrence of null alleles, as indicated by analysis in Micro-Checker. At the loci without the effect of null alleles, deviations from HWE could have had biological causes. The most meaningful of these causes could be non-random mating (that is, the existence of population substructure and inbreeding) and selection. Hedgehogs do not display a pronounced territorial behaviour or a semifossorial life strategy (Reeve, 1982), which are two conditions that often lead to genetic differentiation on small geographic scales. With regard to the relatively limited dispersal capacity of hedgehogs (Reeve, 1982) and habitat fragmentation (especially in areas affected by humans such as urban or agricultural landscapes), the organisation of the gene pool into more or less isolated demes and a subsequent Wahlund effect is probable. The existence of population substructure within a study area was also proposed by landscape genetic approach. The discontinuous distribution of E. europaeus has been studied using both ecological and genetic approaches. The correlation of a patchy distribution with resource availability (Micol et al., 1994) and predator density (Doncaster, 1992) has also been reported. Becher and Griffiths (1998) detected a restriction in gene flow among geographically close populations from a fragmented landscape in Oxfordshire, Great Britain. Radio-tracking studies have shown that main roads and larger fields play a role as a barrier (Rondinini and Doncaster, 2002).
The average home range for males is significantly larger than the range for females in E. europaeus (Reeve, 1982). Because the size of the home range depends on many ecological factors, there is considerable variability depending on geographic position and the type of environment. There is a gradient from the smallest home range size in the woods (Morris, 1988) to the largest home range size in open areas (Boitani and Reggiani, 1984). More intense movement of males has been detected, especially during mating periods (Reeve, 1982). A juvenile dispersal phase has also been reported (Reeve, 1994). In E. roumanicus, information with regard to spatial behaviour is scarce. Our data indicated less sex-biased dispersal and higher levels of gene flow in this species. These findings are in agreement with an observed preference for lowlands, which are characterised by open habitats in Central Europe. These outcomes could be interpreted as a result of the need for larger home ranges due to less concentrated sources, as well as more intense movements necessary for seeking a mate due to lower densities of the species, perhaps contributing to decreased sex specificities in spatial behaviour. There are also fewer barriers to dispersal in lowlands, which may serve as corridors for preferential range expansion.
In E. europaeus, landscape analysis of mtDNA showed one subpopulation in the west of the country, whereas two others formed a mosaic in the eastern part. We interpreted the latter part of the range to be the result of a relatively recent postglacial recolonisation with enough time to evolve moderate genetic substructures. Moreover, the border between the first group and the other groups coincided with the big rivers Vltava and Elbe that intersect the Czech Republic from north to south. The nuclear data showed a homogenous population and reflected a lower sensitivity of nuclear markers to recent demographic processes as well as a more intense male-mediated gene flow. Although the average variability of the mtDNA pool in E. roumanicus was low, it was divided into two clusters. We presumed that the more genetically homogenous western group was the result of a recent and relatively quick expansion during the formation of the sympatric zone. The intensity of the resulting demographic processes also affected nuclear DNA with similar east–west structuring. The western position of the transition zone between both nuclear clusters compared to mtDNA could be interpreted as the result of lineage sorting of diploid nuclear markers in later phases of bottlenecking that were caused by shifting of the range margin. The similarity between mitochondrial and nuclear patterns is further proof of the similarity in gene flow patterns for both sexes of E. roumanicus.
Reproductive isolation
E. europaeus and E. roumanicus have compatible karyotypes with 2n=48 chromosomes as well as differences in heterochromatin localisation (Sokolov et al., 1991). Interspecific mating and obtaining F1 litter in captivity has been conducted (Poduschka and Poduschka, 1983). The same authors were also able to obtain backcrosses with E. roumanicus, but attempts to obtain backcrosses with E. europaeus were not successful. The hybrids showed intermediate states of external characteristics and considerable variability. This phenomenon was used to seek hybrids in nature where the frequency of gene flow between both species remained unclear. Based on morphometric analyses, the occurrence of hybrids in nature was rare (Ruprecht, 1972). Using multivariate analysis of the skulls of captive-bred hybrids (Holz, 1978), the status of wild populations has been assessed, thereby indicating limited crossbreeding of the species (Corbet, 1988).
