Abstract
There has been intense interest in leucine-rich repeat kinase 2 (LRRK2) since 2004, when mutations in the LRRK2 gene were discovered to cause dominantly inherited Parkinson’s disease (PD). This article will address six basic questions about LRRK2 biology as it relates to PD, with particular emphasis on its discovery, current concepts of its physiological and pathological functions, and the strategies being used to discover how LRRK2 dysfunction causes PD.
Mutations in the leucine-rich repeat kinase 2 (LRRK2) gene cause dominantly inherited Parkinson’s disease. A greater understanding of LRRK2 biology will help define pathogenic pathways.
Parkinson’s disease (PD) was long believed to result from environmental insults, with little or no role for genetic factors. This notion began to change in 1997 with the discovery of PD-causing mutations in the SNCA gene that encodes α-synuclein, and the demonstration that α-synuclein is a major component of Lewy bodies, the proteinaceous aggregates that accumulate in the disease. Subsequently, several other “PD genes” that cause familial forms of PD were discovered; these genes implicate protein quality control and mitochondrial dysfunction in the pathogenesis of PD. Mutations in LRRK2 are a major genetic risk factor for familial and sporadic PD, but it remains unclear how mutations in LRRK2 cause PD-related neurodegeneration. Based on the striking similarity of LRRK2 and sporadic PD (see below), it is hoped that a greater understanding of LRRK2 biology will further define pathogenic pathways in sporadic PD and highlight novel therapeutic targets. To summarize current understanding of LRRK2 biology, this article will address six key questions (i.e., a LRRK2 FAQ), ranging from how LRRK2 was initially discovered to what is known about its physiological and pathological functions. We will also attempt to highlight major unanswered questions to be addressed in future studies.
QUESTION 1: HOW WAS LRRK2 DISCOVERED?
The discovery of LRRK2 PD began with the identification of several families suffering from PD with clear genetic inheritance that did not carry any of the then known PD genetic mutations. Prior to the discovery of LRRK2, known Mendelian forms of PD were early-onset diseases, typically beginning in the 30s, with autosomal recessive [PARK2 (parkin), PARK6 (PINK-1), PARK7 (DJ-1)] or dominant [PARK1 (α-synuclein)] patterns of inheritance. In contrast, several families in Asia, the United States, and Europe were found to develop a dominantly inherited disease that, similar to idiopathic PD, began in the 50s or 60s and followed a course typical of the sporadic disease (see below for a clinical description of LRRK2 PD). In 2002, genome-wide linkage analysis of a large Japanese kindred led to the designation of the PARK8 locus on chromosome 12p11.2-q13.1 (Funayama et al. 2002). This work led to the description of several additional PARK8 families. In 2004, two independent groups identified mutations in LRRK2 as the cause of PARK8 PD (Paisan-Ruiz et al. 2004; Zimprich et al. 2004).
Zimprich et al. successfully identified LRRK2 mutations in two families (Zimprich et al. 2004). Both families had been followed since the early 1990s and had been linked to the PARK8 locus (Wszolek et al. 1995, 1997, 2004; Zimprich et al. 2004). The first family, “Family A” of German-Canadian descent, carried the 5096A > G missense mutation causing a change from tyrosine to cysteine at amino acid 1699 (herein “Y1699C”). The second family, “Family D,” carried the 4321C > T missense mutation causing a change from arginine to cysteine at position 1441 (herein “R1441C”) (Zimprich et al. 2004). This identification built on years of clinical observation, neuropathology, and linkage analysis. At the same time, Paisan-Ruiz et al. identified LRRK2 mutations in an additional five families. Four of these families were from the Basque region of Spain and carried a different missense mutation at codon 1441, causing a change from arginine to glycine (herein “R1441G”), likely from the same founder. The fifth family, termed “PL,” was of English ancestry and carried the same Y1699C mutation identified in Family A (Paisan-Ruiz et al. 2004). Subsequently, the original Japanese family was found to carry a missense mutation in the activation loop of the kinase domain (isoleucine to threonine; “I2020T”) (Funayama et al. 2005). Surprisingly, what has since become recognized as the overwhelmingly most common LRRK2 mutation was the last identified. In 2005, another missense mutation in the activation loop of the LRRK2 kinase was reported in 13 families of North American and European origin (Kachergus et al. 2005). This mutation changes residue 2019 from glycine to serine (“G2019S”). G2019S mutations were also identified in patients with no family history of disease (Kachergus et al. 2005). In addition to these five mutations, an additional variant, R1441H, is located at the same codon as two confirmed mutations (R1441C, R1441G) and has been identified in PD patients (Ross et al. 2009). In addition, a variant in the WD40 domain, G2385R, has been reported as a risk factor for PD in Asian populations (Farrer et al. 2007; Funayama et al. 2007; Tan et al. 2009). Although many additional mutations have been suggested to modify disease risk, the five initially described mutations (R1441C, R1441G, Y1699C, G2019S, and I2020T) are the only ones to segregate with disease in large families.
