Abstract
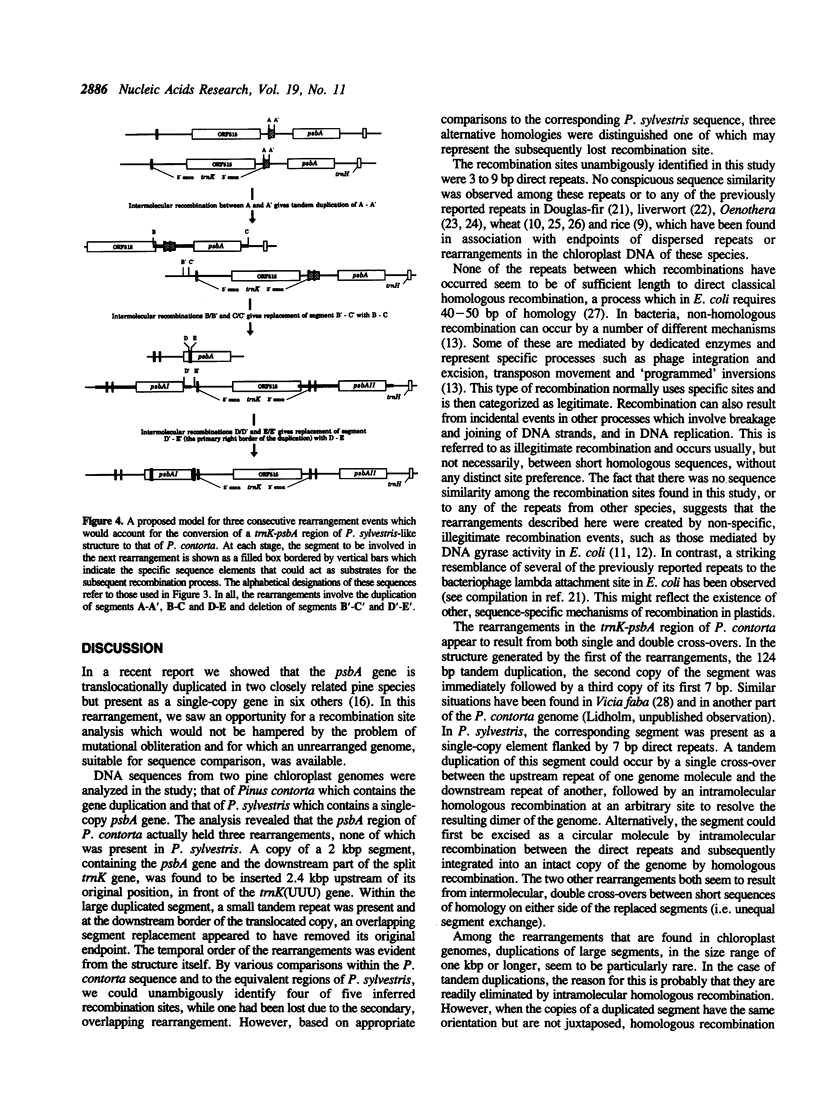
A region of the Pinus contorta chloroplast genome which contains a duplication of the psbA gene was characterized. From previous experiments it was known that the two copies of the psbA gene were located approximately 3.3 kilobase pairs (kbp) apart, that they had the same orientation and that one endpoint of the duplication was 19 base pairs (bp) downstream of the psbA stop codon. In order to determine the size and additional genetic content of the duplicated segment, both copies as well as the intervening DNA were sequenced completely. It was found that the duplicated segment was 1969 bp long, that the two copies were completely identical and were separated by 2431 bp. The duplicated segment carried, in addition to psbA, the 3' exon of the trnK gene, which was partially included in a 124 bp direct repeat. The translocated copy of the duplicated segment was found to be inserted upstream of the trnK(UUU) gene and was immediately followed by a repeated 41 bp stretch from the psbA coding region. The trnK gene was split by a 2509 bp intron which contained an open reading frame of 515 codons. Sequence comparisons of the duplicated segment and its flanking DNA to the corresponding regions of P. sylvestris, a species which lacks the rearrangements found in P. contorta, made it possible to identify 3-9 bp homologies within which recombinations had occurred. A model was derived which would accommodate the conversion of a trnK-psbA locus of the ancestral P. sylvestris-like organization into the rearranged structure found in P. contorta.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonnard G., Weil J. H., Steinmetz A. The intergenic region between the Vicia faba chloroplast tRNA(CAALeu) and tRNA(UAALeu) genes contains a partial copy of the split tRNA(UAALeu) gene. Curr Genet. 1985;9(5):417–422. doi: 10.1007/BF00421614. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Howe C. J., Barker R. F., Bowman C. M., Dyer T. A. Common features of three inversions in wheat chloroplast DNA. Curr Genet. 1988 Apr;13(4):343–349. doi: 10.1007/BF00424430. [DOI] [PubMed] [Google Scholar]
- Howe C. J. The endpoints of an inversion in wheat chloroplast DNA are associated with short repeated sequences containing homology to att-lambda. Curr Genet. 1985;10(2):139–145. doi: 10.1007/BF00636479. [DOI] [PubMed] [Google Scholar]
- Marvo S. L., King S. R., Jaskunas S. R. Role of short regions of homology in intermolecular illegitimate recombination events. Proc Natl Acad Sci U S A. 1983 May;80(9):2452–2456. doi: 10.1073/pnas.80.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A., Naito S., Ikeda H. Homology is not required for recombination mediated by DNA gyrase of Escherichia coli. Mol Gen Genet. 1984;193(2):238–243. doi: 10.1007/BF00330674. [DOI] [PubMed] [Google Scholar]