In contrast to the morphometry analyses, our genetic study did not reveal recent hybridisation or introgression. Thus, interspecific gene flow was either absent or its frequency was very low. If the absence of hybrids from two hybridisable species in nature is taken into consideration, it is reasonable to deduce that either prezygotic reproductive isolating barriers or selection against hybrids is a factor in the wild. The areas of coexistence for both species were delimited by the mosaic distribution of E. roumanicus and were localised in regions with lower altitudes. Our results indicate relative abundance of both species may vary in particular regions. The probability of interspecies contacts could be higher in populated areas where individuals of both species could remain ‘trapped' in isolated habitat patches of fragmented environments such as gardens. In a study by Suchentrunk et al. (1998), the contact zone and adjacent regions in Austria were studied using allozymes and yielded no evidence of hybridisation. This result, however, was based on limited geographical coverage, especially for E. europaeus (four localities within the contact zone).
In contrast to these results, the situation in the Eastern European contact zone might be different. Genetic proof of naturally occurring hybrids has been found in the Moscow region in Russia (Bogdanov et al., 2009). Given that the authors found one hybrid among five of the individuals that were studied, the frequency of hybridisation events in the contact zone in Russia might be higher than in Central Europe. The geographic setting and extent of the sympatric distribution within this zone, however, are less well known, and further investigations are needed.
Speciation and evolutionary history
The support of phylogeographic scenarios by palaeontological data is complicated by difficulty to find reliable discrimination criteria applicable to fragmented fossil material; thus, many published conclusions are based on presumptive (range-based) species identification. Sommer (2007) hypothesised that the meeting of both species in Central Europe occurred during the Boreal age and that the absence of younger E. roumanicus records was an artefact of scarcity in the fossil record. However, comparison of genetic structure of both species suggests that E. roumanicus really occurred in central Europe later than E. europaeus. This event could be synchronous with the large-scale deforestation at the Neolithic period. It cannot be excluded that south of the Alps the contact between the species was of much earlier date owing to proximity to refugia. More detailed sampling in adjacent regions will be necessary to reconstruct precisely the routes of particular populations during colonising the zone of sympatry.
The contact zone in Russia was established later, as the Central European population required some time to spread to the northeast destinations (Seddon et al., 2001). In the older mid-European zone, the reinforcement process may have resulted in the formation of complete reproductive isolation mechanisms that are still incomplete in the Russian zone. After the completion of the reproductive isolation in Central Europe, formation of the zone of sympatry (which was realised by a recent range expansion of E. roumanicus) was possible. In addition, this process might have been facilitated by human activity that disintegrated natural barriers between both species, for example, the climax forests of the western Carpathians and wetlands along big rivers and the spread of the cultural steppe. In many patterns that resulted from the postglacial recolonisation of Europe by Mediterranean-born lineages, ranging from tension hybrid zones (for example, in mice Mus musculus and M. domesticus; Selander et al., 1969) to broad sympatry (for example, in bats Pipistrellus pipistrellus and P. pygmaeus; Hulva et al., 2010), European hedgehogs are an interesting intergrade situation of nascent sympatry.
Conclusions
In our study, we addressed patterns of gene flow at a landscape level, demography and reproductive isolation for E. europaeus and E. roumanicus in a transect of mid-European sympatric zone. We have described and interpreted differences in distribution and genetic structure in both species. We did not detect interspecific hybridisation or introgression; thus, the premating isolating mechanisms were completely formed. The possible matter of differences in ecology of both species and their reproductive isolating mechanisms (habitat preference, allochrony, courtship rituals, acoustic and olfactory communication, and so on) should be addressed by future research.