QUESTION 2: HOW COMMON ARE LRRK2 MUTATIONS?
LRRK2 mutations are the most common genetic cause of PD, accounting for 4% of familial PD and 1% of sporadic PD across all populations (Healy et al. 2008). Furthermore, in certain populations, notably North African Arabs and Ashkenazi Jews, the prevalence of LRRK2 mutations is even higher, accounting for up to 40% of all PD cases (Lesage et al. 2006; Ozelius et al. 2006).
Of the LRRK2 mutations, G2019S is clearly the most prevalent in North American and European populations; it is present in more than 2% of general North American and English PD Brain Bank specimens (Gilks et al. 2005; Nichols et al. 2005). A large multinational study involving 1045 patients with LRRK2 mutations (from 133 families) found penetrance of the G2019S mutation to be 28% at 59 years, 51% at 69 years, and 74% at 79 years (Healy et al. 2008). The other four LRRK2 mutations (R1441C, R1441G, Y1699C, and I2020T) are much less common, having been identified in only a small number of patients.
In addition to the five mutations that clearly segregate with familial PD, variation in the LRRK2 gene has also been implicated as a risk factor for idiopathic PD. Two genome-wide association studies found that common genetic variants in LRRK2, as well as variations in the SNCA gene and the MAPT locus, are associated with an increased risk for sporadic PD (Satake et al. 2009; Simon-Sanchez et al. 2009). These studies, and the clinical and pathological similarity of PD patients with and without LRRK2 mutations, suggest that research into the biological and pathological functions of LRRK2 are likely to apply to sporadic disease as well. It must be cautioned, however, that these association studies do not prove causation, nor do they suggest how changes to the LRRK2 protein might contribute to sporadic PD. Conclusions about the role of LRRK2 function in sporadic disease await formal testing of these polymorphic forms of LRRK2 in vitro, and ultimately, a determination of whether strategies that benefit PD patients with LRRK2 mutations (e.g., kinase inhibitors) are also beneficial in patients with idiopathic PD. An improved understanding of signaling events downstream from LRRK2 would also allow studies exploring a potential link between LRRK2 and sporadic PD.
QUESTION 3: DO PATIENTS WITH LRRK2 MUTATIONS LOOK LIKE PATIENTS WITH “TYPICAL” PD?
Clinically, LRRK2 PD has been described as indistinguishable from idiopathic PD (Healy et al. 2008; Marras et al. 2011). Clinical research is focused on following patients with LRRK2 mutations to establish any differences in clinical presentation, motor symptoms, or disease progression. Some reports do indicate that there may be subtle differences in symptoms in patients with and without LRRK2 mutations. For example, LRRK2-mutant patients are more likely than idiopathic PD patients to present initially with a tremor (Healy et al. 2008; Marras et al. 2011). But there is large phenotypic overlap between the two groups, and such differences actually highlight the overwhelming similarity in clinical presentation between PD patients with and without LRRK2 mutations.
Interestingly, studies of human postmortem brain indicate that different LRRK2 mutations appear to correlate with two broad patterns of neuropathology. The G2019S mutation produces a relatively consistent pattern of neurodegeneration typical of idiopathic PD and accompanied by Lewy bodies and Lewy neurites (Giasson et al. 2006; Ross et al. 2006; Taylor et al. 2006). In contrast, three others (R1441C, Y1699C, and I2020T) often lack Lewy body pathology and are associated with “pure nigral degeneration” or pleomorphic neuropathological findings, including neurofibrillary tangles, atypical cytosolic fibrils, or motor neuron degeneration (Wszolek et al. 1997, 2004; Funayama et al. 2002; Zimprich et al. 2004; Taylor et al. 2006; Gaig et al. 2009). The notion that different pathological phenotypes may be associated with distinct LRRK2 mutations was highlighted in an excellent review of LRRK2 autopsies, which noted that of 26 LRRK2 PD cases examined (encompassing several LRRK2 mutations), only 14 (54%) had Lewy bodies (Cookson et al. 2008). Importantly, the great majority of Lewy body positive cases were from patients with the G2019S mutation (11 of 14 Lewy body cases) and these cases represented ∼65% of all G2019S cases (11 of 17 G2019S cases). In contrast, only one-third of “non-G2019S” cases (3 of 9 cases) had Lewy body pathology (Cookson et al. 2008). These observations suggest that whereas the G2019S mutation is very likely to cause neurotoxicity through mechanisms that include the accumulation of aggregated α-synuclein, it is possible that other LRRK2 mutations cause neurodegeneration through distinct, α-synuclein-independent pathways. One caveat to these potential distinctions is that the total number of non-G2019S mutants is quite small. Additionally, even in cases without aggregated α-synuclein, it is possible that soluble (and undetected) yet toxic oligomers of α-synuclein are present.
