Abstract
RNA interference (RNAi) describes a highly conserved pathway, present in eukaryotic cells, for regulating gene expression. Small stretches of double-stranded RNA, termed small interfering RNAs (siRNAs), utilize this pathway to bind homologous mRNA, resulting in site-specific mRNA cleavage and subsequent protein degradation. The ubiquitous presence of the RNAi machinery, combined with its specificity and efficacy, makes it an attractive mechanism for reducing aberrant gene expression in therapeutic settings. However, a major obstacle to utilizing RNAi in the clinic is siRNA delivery. Administered siRNAs must make contact with the appropriate cell types and, following internalization, gain access to the cytosol where the RNAi machinery resides. This must be achieved so that silencing is maximized, whilst minimizing any undesirable off-target effects. Recently, the utility of siRNAs as a microbicide, usually applied to the genital mucosa for preventing transmission of sexually transmitted diseases including HIV-1 and HSV-2, has been investigated. In this review we will describe these studies and discuss potential strategies for improving gene silencing.
Introduction
Sexually transmitted diseases of viral etiology, in particular HIV-1 and HSV-2, affect millions of people worldwide. A recent UNAIDS report lists the total number of people living with HIV-1 at over 33 million (UNAIDS, 2009). Sub-Saharan Africa accounts for over 60% of infections and women are particularly susceptible as they are often unable to negotiate safe sex (Garcia-Calleja et al., 2006; Pilcher, 2004). HSV-2 is a major co-factor for HIV-1 (Freeman et al., 2006; Wald and Link, 2002). In the U.S. more than 20% of the population is infected with HSV-2, and the infection rate in some areas of sub-Saharan Africa has been recorded to be as high as 90% (Corey et al., 2004). HSV-2 infection is the leading cause of genital ulcers. The ulcers, even when healed, are a major source of inflammatory cells, and it is thought that this creates a permissive environment for transmission of HIV-1 (Celum et al., 2004). There is no cure for HIV-1 or HSV-2, therefore a method to prevent transmission would make a significant impact on slowing the spread of these diseases. Furthermore, a successful strategy that prevents HSV-2 infection could result in a concomitant decrease in HIV-1 transmission. A vaccine that confers long-lived protection would be an ideal strategy. There are several vaccine candidates for HIV-1 and HSV-2. However, these have shown no protection, limited protection, and in some cases resulted in increased infection (Dudek and Knipe, 2006; McElrath et al., 2008; Rerks-Ngarm et al., 2009).
In the absence of an effective vaccine, microbicides are being developed to prevent HIV-1 and HSV-2 transmission. Microbicides are compounds that, when applied to the genital mucosa, decrease viral transmission by inhibiting viral uptake or replication. There are more than 50 microbicides under development for HIV-1 (complete listing on www.microbicide.org). Disappointingly, most of the trials that have proceeded through phase III testing have not shown any significant decrease in the rate of HIV-1 transmission. However, one recent study reported decreased HIV-1 transmission rates of almost 40% when an HIV-1 inhibitor was vaginally applied in a gel formulation (Abdool Karim et al., 2010). This result is encouraging and demonstrates that an effective microbicide is an achievable goal, not merely an unproven concept.
We initiated studies to determine whether RNA interference (RNAi) could be utilized to prevent HSV-2 transmission. RNAi is a pathway, found in many species, that regulates gene expression. Originally described in plants and Caenorhabditis elegans, RNAi is also functional in mammalian cells (Elbashir et al., 2001; Fire et al., 1998). The RNAi pathway is mediated through small non-coding double stranded RNA species, termed small interfering RNAs (siRNAs). The siRNA binds to homologous target mRNA, resulting in mRNA cleavage and subsequent protein degradation. Due to the specificity of siRNA-mediated mRNA cleavage, gene silencing via the RNAi pathway has become an attractive method to target diseases, including those of viral etiology. We have used siRNAs that target either HSV-2-specific viral genes or host-encoded viral entry receptors to prevent transmission in a mouse model of vaginal HSV-2 infection (Palliser et al., 2006; Wu et al., 2009). We will review the studies that demonstrate the feasibility of using siRNAs in a topical microbicide. We will also discuss how a detailed knowledge of the mechanisms involved in delivery and uptake of a vaginal microbicide will likely be required to allow judicious optimization of siRNAs formulated for vaginal delivery.