Acknowledgments
The project was supported by Ministry of Education, Youth and Sports of the Czech Republic (MSMT 0021620828). The study of BB was supported by stipend from municipal of Ostrava City. We thank all of the providers mentioned in Supplementary Appendix A for their assistance in obtaining samples. Three anonymous reviewers gave valuable comments to previous versions of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulagnier S, Haffner P, Mitchell-Jones AJ, Moutou F, Zima J. Mammals of Europe, North Africa and the Middle East. A&C Black Publisher Ltd: London; 2009. [Google Scholar]
- Avise JC, Arnold J, Baal RM. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Ann Rev Ecol Syst. 1987;18:489–522. [Google Scholar]
- Bandelt HJ, Foster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Becher SA, Griffiths R. Isolation and characterization of six polymorphic microsatellite loci in the European hedgehog Erinaceus europaeus. Mol Ecol. 1997;6:89–90. doi: 10.1046/j.1365-294x.1997.00159.x. [DOI] [PubMed] [Google Scholar]
- Becher SA, Griffiths R. Genetic differentiation among local populations of the European hedgehog (Erinaceus europaeus) in mosaic habitats. Mol Ecol. 1998;7:1599–1604. doi: 10.1046/j.1365-294x.1998.00457.x. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Logiciel sous Windows (tm) pour la Génétique des Populations. Laboratoire Génome, Populations, Interactions: CNRS UMR. 5000. Université de Montpellier II: Montpellier; 2004. [Google Scholar]
- Berggren KT, Ellegren H, Hewitt GM, Seddon JM. Understanding the phylogeographic patterns of European hedgehogs, Erinaceus concolor and E. europaeus using the MHC. Heredity. 2005;95:84–90. doi: 10.1038/sj.hdy.6800694. [DOI] [PubMed] [Google Scholar]
- Bogdanov AS, Bannikova AA, Pirusskii YuM, Formozov NA. Genetic evidence of hybridization between West European and Northern white-breasted Hedgehogs (Erinaceus europaeus and E. roumanicus) in Moscow Region. Biol Bull. 2009;36:647–651. [PubMed] [Google Scholar]
- Boitani L, Reggiani G. Movements and activity patterns of hedgehogs Erinaceus europeaus in Mediterranean coastal habitats. Zool Anz. 1984;49:193–206. [Google Scholar]
- Corbet GB. The family 539 Erinaceidae: a synthesis of its taxonomy, phylogeny, ecology and zoogeography. Mammal Rev. 1988;18:117–172. [Google Scholar]
- Coyne J, Orr HA. Speciation. Sinauer: Sunderland, MA; 2004. [Google Scholar]
- Doncaster CP. Testing the role of intraguild predation in regulating hedgehog populations. P R Soc London B. 1992;249:113–117. doi: 10.1098/rspb.1992.0092. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;5:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT: A Program to Estimate and Test Gene Diversities and Fixation Indices, Version 2.9.3. Lausanne University: Lausanne, Switzerland; 2001. [Google Scholar]
- Guillot G, Mortier F, Estoup A. GENELAND: a computer package for landscape genetics. Mol Ecol Notes. 2005;5:712–715. [Google Scholar]
- Hall TA. BioEdit: a user—friendly 563 biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Henderson M, Becher SA, Doncaster CP, Maclean N. Five new polymorphic microsatellite loci in the European hedgehog, Erinaceus europaeus. Mol Ecol. 2000;9:1949–1950. doi: 10.1046/j.1365-294x.2000.01098-18.x. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Ser B. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz H. Studien an europäischen Igeln. Z Zool Syst Evol. 1978;16:148–165. [Google Scholar]
- Holz H, Niethammer J.1990aErinaceus europaeus Linnaeus, 1758–Braunbrustigel, WestigelIn: Niethammer J, Krapp F (eds).Handbuch der Säugetiere Europas. Vol. 3/1: Insektenfresser, Herrentiere Aula-Publisher: Wiesbaden; 26–49. [Google Scholar]
- Holz H, Niethammer J.1990bErinaceus roumanicus Martin, 1838—Weißbrustigel, OstigelIn: Niethammer J, Krapp F (eds).Handbuch der Säugetiere Europas. Vol. 3/1: Insektenfresser, Herrentiere Aula-Publisher: Wiesbaden; 50–64. [Google Scholar]