In summary, the great majority of LRRK2 patients carry the G2019S mutation and show clinical and neuropathological features that are essentially indistinguishable from those of idiopathic PD patients. Other LRRK2 mutations may lead to unique downstream events that produce different clinical features, but elucidating the potentially novel effects of these mutations requires a more detailed understanding of LRRK2 function and the effect of disease mutations.
QUESTION 4: WHAT IS THE NORMAL FUNCTION OF LRRK2?
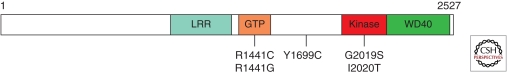
LRRK2 is a 280 kDa protein of the ROCO family that contains several domains. Most research has focused on the two central enzymatic domains, which are the site of all five mutations associated with disease. These enzymatic domains are the Ras of complex (Roc) GTPase protein domain and the kinase domain. In addition, a short carboxy-terminal of the Ras (COR) sequence links the kinase and GTPase. Flanking the two enzymatic domains are multiple protein–protein interaction domains, including amino-terminal leucine-rich repeat domain and a carboxy-terminal WD40 domain (Fig. 1). LRRK2 appears to function as a dimer (Deng et al. 2008; Greggio et al. 2008; Jorgensen et al. 2009) and studies of fragments of LRRK2 or its prokaryotic homolog suggest that dimerization occurs in the Roc-COR region (Deng et al. 2008).
Figure 1.
Domain structure and Parkinson’s disease mutations of LRRK2. LRR, leucine-rich repeat; GTP, GTPase domain (also called Roc domain, Ras of complex proteins). Five PD-causing missense mutations are shown.
The best-characterized LRRK2 interacting proteins are the 14-3-3 proteins (Dzamko et al. 2010; Nichols et al. 2010), which regulate protein–protein interactions typically by binding to phosphoserine/threonine-containing motifs (Tzivion and Avruch 2002; Bridges and Moorhead 2005). 14-3-3 appears to bind to LRRK2 via two phosphorylated serine residues in the amino terminus (phosphorylated Ser910 and Ser 935) (Dzamko et al. 2010; Nichols et al. 2010). Similar to LRRK2 toxicity, this binding is kinase-dependent, and is disrupted by three PD-associated mutants (R1441G, Y1699C, and I2020T) (Dzamko et al. 2010; Nichols et al. 2010). Definitive substrates for the LRRK2 kinase have yet to be discovered, although putative substrates include microtubule-associated proteins (Lee et al. 2010b), Akt1 (Ohta et al. 2011), and LRRK2 itself (Kumar et al. 2010; Webber et al. 2011).
Membrane Trafficking
The normal function of LRRK2 is still largely unknown, with evidence implicating remarkably diverse pathways, including regulation of transcription (Kanao et al. 2010), translation (Imai et al. 2008), apoptosis (Ho et al. 2009), and mitochondrial function (Smith et al. 2005). In contrast to these divergent observations, LRRK2 is consistently observed localized at intracellular membranous structures including mitochondria (West et al. 2005; Biskup et al. 2006; Gloeckner et al. 2006; Hatano et al. 2007), the endolysosomal system (Alegre-Abarrategui et al. 2009), the endoplasmic reticulum (ER) (Gloeckner et al. 2006; Vitte et al. 2010), and Golgi (Biskup et al. 2006; Gloeckner et al. 2006; Hatano et al. 2007). Loss-of-function studies are typically used to infer a protein’s normal function, and multiple lines of evidence from in vivo loss-of-function studies provide support for the possibility that LRRK2 participates in membrane-trafficking reactions. Tong et al. reported kidney pathology in aged LRRK2 null mice, which appeared to be a manifestation of an impaired autophagic-lysosomal pathway, with consequent accumulation of p62 staining, decreased levels of the autophagosome marker LC3-II, and accumulation of α-synuclein and ubiquitin (Tong et al. 2010). These results were recently confirmed by a second group that reported autophagic-lysosomal dysfunction in the kidneys and lungs of LRRK2 null animals as young as 6 weeks old (Herzig et al. 2011). Kidneys and lungs both show higher expressions of LRRK2 than brain (Biskup et al. 2007; Westerlund et al. 2008), and so may be most prone to phenotypic changes in LRRK2 null animals. Notably, LRRK2 null mice are viable and survive to at least 20 months (Tong et al. 2010; Herzig et al. 2011) and do not show any clear in vivo central nervous system (CNS) phenotypes (membrane-related or otherwise) (Tong et al. 2010; Herzig et al. 2011).