An Unmet Need
Although there are many microbicides in clinical trials, only one has demonstrated efficacy in reducing HIV-1 transmission (summarized in Hladik and Doncel, 2010). Microbicides can be categorized by their mode of action. Non-specific compounds have the potential to inhibit multiple types of virus. However, to date, these microbicides have not proven to be effective. Nonoxynol-9 (N9), a detergent that disrupts the viral envelope, was found to effectively inhibit HIV-1 replication in vitro. However, in clinical trials, vaginal application of N9 resulted in increased HIV-1 transmission. This enhanced susceptibility was attributed to N9 causing disruption of the vaginal epithelium, resulting in inflammation (Van Damme et al., 2002). Recently, glycerol monolaurate (GML; also a detergent) was reported to protect macaques from vaginal SIV infection (Li et al., 2009). GML was evaluated in a macaque model for safety (Schlievert et al., 2008). Multiple criteria were reported, including normal mucosal integrity and no induction of inflammation. Although these results are encouraging, a study using a murine HSV-2 susceptibility model reported that vaginal application of GML resulted in increased levels of HSV-2 virus in the vaginal cavity (Moench et al., 2010). Which safety models are most predictive of an outcome in humans is not known and will need to be determined.
Specific microbicides are designed to inhibit a distinct viral interaction, e.g., CCR5 antagonists that prevent HIV-1 binding its co-receptor. Tenofovir is an adenosine nucleotide analogue that is used for treating HIV-1 infection. A clinical trial tested the efficacy of a 1% concentration of tenofovir in a gel formulation as a vaginal microbicide and found that HIV transmission was reduced by 39% (Abdool Karim et al., 2010). This figure rose to 54% in women who better adhered to the gel application regimen. A 51% reduction in HSV-2 infection was also reported. However, HIV-1 acquisition did not correlate with HSV-2 infection — the efficacy of tenofovir gel was similar, irrespective of HSV-2 status. This study suggests that a topically applied gel can effectively reduce transmission of HIV-1 and HSV-2. These results are encouraging; however, issues regarding dosing, long-term tolerability, and cost need to be addressed. It may be that other compounds, or combinations of compounds, may yield better results and should be evaluated.
siRNAs as a Vaginal Microbicide
siRNAs are considered attractive therapeutic agents because of their ability to potentially target any disease for which the genetic basis is known. Clinical trials (see http://clinicaltrials.gov/) targeting respiratory syncytial virus (DeVincenzo et al., 2008; DeVincenzo et al., 2010) (Alnylam), liver cancers (Alnylam, 2011) (Alnylam), melanoma (Davis et al., 2010) (Calando), and hypercholesterolemia (Zimmermann et al., 2006) (Tekmira) highlight some of the diverse diseases that are being targeted using RNAi. HIV-1 was one of the first viruses found to be amenable to siRNA-mediated cleavage in mammalian cells (Capodici et al., 2002; Coburn and Cullen, 2002; Jacque et al., 2002; Lee et al., 2002; Novina et al., 2002; Surabhi and Gaynor, 2002). Subsequent studies showed that the RNAi pathway could be used to target viruses including polio, influenza, and HSV (Bhuyan et al., 2004; Ge et al., 2004; Gitlin et al., 2002). Therefore, siRNAs can effectively inhibit viral replication in vitro, following transfection of either chemically synthesized siRNAs or vector-encoded short hairpin RNA (shRNA). However, to be effective as a topical microbicide, siRNAs must be formulated so that they remain available for cellular uptake. They must be designed to resist degradation by nucleases, inactivation due to the low pH vaginal environment, or entrapment in the mucosal layer. siRNAs must gain access to the appropriate cells targeted by the virus, e.g., CD4+ T cells, macrophages, and DCs for HIV-1 and epithelial cells and neurons for HSV-2. The mechanism of cellular uptake must result in delivery of siRNAs into the cytoplasm, where the RNA induced silencing complex (RISC) resides. As an siRNA must circumvent such a large and diverse set of obstacles following vaginal application, siRNA formulation is critical in determining how effectively viral transmission is inhibited (summarized in Figure 1; for a general review discussing siRNA delivery, see Manjunath and Dykxhoorn, 2010).
Figure 1.