- Hulva P, Fornuskova A, Chudarkova A, Evin A, Allegrini B, Benda P, et al. Mechanisms of radiation in a bat group from the genus Pipistrellus inferred by phylogeography, demography and population genetics. Mol Ecol. 2010;19:5417–5431. doi: 10.1111/j.1365-294X.2010.04899.x. [DOI] [PubMed] [Google Scholar]
- Kratochvíl J. Zur Kenntnis der Igel der Gattung Erinaceus in der ÈSSR (Insectivora, Mamm.) Zool Listy. 1975;24:297–312. [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 2003;18:189–197. [Google Scholar]
- Micol T, Doncaster CP, Mackinlay LA. Correlates of local variation in the abundance of hedgehogs Erinaceus europaeus. J Anim Ecol. 1994;63:851–860. [Google Scholar]
- Morris PA. A study of home range and movements in the hedgehog (Erinaceus europaeus) J Zool. 1988;214:433–449. [Google Scholar]
- Poduschka W, Poduschka C. Kreuzungsversuche an mitteleuroäischen. Igeln Säugetierkdl Mitt. 1983;3:1–12. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnely PJ. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve NJ. The home range of the hedgehogs as revealed by a radio tracking study. Sym Zool S. 1982;49:207–230. [Google Scholar]
- Reeve NJ. Hedgehogs. T & AD Poyser (Natural History): London; 1994. [Google Scholar]
- Rondinini C, Doncaster P. Roads as barriers to movement for hedgehogs. Funct Ecol. 2002;16:504–509. [Google Scholar]
- Rousset F. GENEPOP ‘ 007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DNASP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Ruprecht AL. Correlation structure of the skull dimensions in European hedgehogs. Acta Theriol. 1972;17:419–442. [Google Scholar]
- Santucci F, Emerson BC, Hewitt GM. Mitochondrial DNA phylogeography of European hedgehogs. Mol Ecol. 1998;7:1163–1172. doi: 10.1046/j.1365-294x.1998.00436.x. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Santucci F, Reeve N, Hewitt GM. Caucasus mountains divide postulated postglacial colonization routes in the white-breasted hedgehog, Erinaceus concolor. J Evol Biol. 2002;15:463–467. [Google Scholar]
- Seddon JM, Santucci F, Reeve NJ, Hewitt GM. DNA footprints of European hedgehogs, Erinaceus europaeus and E. concolor: Pleistocene refugia, postglacial expansion and colonization routes. Mol Ecol. 2001;10:2187–2198. doi: 10.1046/j.0962-1083.2001.01357.x. [DOI] [PubMed] [Google Scholar]
- Selander RK, Hunt WG, Yang SY. Protein polymorphism and genic heterozygosity in two European subspecies of the house mouse. Evolution. 1969;23:379–390. doi: 10.1111/j.1558-5646.1969.tb03522.x. [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates very among sites: application to human mitochondrial DNA. Genetics. 1999;3:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov VY, Aniskin VM, Lukyanova IV. Karyological differentiation of two hedgehog species in genus Erinaceus (Insectivora, Erinaceidae) in the USSR. Zool Zh. 1991;70:111–120. [Google Scholar]
- Sommer RS. When east met west: the sub-fossil footprints of the west European hedgehog and the northen white-breasted hedgehog during the Late Quaternary in Europe. J Zool. 2007;273:82–89. [Google Scholar]
- Storfer A, Murphy MA, Spear SF, Holderegger R, Waits L. Landscape genetics: where are we now. Mol Ecol. 2010;19:3496–3514. doi: 10.1111/j.1365-294X.2010.04691.x. [DOI] [PubMed] [Google Scholar]
- Suchentrunk F, Haiden A, Hartl GB. On biochemical genetic variability and divergence of the two hedgehog species Erinaceus europaeus and E. concolor in central Europe. Z Saugetierkd. 1998;63:257–265. [Google Scholar]
- Swindell SR, Plasterer TN. SEQMAN: contig assembly. Methods Mol Biol. 1996;70:75–89. [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Wang IJ. Recognizing the temporal distinctions between landscape genetics and phylogeography. Mol Ecol. 2010;19:2605–2608. doi: 10.1111/j.1365-294X.2010.04715.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.