Loss-of-function studies in invertebrate models are also consistent with a potential role for LRRK2 in membrane dynamics. Caenorhabditis elegans and Drosophila have one form of LRRK, whereas humans have two (LRRK1 and LRRK2). In flies, Drosophila LRRK (dLRRK) shares 24% identity and 38% similarity at the amino acid level to human LRRK2 (Wang et al. 2008). Loss of dLRRK causes the membrane phenotype of synaptic overgrowth at neuromuscular junctions, an effect that may relate to its interaction with the translational regulator 4E-BP and altered microtubule dynamics (Lee et al. 2010b). Interestingly, this membrane overgrowth phenotype has been observed in several other Drosophila mutants with disrupted membrane trafficking. In C. elegans, LRK-1 null animals are also viable, but show deficits in the normal sorting and localization of synaptic vesicles (Sakaguchi-Nakashima et al. 2007).
In vivo loss-of-function studies in zebrafish have not provided evidence in support of membrane-trafficking defects and have yielded conflicting findings. In marked contrast to the findings of LRRK2 null mice, Sheng et al. reported embryonic lethality with injection of morpholinos against the zebrafish homolog of LRRK2 (zLRRK2), whereas disruption of the WD40 domain enabled normal development but loss of dopaminergic neurons (Sheng et al. 2010). However, this result was not replicated in a similar study using the same morpholinos (Ren et al. 2011). Importantly, work in C. elegans, Drosophila, and zebrafish must be interpreted with caution, because phenotypes resulting from deletion of the single LRRK2 protein in these organisms may reflect functions related to mammalian LRRK1 rather than LRRK2.
Cytoskeleton
Another emerging theme in the biology of LRRK2 is a potential role at cytoskeletal structures. This function may be related to membrane-trafficking events noted above, because membrane traffic occurs on the cytoskeleton. Several in vitro studies suggest that LRRK2 interacts with the cytoskeleton and indicate a potential role in cytoskeletal dynamics (Jaleel et al. 2007; Gandhi et al. 2008; Gillardon 2009; Parisiadou et al. 2009; Lin et al. 2010). Potentially related to these findings, LRRK2 has been consistently observed to alter neural processes; overexpression of wild-type or mutant LRRK2 causes neurite retraction (in primary neuronal cultures) and knock-down of LRRK2 causes neurite overgrowth (MacLeod et al. 2006; Parisiadou et al. 2009; Dachsel et al. 2010). Additional evidence indicates that LRRK2 may interact with cytoskeletal structures, with both the actin and microtubule cytoskeleton being implicated. In vitro, LRRK2 binds α/β tubulin heterodimers (Gandhi et al. 2008) and promotes tubulin polymerization in the presence of microtubule-associated proteins (Gillardon 2009). The role of LRRK2 at microtubules is unclear, however. It may act directly on microtubules or microtubule-associated proteins. Alternatively, microtubules may function to localize LRRK2 signaling within the cell. For example, LRRK2 has been implicated in the FADD-caspase-8 apoptotic signaling pathway (Ho et al. 2009) and activated caspase-8 is transported retrogradely along microtubules in olfactory receptor neuron apoptosis (Carson et al. 2005). Microtubules might therefore serve as a scaffold for LRRK2 signaling via the caspase-8 pathway, or other microtubule-based signaling complexes. Further investigation is needed to clarify the role of LRRK2 both in microtubule dynamics and in neurite outgrowth to understand the normal function of LRRK2 and how this may be disrupted with disease mutations.
In summary, an emerging body of literature supports a role for LRRK2 function in vesicle trafficking and microtubule dynamics, cellular functions that themselves are closely intertwined. For example, microtubule transport is critical for the transport of membrane to distinct cellular locations in autophagy and synaptic vesicle trafficking. The membrane and microtubule effects of LRRK2 loss-of-function may therefore reflect a single LRRK2 function that becomes manifest in both of these systems, and current studies of LRRK2 function are aimed at determining whether LRRK2 functions at membranous vesicles on the cytoskeleton, or at the interface of these structures.
QUESTION 5: HOW DO PD MUTATIONS AFFECT LRRK2 FUNCTION?
Mutations can affect proteins by decreasing (“loss-of-function”) or increasing (“gain-of-function”) their function. For gain-of-function mutations, a mutation may either increase a protein’s normal function, or confer a novel function to the protein. Importantly, these effects are not mutually exclusive. One can imagine a scenario where a mutation impairs binding to one partner (loss-of-function), leaving a protein free to bind with normal or novel partners (gain-of-function). In general, dominantly inherited mutations act via gain-of-function mechanisms, whereas recessive mutations act via loss-of-function. However, there are exceptions, such as dominantly inherited mutations that impair the function of the wild-type protein—so-called dominant-negative mutations.
All LRRK2 PD mutations fall in or near the “enzymatic core” of the protein. R1441C, R1441G, and R1441H fall within the GTPase domain, and the G2019S and I2020T mutations are located in adjacent residues in the kinase domain. These mutations fall in the “activation loop” of the kinase that is critical in coordinating the kinase phosphotransferase reaction. In addition, the Y1699C mutation falls in the region between the GTPase and kinase domains.