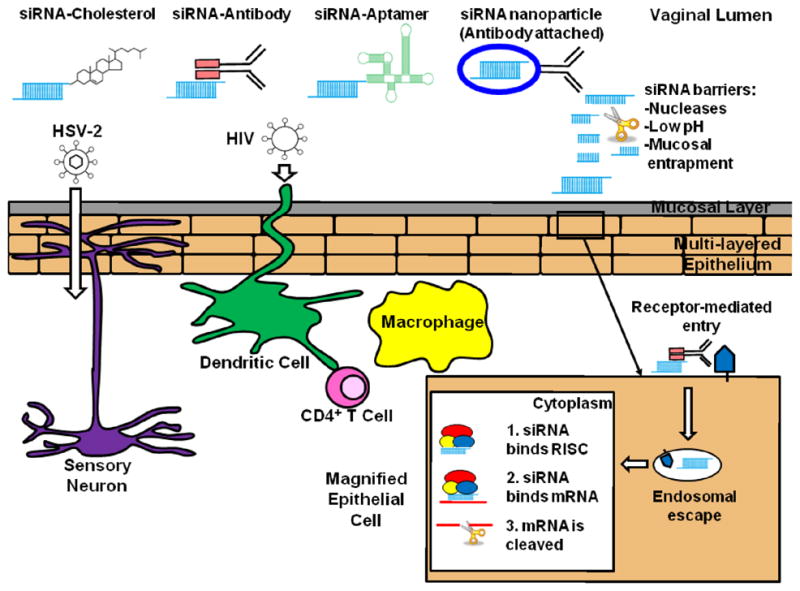
Initial cellular targets for viral entry of HSV-2 and HIV-1 and methods of delivering siRNAs that could be used to prevent transmission of these viruses. HSV-2 infects epithelial cells and neurons. HIV-1 infects CD4+ T cells, macrophages, and dendritic cells. siRNAs delivered to the vaginal lumen must be protected from nucleases, low pH environment, and avoid mucosal entrapment. Upon uptake by a cell, such as an epithelial cell, siRNAs must escape the endosome and gain entry to the cytoplasm. Once inside the cytoplasm, siRNA is incorporated into the RNA-induced silencing complex (RISC). The siRNA then binds, via sequence homology, to its target mRNA, resulting in site-specific cleavage of the mRNA by the RNase III enzyme argonaute 2.
In initial studies we used a mouse model of HSV-2 transmission as a proof-of-concept to determine whether siRNAs could inhibit viral infection across the genital mucosa (Palliser et al., 2006). siRNAs targeting viral genes, essential for viral viability, were complexed with a cationic lipid and applied to the vaginal mucosa both prior to and following challenge with HSV-2 virus. Mice were protected from HSV-2 transmission. However, protection was transient; siRNAs had to be administered within a few hours of viral challenge for mice to be protected. siRNAs were observed in the vaginal epithelial cells and were detectable deep into the lamina propria. Furthermore, targeting an endogenous gene resulted in more durable silencing, with reduced expression of mRNA and protein observed for at least 7 days (Palliser et al., 2006; Zhang et al., 2006). A similar observation had been reported for siRNA-mediated inhibition of HIV-1 replication in macrophages in vitro. Knockdown of the HIV co-receptor, CCR5, was sustained for several days and coincided with decreased levels of HIV replication, whereas silencing HIV viral genes inhibited viral replication only if administered within hours of viral challenge (Song et al., 2003).
Although the preliminary results are encouraging, the transient nature of the protection observed is not optimal for a microbicide. The CAPRISA 004 trial reported low adherence rates for tenofovir gel, with compliance possibly decreasing further over time, concomitant with a reduction in gel efficacy (Abdool Karim et al., 2010). A microbicide that confers more durable protection could circumvent this problem.
Obstacles Along the Way
Histological analysis of vaginal tissue isolated from mice treated with lipid-complexed siRNAs showed no evidence of inflammatory infiltrates and induction of interferon-related genes was not detected (Palliser et al., 2006). However, in follow-up studies using additional criteria, lipid-related toxicity was observed (Woodrow et al., 2009; Wu et al., 2009). These results emphasize the need to use assays that accurately determine whether a microbicide, both the active component and the carrier, can induce undesirable responses. As discussed above, this is not a trivial undertaking. When microbicides such as N9, originally reported to be safe, were evaluated in a mouse model, disruption of the vaginal epithelium, significant inflammatory responses, and increased susceptibility to HSV-2 were reported (Mesquita et al., 2009; Moench et al., 2010).
