Abstract
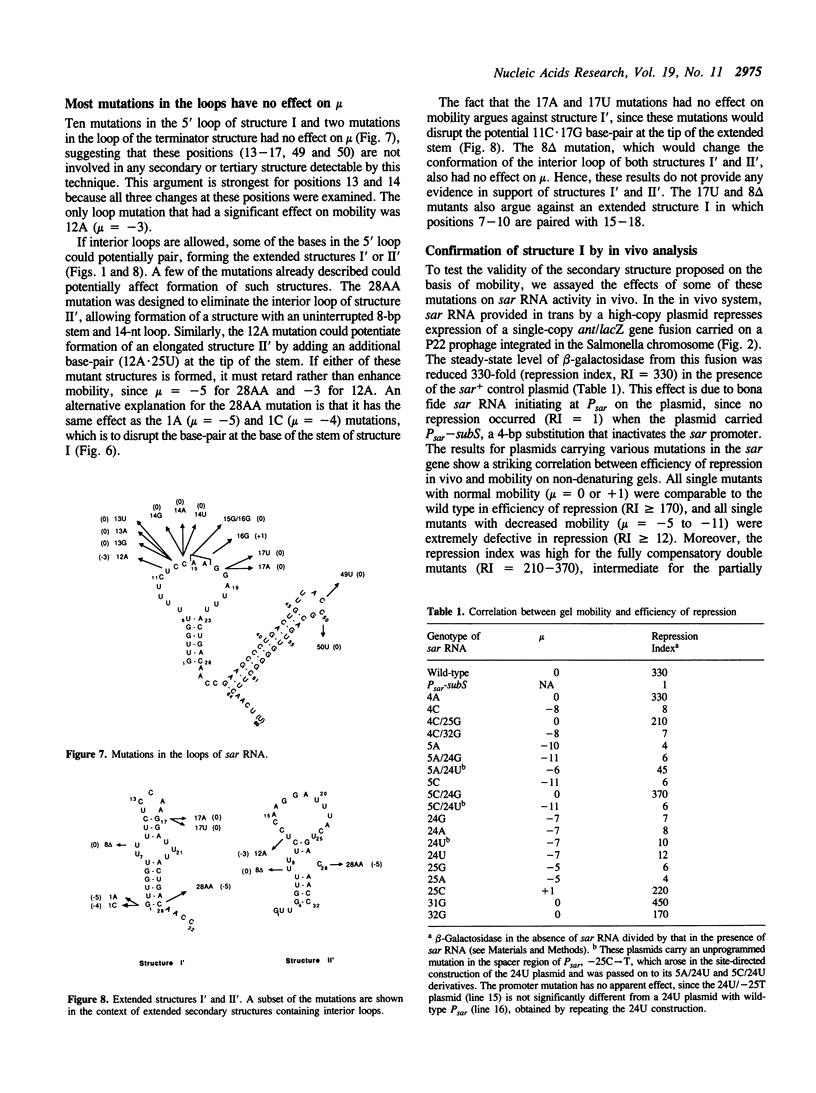
Natural antisense RNAs have stem-loop (hairpin) secondary structures that are important for their function. The sar antisense RNA of phage P22 is unusual: the 3' half of the molecule forms an extensive stem-loop, but potential structures for the 5' half are not predicted to be thermodynamically stable. We devised a novel method to determine the secondary structure of sar RNA by examining the electrophoretic mobility on non-denaturing gels of numerous sar mutants. The results show that the wild-type RNA forms a 5' stem-loop that enhances electrophoretic mobility. All mutations that disrupt the stem of this hairpin decrease mobility of the RNA. In contrast, mutations that change the sequence of the stem without disrupting it (e.g. change G.U to A.U) do not affect mobility. Nearly all mutations in single-stranded regions of the structure also have no effect on mobility. Confirmation of the proposed 5' stem-loop was obtained by constructing and analyzing compensatory double mutants. Combinations of mutations that restore a base-pair of the stem also restore mobility. The genetic phenotypes of sar mutants confirm that the proposed secondary structure is correct and is essential for optimal activity of the antisense RNA in vivo.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhattacharyya A., Lilley D. M. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 1989 Sep 12;17(17):6821–6840. doi: 10.1093/nar/17.17.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Case C. C., Roels S. M., Jensen P. D., Lee J., Kleckner N., Simons R. W. The unusual stability of the IS10 anti-sense RNA is critical for its function and is determined by the structure of its stem-domain. EMBO J. 1989 Dec 20;8(13):4297–4305. doi: 10.1002/j.1460-2075.1989.tb08616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Graña D., Gardella T., Susskind M. M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988 Oct;120(2):319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graña D., Youderian P., Susskind M. M. Mutations that improve the ant promoter of Salmonella phage P22. Genetics. 1985 May;110(1):1–16. doi: 10.1093/genetics/110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques J. P., Susskind M. M. Pseudo-templated transcription by Escherichia coli RNA polymerase at a mutant promoter. Genes Dev. 1990 Oct;4(10):1801–1810. doi: 10.1101/gad.4.10.1801. [DOI] [PubMed] [Google Scholar]
- Knight K. L., Bowie J. U., Vershon A. K., Kelley R. D., Sauer R. T. The Arc and Mnt repressors. A new class of sequence-specific DNA-binding protein. J Biol Chem. 1989 Mar 5;264(7):3639–3642. [PubMed] [Google Scholar]
- Kramer F. R., Mills D. R. RNA sequencing with radioactive chain-terminating ribonucleotides. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5334–5338. doi: 10.1073/pnas.75.11.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. M., Wu T. H., Chiang C. H., Susskind M. M., McClure W. R. Control of gene expression in bacteriophage P22 by a small antisense RNA. I. Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev. 1987 Apr;1(2):197–203. doi: 10.1101/gad.1.2.197. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J. A., Crothers D. M. DNA bending by the bulge defect. Biochemistry. 1989 May 16;28(10):4512–4516. doi: 10.1021/bi00436a058. [DOI] [PubMed] [Google Scholar]
- SMITH H. O., LEVINE M. TWO SEQUENTIAL REPRESSIONS OF DNA SYNTHESIS IN THE ESTABLISHMENT OF LYSOGENY BY PHAGE P22 AND ITS MUTANTS. Proc Natl Acad Sci U S A. 1964 Aug;52:356–363. doi: 10.1073/pnas.52.2.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Wu T. H., Liao S. M., McClure W. R., Susskind M. M. Control of gene expression in bacteriophage P22 by a small antisense RNA. II. Characterization of mutants defective in repression. Genes Dev. 1987 Apr;1(2):204–212. doi: 10.1101/gad.1.2.204. [DOI] [PubMed] [Google Scholar]