While our understanding of the effect of LRRK2 mutations is at an early stage, current evidence indicates that these mutations act through a gain-of-function mechanism. First, all LRRK2 mutations are dominantly inherited. Second, multiple reports indicate that the G2019S mutations increases LRRK2 kinase activity (West et al. 2005; Greggio et al. 2006; Jaleel et al. 2007), and kinase inhibition is reported to block neurotoxicity both in vitro and in vivo (Greggio et al. 2006; Smith et al. 2006; Lee et al. 2010a). Third, transgenic overexpression of mutant (but not wild-type) LRRK2 is reported to cause PD-related neuropathology (Li et al. 2009; Ramonet et al. 2011), whereas LRRK2 null mice have no clear CNS phenotype (Andres-Mateos et al. 2009; Lin et al. 2009; Herzig et al. 2011). Lastly, as noted above, LRRK2 loss-of-function and overexpression of PD-mutant LRRK2 have opposite effects on neurite outgrowth (MacLeod et al. 2006; Parisiadou et al. 2009; Dachsel et al. 2010; Lin et al. 2010; Ramonet et al. 2011).
While compelling, these observations leave much unknown about the mechanisms whereby LRRK2 mutations promote toxicity. There are no well-validated LRRK2 substrates, so the downstream effects of increased kinase activity are unknown. In fact, it is possible that the LRRK2 GTPase domain is a target of the LRRK2 kinase, raising the possibility that altered GTPase function mediates neurotoxic LRRK2 signals (Xiong et al. 2010). GTPase-binding partners and the effects of mutations on this domain are less well studied, although some mutations have been reported to decrease GTPase function (Guo et al. 2007; Lewis et al. 2007). It is also unknown how the other PD mutations promote toxicity, as these do not consistently increase kinase activity (Greggio et al. 2006; MacLeod et al. 2006; Smith et al. 2006; Jaleel et al. 2007; West et al. 2007). One possibility may be the ability of these mutations to enhance binding to FADD and promote caspase-8-dependent apoptosis, an effect that is also kinase-dependent (Ho et al. 2009).
QUESTION 6: DOES LRRK2 FUNCTION IN PATHWAYS COMMON TO OTHER PD GENES?
Perhaps the most compelling evidence that LRRK2 functions in pathways common to other PD genes is the human neuropathological data. As detailed above, the majority of autopsies performed on patients with LRRK2 mutations display α-synuclein-positive Lewy bodies, tau tangles, or both (Cookson et al. 2008), suggesting that mutations in LRRK2 result in the accumulation of α-synuclein and tau. Yet, some LRRK2 cases do not display overt protein accumulation, instead having pure nigral degeneration, raising the possibility that some aspects of LRRK2 neurotoxicity may occur independently of α-synuclein or tau.
Mouse genetic studies provide some support for the possibility that LRRK2 and α-synuclein operate in a common molecular pathway. Crossing LRRK2 transgenic mice to mice overexpressing mutant α-synuclein results in worsened neuropathology, both in terms of increased protein aggregation and increased loss of neurons in the dorsal striatum (Lin et al. 2009). Importantly, crossing LRRK2 null animals to α-synuclein overexpressing mice has the opposite effect, causing decreased aggregation and enhanced neuronal survival. As noted above, LRRK2 null mice reportedly show α-synuclein accumulation in the kidneys of 20-month-old mice (Tong et al. 2010), providing some additional support for an interaction between LRRK2 and α-synuclein.
It is unknown whether LRRK2 and α-synuclein interact with other proteins implicated from PD genetics. As discussed above, LRRK2 PD presents with a distinct clinical picture from the earlier-onset PD associated with mutations in PINK-1, DJ-1, and parkin. PINK1, DJ-1, and parkin appear to result in disease pathogenesis through mitochondrial dysfunction and oxidative stress. LRRK2 has been reported to localize to mitochondria (Biskup et al. 2006; Gloeckner et al. 2006; Hatano et al. 2007; West et al. 2007) and can immunoprecipitate with parkin following overexpression in cell lines (Smith et al. 2005). However, several important caveats limit the relevance of these studies. First, they were performed before the development of highly specific LRRK2 antibodies. Second, cell studies were performed with overexpression of LRRK2, which may not reflect localization of the protein at endogenous levels. Finally, the same studies showed LRRK2 localization in many other intracellular membranous structures and subsequent cell biological experiments and animal models implicate LRRK2 in the endolysosomal pathway more so than mitochondria. Future studies will need to clarify if and how the proteins associated with autosomal recessive PD interact with LRRK2 and α-synuclein.