Due to their short length, siRNAs were originally believed not to induce interferon-related responses. However, activation of immune responses via toll-like receptors (TLR) and retinoic acid inducible gene-I (RIG-I) by siRNAs has been reported (Robbins et al., 2009). Some of these responses are sequence or structure dependent and can therefore be avoided when designing siRNAs. Chemical modification of siRNAs is routinely used to prevent induction of immune responses. 2′-O-methyl and 2′-fluoro substitutions effectively negate immune stimulation. Not all ribonucleotides need to be replaced with 2′-O-methyl or 2′-fluoro. Two substitutions in the siRNA sense strand can be sufficient to ameliorate immunostimulatory activity. Minimal alterations to the siRNA backbone are preferable to maintain silencing efficiency of the siRNA (Robbins et al., 2009).
In vivo, degradation of siRNAs by nucleases shortens the half-life of unmodified siRNAs to minutes. Unmodified siRNAs can be protected from nucleases, for example, by complexation in a lipid or nanoparticle (Wu et al., 2009). However, as these carriers can induce toxic effects, it may be preferable to use siRNAs that are nuclease-resistant. In addition to decreasing immune stimulation, siRNAs are stabilized by modification with 2′-O-methyl or 2′-fluoro (Robbins et al., 2009). Incorporation of phosphorothioate (PS) groups also extends siRNA half-life (Soutschek et al., 2004; Vaishnaw et al., 2010).
miRNA-like off-target effects have been reported following siRNA treatment (Birmingham et al., 2006; Jackson et al., 2003). This occurs if the siRNA contains sufficient homology to non-targeted mRNA in the open reading frame or the 32 untranslated region. Algorithms used to identify active sequences usually exclude siRNA that share a degree of homology with non-targeted mRNAs. If homologous sequences need to be chosen, the sense and/or antisense strands can be chemically modified to minimize off-target silencing (Jackson et al., 2006). In vitro microarray data has shown down-regulation of a large number of transcripts following siRNA transfection (Jackson and Linsley, 2010). Whether the decrease in mRNA reported is sufficient to significantly modulate protein expression is not clear. Furthermore, miRNA-like off-target effects are often minimal at low concentrations of siRNA (picomolar to low nanomolar range), the dose at which potent siRNAs are effective (Vaishnaw et al., 2010).
Saturation of the RNAi machinery can also mediate unwanted effects. Overexpression of shRNA in mouse hepatocytes following intravenous infusion resulted in liver injury and, in some cases, death. Morbidity correlated with decreased levels of liver-associated miRNAs, and saturation of exportin-5 was observed (Grimm et al., 2006). Administration of siRNAs in vivo has been reported not to interfere with the miRNA pathway, even when multiple siRNAs were injected (John et al., 2007). However, some studies report that, just as observed for shRNAs, high concentrations of siRNAs can perturb miRNA function (Khan et al., 2009). Furthermore, injection of multiple siRNAs can lead to competition for RISC resulting in less efficient silencing of one of the targeted genes (Bitko et al., 2005; Castanotto et al., 2007).
Improving siRNA Delivery
As the lipid reagent used in our initial studies displayed some toxicity, an alternative method to deliver the siRNAs was needed. Studies looking at siRNA delivery at another mucosal surface, the lung, reported siRNA uptake and gene-specific silencing using unmodified siRNAs formulated in saline, delivered intranasally (Alvarez et al., 2009; Bitko et al., 2005). However, we found that unmodified siRNAs were degraded within minutes following incubation with vaginal washes (Wu et al., 2009). Substitution of one PS group on the sense and antisense strand of the siRNA was sufficient for protection from nucleases. However, the PS-siRNAs were not effective for gene silencing following vaginal delivery. Further modification of the siRNA sense strand by adding a cholesterol moiety resulted in siRNA uptake and silencing of the targeted gene (Soutschek et al., 2004; Wu et al., 2009).
Previous studies, including our own, suggested that targeting endogenous genes results in long-lived gene silencing (Palliser et al., 2006; Song et al., 2003; Wu et al., 2009). Therefore, to increase durability of protection in the HSV-2 infection model, we targeted HSV-2 viral entry receptor genes. We used siRNAs modified for stability (PS-siRNA) that had a cholesterol group attached to the sense strand (C-siRNA) to circumvent a requirement for complexing the siRNA with a lipid transfection reagent (Soutschek et al., 2004). When C-siRNA specific for a putative HSV-2 entry receptor, nectin-1, was vaginally administered, mice were protected from viral transmission for at least 7 days following C-siRNA application. Protection correlated with receptor knockdown, with no protection observed when C-siRNAs were given within hours of viral challenge. As observed in our previous study, application of C-siRNA specific for viral genes protected mice only when administered close to the time of viral challenge. When C-siRNAs specific for both viral genes and nectin-1 were combined, mice were protected from viral infection for 7 days, irrespective of time of challenge (Wu et al., 2009).