- Neuhaus H., Link G. The chloroplast tRNALys(UUU) gene from mustard (Sinapis alba) contains a class II intron potentially coding for a maturase-related polypeptide. Curr Genet. 1987;11(4):251–257. doi: 10.1007/BF00355398. [DOI] [PubMed] [Google Scholar]
- Ogihara Y., Terachi T., Sasakuma T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8573–8577. doi: 10.1073/pnas.85.22.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K., Fukuzawa H., Kohchi T., Sano T., Sano S., Shirai H., Umesono K., Shiki Y., Takeuchi M., Chang Z. Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification. J Mol Biol. 1988 Sep 20;203(2):281–298. doi: 10.1016/0022-2836(88)90001-0. [DOI] [PubMed] [Google Scholar]
- Quigley F., Weil J. H. Organization and sequence of five tRNA genes and of an unidentified reading frame in the wheat chloroplast genome: evidence for gene rearrangements during the evolution of chloroplast genomes. Curr Genet. 1985;9(6):495–503. doi: 10.1007/BF00434054. [DOI] [PubMed] [Google Scholar]
- Shimada H., Sugiura M. Pseudogenes and short repeated sequences in the rice chloroplast genome. Curr Genet. 1989 Oct;16(4):293–301. doi: 10.1007/BF00422116. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in procaryotes. Microbiol Rev. 1988 Mar;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss S. H., Palmer J. D., Howe G. T., Doerksen A. H. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3898–3902. doi: 10.1073/pnas.85.11.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. H., Strauss S. H. Dispersed repetitive sequences in the chloroplast genome of Douglas-fir. Curr Genet. 1989 Sep;16(3):211–218. doi: 10.1007/BF00391479. [DOI] [PubMed] [Google Scholar]
- Umesono K., Inokuchi H., Shiki Y., Takeuchi M., Chang Z., Fukuzawa H., Kohchi T., Shirai H., Ohyama K., Ozeki H. Structure and organization of Marchantia polymorpha chloroplast genome. II. Gene organization of the large single copy region from rps'12 to atpB. J Mol Biol. 1988 Sep 20;203(2):299–331. doi: 10.1016/0022-2836(88)90002-2. [DOI] [PubMed] [Google Scholar]
- Zhou D. X., Massenet O., Quigley F., Marion M. J., Monéger F., Huber P., Mache R. Characterization of a large inversion in the spinach chloroplast genome relative to Marchantia: a possible transposon-mediated origin. Curr Genet. 1988 May;13(5):433–439. doi: 10.1007/BF00365665. [DOI] [PubMed] [Google Scholar]
- vom Stein J., Hachtel W. Deletions/insertions, short inverted repeats, sequences resembling att-lambda, and frame shift mutated open reading frames are involved in chloroplast DNA differences in the genus Oenothera subsection Munzia. Mol Gen Genet. 1988 Aug;213(2-3):513–518. doi: 10.1007/BF00339624. [DOI] [PubMed] [Google Scholar]