CONCLUSIONS
Mutations in LRRK2 contribute to both familial and sporadic PD. Therefore, increased knowledge of LRRK2 biology is likely to have profound implications to a better understanding of PD progression and prevention. In the 7 years since LRRK2 was first identified, much has been learned about it clinically and biologically. Although many questions have been answered, many more remain to be explored. Of immediate concern is the identification of substrates for the kinase and GTPase to gain a better understanding of the cell signaling pathways involved. Similarly, it appears possible that LRRK2 may function upstream of α-synuclein, but how these proteins are connected remains to be elucidated. Finally, and perhaps most pressing, is the question of how relevant LRRK2 function is in sporadic PD. The best test of this question will come from the creation of specific LRRK2 therapies that can be used clinically in PD patients with and without mutations in LRRK2.
Footnotes
Editor: Serge Przedborski
Additional Perspectives on Parkinson’s Disease available at www.perspectivesinmedicine.org
REFERENCES
- Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R 2009. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet 18: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Mateos E, Mejias R, Sasaki M, Li X, Lin BM, Biskup S, Zhang L, Banerjee R, Thomas B, Yang L, et al. 2009. Unexpected lack of hypersensitivity in LRRK2 knock-out mice to MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine). J Neurosci 29: 15846–15850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, et al. 2006. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol 60: 557–569 [DOI] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Rea A, Lorenz-Deperieux B, Coombes CE, Dawson VL, Dawson TM, West AB 2007. Dynamic and redundant regulation of LRRK2 and LRRK1 expression. BMC Neurosci 8: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges D, Moorhead GB 2005. 14-3-3 Proteins: A number of functions for a numbered protein. Sci STKE 2005: re10. [DOI] [PubMed] [Google Scholar]
- Carson C, Saleh M, Fung FW, Nicholson DW, Roskams AJ 2005. Axonal dynactin p150Glued transports caspase-8 to drive retrograde olfactory receptor neuron apoptosis. J Neurosci 25: 6092–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson MR, Hardy J, Lewis PA 2008. Genetic neuropathology of Parkinson’s disease. Int J Clin Exp Pathol 1: 217–231 [PMC free article] [PubMed] [Google Scholar]
- Dachsel JC, Behrouz B, Yue M, Beevers JE, Melrose HL, Farrer MJ 2010. A comparative study of Lrrk2 function in primary neuronal cultures. Parkinsonism Relat Disord 16: 650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Lewis PA, Greggio E, Sluch E, Beilina A, Cookson MR 2008. Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proc Natl Acad Sci 105: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzamko N, Deak M, Hentati F, Reith AD, Prescott AR, Alessi DR, Nichols RJ 2010. Inhibition of LRRK2 kinase activity leads to dephosphorylation of Ser(910)/Ser(935), disruption of 14-3-3 binding and altered cytoplasmic localization. Biochem J 430: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer MJ, Stone JT, Lin CH, Dachsel JC, Hulihan MM, Haugarvoll K, Ross OA, Wu RM 2007. Lrrk2 G2385R is an ancestral risk factor for Parkinson’s disease in Asia. Parkinsonism Relat Disord 13: 89–92 [DOI] [PubMed] [Google Scholar]
- Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F 2002. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol 51: 296–301 [DOI] [PubMed] [Google Scholar]
- Funayama M, Hasegawa K, Ohta E, Kawashima N, Komiyama M, Kowa H, Tsuji S, Obata F 2005. An LRRK2 mutation as a cause for the parkinsonism in the original PARK8 family. Ann Neurol 57: 918–921 [DOI] [PubMed] [Google Scholar]
- Funayama M, Li Y, Tomiyama H, Yoshino H, Imamichi Y, Yamamoto M, Murata M, Toda T, Mizuno Y, Hattori N 2007. Leucine-rich repeat kinase 2 G2385R variant is a risk factor for Parkinson disease in Asian population. Neuroreport 18: 273–275 [DOI] [PubMed] [Google Scholar]
- Gaig C, Marti MJ, Ezquerra M, Cardozo A, Rey MJ, Tolosa E 2009. G2019S LRRK2 mutation causing Parkinson’s disease without Lewy bodies. BMJ Case Reports 10.1136/bcr.08.2008.0632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL 2008. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res 86: 1711–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Covy JP, Bonini NM, Hurtig HI, Farrer MJ, Trojanowski JQ, Van Deerlin VM 2006. Biochemical and pathological characterization of Lrrk2. Ann Neurol 59: 315–322 [DOI] [PubMed] [Google Scholar]
- Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, et al. 2005. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet 365: 415–416 [DOI] [PubMed] [Google Scholar]
- Gillardon F 2009. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-βisoforms and modulates microtubule stability—A point of convergence in parkinsonian neurodegeneration? J Neurochem 110: 1514–1522 [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O’Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M 2006. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet 15: 223–232 [DOI] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, et al. 2006. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis 23: 329–341 [DOI] [PubMed] [Google Scholar]