From the studies outlined, targeting a combination of genes may confer better protection from viral transmission than knocking down one gene with a single siRNA. Using multiple siRNAs should result in fewer off-target effects and for an error-prone virus, such as HIV-1, targeting multiple genes should prevent emergence of RNAi-resistant variants that harbor mutations in the target sequence. Targeting both viral and endogenous genes involved in viral replication may be a good strategy for achieving durable protection that is effective within hours of application as well as several days later. However, targeting endogenous genes may not be feasible if knockdown results in altered cell function. As siRNAs targeting viral genes may be short-lived once inside the cell, particles designed for sustained release of siRNAs could be useful for providing sustained protection from viral transmission. In a recent study, nanoparticles were synthesized to release siRNAs over several weeks. Vaginally applied nanoparticles were observed in the epithelium and lamina propria, and the cervix. Endogenous genes could be silenced for 14 days, in the absence of any overt cellular infiltration or epithelial disruption (Woodrow et al., 2009).
What Next?
The initial studies outlined demonstrate that use of siRNAs as part of a microbicide is feasible. To achieve effective and durable protection from viral transmission, siRNA delivery must be optimized. As discussed above, off-target effects can be overcome using potent siRNAs that are chemically modified. Another potential source of toxicity is uptake of siRNAs by bystander cells. This problem could be alleviated by limiting siRNA delivery to cells targeted by virus. For example, the primary cells infected by HIV are macrophages, dendritic cells, and CD4+ T cells. siRNAs have been delivered to these cells using antibody- and aptamer-based modalities. When attached to an antibody, protamine, a positively charged protein that binds nucleic acids, can be used to specifically deliver siRNAs to a receptor-positive cell. An HIV-specific gp120 antibody, fused to protamine, was used to deliver siRNAs to HIV-infected cells (i.e., expressing HIV-gp120) (Song et al., 2005). Growth of a melanoma, stably expressing gp120, was delayed when gp120 antibody-protamine bound to oncogene-specific siRNAs were injected intravenously into a mouse model. Histological analysis of tumors from mice injected with fluorescently-labeled siRNAs bound to gp120-antibody-protamine showed siRNA uptake only in gp120-positive tumors. This method has been used to deliver siRNAs to CD7+ and LFA-1+ cells using CD7− and LFA-1-specific antibodies, respectively (Kumar et al., 2008; Peer et al., 2007). CD7 antibody, fused to nine arginines, was used to deliver siRNAs specific for HIV-1 viral genes and the CCR5 coreceptor to T cells in a humanized mouse model. Intravenous injection of the antibody-bound siRNAs protected mice from HIV-1 challenge. This regimen was also effective for controlling HIV-1 infection and maintaining CD4+ T cells (Kumar et al., 2008).
Similar to antibodies, aptamers bind specific ligands, and have been used to deliver siRNAs to receptor-positive cells. Aptamers are structured nucleic acids and ligand-specific aptamers can be selected following several round of selection, using a protein or cell type of interest (Yan and Levy, 2009). Aptamers have been selected with higher binding affinities, compared with antibodies (picomolar range). In addition, aptamers are easily synthesized and are amenable to chemical modification thereby protecting the nucleic acid from nucleases and limiting off-target responses. An anti-PSMA aptamer (prostate-specific membrane antigen) has been reported to deliver siRNAs to prostate specific membrane antigen (PSMA)-positive cells in vitro and in vivo (Chu et al., 2006; Dassie et al., 2009; McNamara et al., 2006). A gp120 aptamer neutralizes HIV-1. Fusion of this aptamer to anti-tat/rev siRNA resulted in uptake of this construct only by HIV-1 infected cells, decreased expression of tat/rev transcripts, and better suppression of HIV-1, when compared to using gp120 aptamer alone (Zhou et al., 2008). Injection of this construct into an HIV-1 infected humanized mouse model is reported to inhibit HIV-1 replication by several logs and prevent CD4+ T cell depletion normally associated with HIV-1 infection (Neff et al., 2011). Modification of the gp120 aptamer to bind multiple siRNAs (using a GC-rich “sticky bridge”) has been used to deliver siRNAs targeting HIV-1 tat/rev, the HIV-1 receptor CD4, and HIV-1-dependency factor TNPO3 to HIV-1 infected cells in vivo. Competition for RISC entry by the siRNAs was observed. However, silencing was sufficient to suppress HIV-1 viral loads and stabilize CD4+ T cell numbers. As previously stated, for an error-prone virus such as HIV-1, targeting multiple genes should minimize the emergence of viral escape mutants (Zhou and Rossi, 2010; Zhou et al., 2009).