- Greggio E, Zambrano I, Kaganovich A, Beilina A, Taymans JM, Daniels V, Lewis P, Jain S, Ding J, Syed A, et al. 2008. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem 283: 16906–16914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG 2007. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res 313: 3658–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Kubo S, Imai S, Maeda M, Ishikawa K, Mizuno Y, Hattori N 2007. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum Mol Genet 16: 678–690 [DOI] [PubMed] [Google Scholar]
- Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, Bressman S, Brice A, Aasly J, Zabetian CP, Goldwurm S, et al. 2008. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: A case-control study. Lancet Neurol 7: 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig MC, Kolly C, Persohn E, Theil D, Schweizer T, Hafner T, Stemmelen C, Troxler TJ, Schmid P, Danner S, et al. 2011. LRRK2 protein levels are determined by kinase function and are crucial for kidney and lung homeostasis in mice. Hum Mol Genet 20: 4209–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CC, Rideout HJ, Ribe E, Troy CM, Dauer WT 2009. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J Neurosci 29: 1011–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B 2008. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J 27: 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR 2007. LRRK2 phosphorylates moesin at threonine-558: Characterization of how Parkinson’s disease mutants affect kinase activity. Biochem J 405: 307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen ND, Peng Y, Ho CC, Rideout HJ, Petrey D, Liu P, Dauer WT 2009. The WD40 domain is required for LRRK2 neurotoxicity. PLoS One 4: e8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, Gibson JM, Ross OA, Lynch T, Wiley J, et al. 2005. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: Evidence of a common founder across European populations. Am J Hum Genet 76: 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao T, Venderova K, Park DS, Unterman T, Lu B, Imai Y 2010. Activation of FoxO by LRRK2 induces expression of proapoptotic proteins and alters survival of postmitotic dopaminergic neuron in Drosophila. Hum Mol Genet 19: 3747–3758 [DOI] [PubMed] [Google Scholar]
- Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Taymans JM, Wolozin B, Cookson MR 2010. The Parkinson’s disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One 5: e8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, et al. 2010a. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med 16: 998–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu HP, Lin WY, Guo H, Lu B 2010b. LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction. J Neurosci 30: 16959–16969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Durr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, Pollak P, Brice A 2006. LRRK2 G2019S as a cause of Parkinson’s disease in North African Arabs. New Engl J Med 354: 422–423 [DOI] [PubMed] [Google Scholar]
- Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR 2007. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem Biophys Res Commun 357: 668–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu W, Oo TF, Wang L, Tang Y, Jackson-Lewis V, Zhou C, Geghman K, Bogdanov M, Przedborski S, et al. 2009. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson’s disease. Nat Neurosci 12: 826–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Parisiadou L, Gu XL, Wang L, Shim H, Sun L, Xie C, Long CX, Yang WJ, Ding J, et al. 2009. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson’s-disease-related mutant α-synuclein. Neuron 64: 807–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Tsai PI, Wu RM, Chien CT 2010. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3ss. J Neurosci 30: 13138–13149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A 2006. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron 52: 587–593 [DOI] [PubMed] [Google Scholar]
- Marras C, Schuele B, Munhoz RP, Rogaeva E, Langston JW, Kasten M, Meaney C, Klein C, Wadia PM, Lim SY, et al. 2011. Phenotype in parkinsonian and nonparkinsonian LRRK2 G2019S mutation carriers. Neurology 77: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WC, Pankratz N, Hernandez D, Paisan-Ruiz C, Jain S, Halter CA, Michaels VE, Reed T, Rudolph A, Shults CW, et al. 2005. Genetic screening for a single common LRRK2 mutation in familial Parkinson’s disease. Lancet 365: 410–412 [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, et al. 2010. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson’s disease-associated mutations and regulates cytoplasmic localization. Biochem J 430: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta E, Kawakami F, Kubo M, Obata F 2011. LRRK2 directly phosphorylates Akt1 as a possible physiological substrate: Impairment of the kinase activity by Parkinson’s disease-associated mutations. FEBS Lett 585: 2165–2170 [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, Hunt AL, Klein C, Henick B, Hailpern SM, et al. 2006. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. New Engl J Med 354: 424–425 [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. 2004. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44: 595–600 [DOI] [PubMed] [Google Scholar]
- Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, et al. 2009. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci 29: 13971–13980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonet D, Daher JP, Lin BM, Stafa K, Kim J, Banerjee R, Westerlund M, Pletnikova O, Glauser L, Yang L, et al. 2011. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One 6: e18568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Xin S, Li S, Zhong H, Lin S 2011. Disruption of LRRK2 does not cause specific loss of dopaminergic neurons in zebrafish. PLoS One 6: e20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross OA, Toft M, Whittle AJ, Johnson JL, Papapetropoulos S, Mash DC, Litvan I, Gordon MF, Wszolek ZK, Farrer MJ, et al. 2006. Lrrk2 and Lewy body disease. Ann Neurol 59: 388–393 [DOI] [PubMed] [Google Scholar]
- Ross OA, Spanaki C, Griffith A, Lin CH, Kachergus J, Haugarvoll K, Latsoudis H, Plaitakis A, Ferreira JJ, Sampaio C, et al. 2009. Haplotype analysis of Lrrk2 R1441H carriers with parkinsonism. Parkinsonism Relat Disord 15: 466–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi-Nakashima A, Meir JY, Jin Y, Matsumoto K, Hisamoto N 2007. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Curr Biol 17: 592–598 [DOI] [PubMed] [Google Scholar]
- Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, et al. 2009. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet 41: 1303–1307 [DOI] [PubMed] [Google Scholar]
- Sheng D, Qu D, Kwok KH, Ng SS, Lim AY, Aw SS, Lee CW, Sung WK, Tan EK, Lufkin T, et al. 2010. Deletion of the WD40 domain of LRRK2 in zebrafish causes Parkinsonism-like loss of neurons and locomotive defect. PLoS Genet 6: e1000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, et al. 2009. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 41: 1308–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA 2005. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci 102: 18676–18681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA 2006. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci 9: 1231–1233 [DOI] [PubMed] [Google Scholar]
- Tan EK, Peng R, Wu YR, Wu RM, Wu-Chou YH, Tan LC, An XK, Chen CM, Fook-Chong S, Lu CS 2009. LRRK2 G2385R modulates age at onset in Parkinson’s disease: A multi-center pooled analysis. Am J Med Genet B 150B: 1022–10232 [DOI] [PubMed] [Google Scholar]
- Taylor JP, Mata IF, Farrer MJ 2006. LRRK2: A common pathway for parkinsonism, pathogenesis and prevention? Trends Mol Med 12: 76–82 [DOI] [PubMed] [Google Scholar]
- Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher R Jr, Shen J 2010. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of α-synuclein, and apoptotic cell death in aged mice. Proc Natl Acad Sci 107: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Avruch J 2002. 14-3-3 proteins: Active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem 277: 3061–3064 [DOI] [PubMed] [Google Scholar]
- Vitte J, Traver S, De Paula AM, Lesage S, Rovelli G, Corti O, Duyckaerts C, Brice A 2010. Leucine-rich repeat kinase 2 is associated with the endoplasmic reticulum in dopaminergic neurons and accumulates in the core of Lewy bodies in Parkinson disease. J Neuropathol Exp Neurol 69: 959–972 [DOI] [PubMed] [Google Scholar]
- Wang D, Tang B, Zhao G, Pan Q, Xia K, Bodmer R, Zhang Z 2008. Dispensable role of Drosophila ortholog of LRRK2 kinase activity in survival of dopaminergic neurons. Mol Neurodegener 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber PJ, Smith AD, Sen S, Renfrow MB, Mobley JA, West AB 2011. Autophosphorylation in the leucine-rich repeat kinase 2 (LRRK2) GTPase domain modifies kinase and GTP-binding activities. J Mol Biol 412: 94–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM 2005. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci 102: 16842–16847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, et al. 2007. Parkinson’s disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet 16: 223–232 [DOI] [PubMed] [Google Scholar]
- Westerlund M, Belin AC, Anvret A, Bickford P, Olson L, Galter D 2008. Developmental regulation of leucine-rich repeat kinase 1 and 2 expression in the brain and other rodent and human organs: Implications for Parkinson’s disease. Neuroscience 152: 429–436 [DOI] [PubMed] [Google Scholar]
- Wszolek ZK, Pfeiffer B, Fulgham JR, Parisi JE, Thompson BM, Uitti RJ, Calne DB, Pfeiffer RF 1995. Western Nebraska family (family D) with autosomal dominant parkinsonism. Neurology 45: 502–505 [DOI] [PubMed] [Google Scholar]
- Wszolek ZK, Vieregge P, Uitti RJ, Gasser T, Yasuhara O, McGeer P, Berry K, Calne DB, Vingerhoets FJG, Klein C, et al. 1997. German-Canadian family (Family A) with parkinsonism, amyotrophy, and dementia—Longitudinal observations. Parkinsonism Relat Disord 3: 125–139 [DOI] [PubMed] [Google Scholar]
- Wszolek ZK, Pfeiffer RF, Tsuboi Y, Uitti RJ, McComb AJ, Stoessl AJ, Strongosky A, Zimprich B, Muller-Myhsok MJ, Farrer, et al. 2004. Autosomal dominant parkinsonism associated with variable synuclein and tau pathology. Neurology 62: 1619–1622 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ 2010. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet 6: e1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, et al. 2004. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44: 601–607 [DOI] [PubMed] [Google Scholar]