Although delivery of siRNAs via specific receptors represents a method that should lower effective dose and limit toxicity, siRNA uptake does not necessarily equate with effective silencing. Following internalization, siRNAs must gain access to the cytosol where RISC resides (Zeng and Cullen, 2002). Receptor-mediated endocytosis would generally require siRNAs to cross the endosomal membrane to reach the cytosol. Studies have shown that this process can be very inefficient. However, strategies such as the inclusion of an endosomal escape peptide in an siRNA/ligand complex can facilitate siRNA entry into the cytosol and increase efficiency of silencing (Aouadi et al., 2009; Oliveira et al., 2007).
Conclusion
Initial studies support the feasibility of using siRNAs as part of a microbicide to prevent sexually transmitted diseases, such as HIV-1 and HSV-2. Vaginally applied siRNAs were observed in epithelial cells and deep in the lamina propria. This level of siRNA uptake was sufficient to protect mice from a lethal HSV-2 infection (Palliser et al., 2006; Wu et al., 2009). Lipid-based reagents confer some toxicity, and it is unclear whether lipid-formulated siRNAs are taken up effectively by the initial cells infected by HIV-1. Therefore, receptor-mediated siRNA delivery may represent a feasible strategy for microbicide development. The expense associated with antibody production may limit their utility as part of a microbicide, with aptamer-mediated delivery potentially being more cost-effective. Increased binding affinity, ease of manufacture, ability to modify nucleic acids, and possible lack of immune-stimulating responses are additional attractive features associated with aptamer technology (Yan and Levy, 2009).
In addition to ensuring delivery of siRNAs to appropriate cells, durability of protection is an important factor to consider in terms of overall cost of each application and, as has been reported in microbicide trials, compliance (Abdool Karim et al., 2010). Silencing endogenous genes may result in long-term (days) inhibition of protein expression (Palliser et al., 2006; Song et al., 2003; Wu et al., 2009). If knockdown of host genes is not feasible, reagents such as nanoparticles could be used to release siRNAs over a period of days to weeks (Woodrow et al., 2009). To ensure uptake of nanoparticles by appropriate cells, the particles could be modified to bind antibodies or aptamers (Peer et al., 2008). Such modification should also serve to reduce the effective siRNA dose and limit toxicity.
As has been emphasized throughout this review, determining any toxic effects associated with an siRNA or its carrier is critical for synthesizing a microbicide that does not fail in initial safety trials, due to undesirable side effects. A major problem in evaluating potential microbicides is a lack of models to predict safety. Recent work has identified new biomarkers that are consistent with safety outcomes seen in clinical trials (Mesquita et al., 2009; Moench et al., 2010; Wilson et al., 2009).
Moving forward, increased efficacy and safety of topically applied siRNAs will likely require receptor-mediated delivery and possibly sustained siRNA release. These technologies are available and with the rapid pace with which siRNA-mediated clinical trials are proceeding, an siRNA-based microbicide should be a viable option.
Footnotes
Disclosure
The authors have no financial conflicts of interest to disclose.
References
- Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, Kharsany AB, Sibeko S, Mlisana KP, Omar Z, Gengiah TN, Maarschalk S, Arulappan N, Mlotshwa M, Morris L, Taylor D. Effectiveness and safety of tenofovir gel, an antiretroviral microbicide, for the prevention of HIV infection in women. Science. 2010;329(5996):1168–1174. doi: 10.1126/science.1193748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnylam [Accessed on Feb. 14, 2011];Alnylam demonstrates RNAi in man with systemically delivered RNAi therapeutics. 2011 http://phx.corporate-ir.net/phoenix.zhtml?c=148005&p=irol-newsArticle2&ID=1512322&highlight=
- Alvarez R, Elbashir S, Borland T, Toudjarska I, Hadwiger P, John M, Roehl I, Morskaya SS, Martinello R, Kahn J, Van Ranst M, Tripp RA, DeVincenzo JP, Pandey R, Maier M, Nechev L, Manoharan M, Kotelianski V, Meyers R. RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob Agents Chemother. 2009;53(9):3952–3962. doi: 10.1128/AAC.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, Ostroff GR, Czech MP. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458(7242):1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuyan PK, Kariko K, Capodici J, Lubinski J, Hook LM, Friedman HM, Weissman D. Short interfering RNA-mediated inhibition of herpes simplex virus type 1 gene expression and function during infection of human keratinocytes. J Virol. 2004;78(19):10276–10281. doi: 10.1128/JVI.78.19.10276-10281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3(3):199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11(1):50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- Capodici J, Kariko K, Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J Immunol. 2002;169(9):5196–5201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Sakurai K, Lingeman R, Li H, Shively L, Aagaard L, Soifer H, Gatignol A, Riggs A, Rossi JJ. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35(15):5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celum C, Levine R, Weaver M, Wald A. Genital herpes and human immunodeficiency virus: double trouble. Bull World Health Organ. 2004;82(6):447–453. [PMC free article] [PubMed] [Google Scholar]
- Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34(10):e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn GA, Cullen BR. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J Virol. 2002;76(18):9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L, Wald A, Celum CL, Quinn TC. The effects of herpes simplex virus-2 on HIV-1 acquisition and transmission: a review of two overlapping epidemics. J Acquir Immune Defic Syndr. 2004;35(5):435–445. doi: 10.1097/00126334-200404150-00001. [DOI] [PubMed] [Google Scholar]
- Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, 2nd, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27(9):839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, Toudjarska I, Nechev L, Murugaiah V, Van Vliet A, Vaishnaw AK, Meyers R. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral Res. 2008;77(3):225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- DeVincenzo J, Lambkin-Williams R, Wilkinson T, Cehelsky J, Nochur S, Walsh E, Meyers R, Gollob J, Vaishnaw A. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2010;107(19):8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek T, Knipe DM. Replication-defective viruses as vaccines and vaccine vectors. Virology. 2006;344(1):230–239. doi: 10.1016/j.virol.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Freeman EE, Weiss HA, Glynn JR, Cross PL, Whitworth JA, Hayes RJ. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS. 2006;20(1):73–83. doi: 10.1097/01.aids.0000198081.09337.a7. [DOI] [PubMed] [Google Scholar]
- Garcia-Calleja JM, Gouws E, Ghys PD. National population based HIV prevalence surveys in sub-Saharan Africa: results and implications for HIV and AIDS estimates. Sex Transm Infect. 2006;82(Suppl 3):iii64–70. doi: 10.1136/sti.2006.019901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, Filip L, Bai A, Nguyen T, Eisen HN, Chen J. Inhibition of influenza virus production in virus-infected mice by RNA interference. Proc Natl Acad Sci U S A. 2004;101(23):8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418(6896):430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Hladik F, Doncel GF. Preventing mucosal HIV transmission with topical microbicides: challenges and opportunities. Antiviral Res. 2010;88(Suppl 1):S3–S9. doi: 10.1016/j.antiviral.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12(7):1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418(6896):435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M, Constien R, Akinc A, Goldberg M, Moon YA, Spranger M, Hadwiger P, Soutschek J, Vornlocher HP, Manoharan M, Stoffel M, Langer R, Anderson DG, Horton JD, Koteliansky V, Bumcrot D. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007;449(7163):745–747. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27(6):549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134(4):577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20(5):500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, Reilly CS, Peterson ML, Schultz-Darken N, Brunner KG, Nephew KR, Pambuccian S, Lifson JD, Carlis JV, Haase AT. Glycerol mono-laurate prevents mucosal SIV transmission. Nature. 2009;458(7241):1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath N, Dykxhoorn DM. Advances in synthetic siRNA delivery. Discov Med. 2010;9(48):418–430. [PubMed] [Google Scholar]
- McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372(9653):1894–1905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24(8):1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Mesquita PM, Cheshenko N, Wilson SS, Mhatre M, Guzman E, Fakioglu E, Keller MJ, Herold BC. Disruption of tight junctions by cellulose sulfate facilitates HIV infection: model of microbicide safety. J Infect Dis. 2009;200(4):599–608. doi: 10.1086/600867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moench TR, Mumper RJ, Hoen TE, Sun M, Cone RA. Microbicide excipients can greatly increase susceptibility to genital herpes transmission in the mouse. BMC Infect Dis. 2010;10:331. doi: 10.1186/1471-2334-10-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H, Smith DD, Swiderski P, Rossi JJ, Akkina R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med. 2011;3(66):66ra66. doi: 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8(7):681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- Oliveira S, Van Rooy I, Kranenburg O, Storm G, Schiffelers RM. Fusogenic peptides enhance endosomal escape improving siRNA-induced silencing of oncogenes. Int J Pharm. 2007;331(2):211–214. doi: 10.1016/j.ijpharm.2006.11.050. [DOI] [PubMed] [Google Scholar]
- Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronson RT, Knipe DM, Lieberman J. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439(7072):89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319(5863):627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer D, Zhu P, Carman CV, Lieberman J, Shimaoka M. Selective gene silencing in activated leukocytes by targeting siRNAs to the integrin lymphocyte function-associated antigen-1. Proc Natl Acad Sci U S A. 2007;104(10):4095–4100. doi: 10.1073/pnas.0608491104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher H. Starting to gel. Nature. 2004;430(6996):138–140. doi: 10.1038/430138a. [DOI] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, De Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19(2):89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- Schlievert PM, Strandberg KL, Brosnahan AJ, Peterson ML, Pambuccian SE, Nephew KR, Brunner KG, Schultz-Darken NJ, Haase AT. Glycerol monolaurate does not alter rhesus macaque (Macaca mulatta) vaginal lactobacilli and is safe for chronic use. Antimicrob Agents Chemother. 2008;52(12):4448–4454. doi: 10.1128/AAC.00989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Lee SK, Dykxhoorn DM, Novina C, Zhang D, Crawford K, Cerny J, Sharp PA, Lieberman J, Manjunath N, Shankar P. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J Virol. 2003;77(13):7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Surabhi RM, Gaynor RB. RNA interference directed against viral and cellular targets inhibits human immunodeficiency virus type 1 replication. J Virol. 2002;76(24):12963–12973. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS. [Accessed on Feb. 14, 2011];AIDS epidemic update. 2009 http://www.unaids.org/en/data-analysis/epidemiology/2009aidsepidemicupdate/
- Vaishnaw AK, Gollob J, Gamba-Vitalo C, Hutabarat R, Sah D, Meyers R, De Fougerolles T, Maraganore J. A status report on RNAi therapeutics. Silence. 2010;1(1):14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme L, Ramjee G, Alary M, Vuylsteke B, Chandeying V, Rees H, Sirivongrangson P, Mukenge-Tshibaka L, Ettiegne-Traore V, Uaheowitchai C, Karim SS, Masse B, Perriens J, Laga M. Effectiveness of COL-1492, a nonoxynol-9 vaginal gel, on HIV-1 transmission in female sex workers: a randomised controlled trial. Lancet. 2002;360(9338):971–977. doi: 10.1016/s0140-6736(02)11079-8. [DOI] [PubMed] [Google Scholar]
- Wald A, Link K. Risk of human immunodeficiency virus infection in herpes simplex virus type 2-seropositive persons: a meta-analysis. J Infect Dis. 2002;185(1):45–52. doi: 10.1086/338231. [DOI] [PubMed] [Google Scholar]
- Wilson SS, Cheshenko N, Fakioglu E, Mesquita PM, Keller MJ, Herold BC. Susceptibility to genital herpes as a biomarker predictive of increased HIV risk: expansion of a murine model of microbicide safety. Antivir Ther. 2009;14(8):1113–1124. doi: 10.3851/IMP1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow KA, Cu Y, Booth CJ, Saucier-Sawyer JK, Wood MJ, Saltzman WM. Intravaginal gene silencing using biodegradable polymer nanoparticles densely loaded with small-interfering RNA. Nat Mater. 2009;8(6):526–533. doi: 10.1038/nmat2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Navarro F, Lal A, Basar E, Pandey RK, Manoharan M, Feng Y, Lee SJ, Lieberman J, Palliser D. Durable protection from Herpes Simplex Virus-2 transmission following intravaginal application of siRNAs targeting both a viral and host gene. Cell Host Microbe. 2009;5(1):84–94. doi: 10.1016/j.chom.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan AC, Levy M. Aptamers and aptamer targeted delivery. RNA Biol. 2009;6(3):316–320. doi: 10.4161/rna.6.3.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. RNA interference in human cells is restricted to the cytoplasm. RNA. 2002;8(7):855–860. doi: 10.1017/s1355838202020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cristofaro P, Silbermann R, Pusch O, Boden D, Konkin T, Hovanesian V, Monfils PR, Resnick M, Moss SF, Ramratnam B. Engineering mucosal RNA interference in vivo. Mol Ther. 2006;14(3):336–342. doi: 10.1016/j.ymthe.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16(8):1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rossi JJ. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides. 2010 Dec 23; doi: 10.1089/oli.2010.0264. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res. 2009;37(9):3094–3109. doi: 10.1093/nar/gkp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]


