Abstract
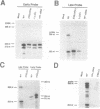
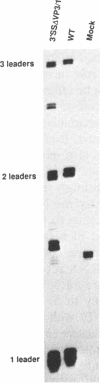
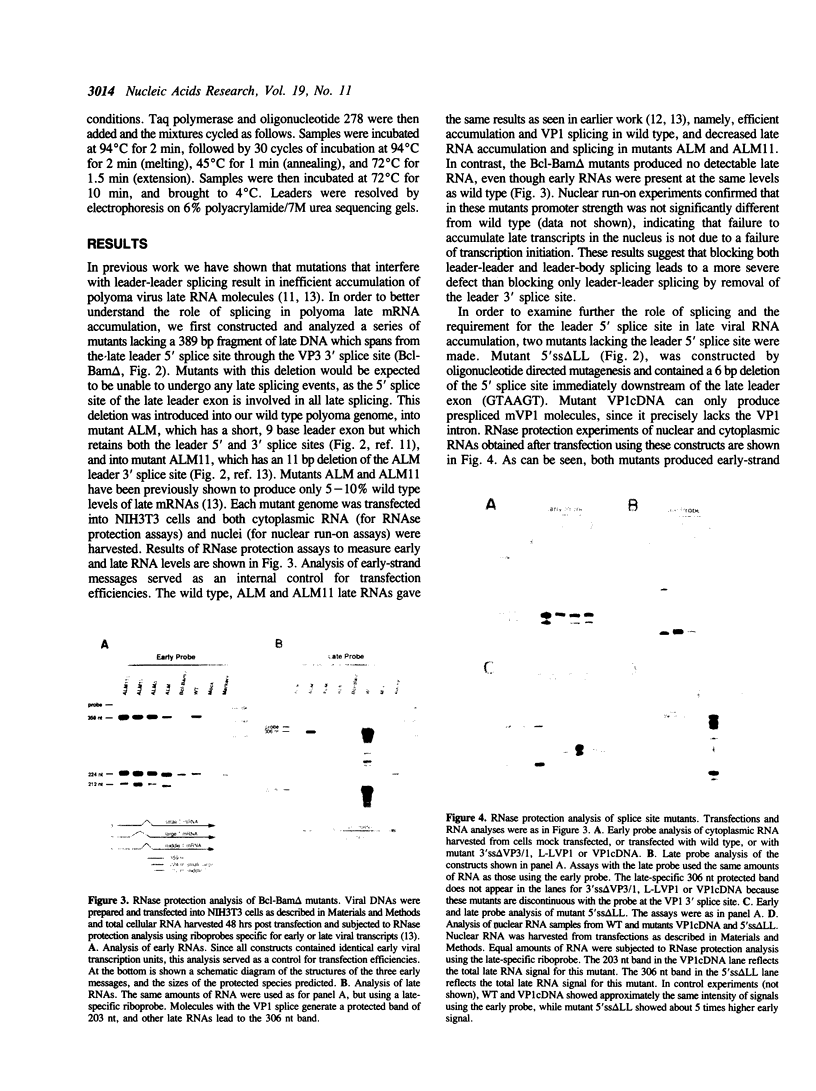
Polyoma virus late nuclear primary transcripts are giant and heterogeneous, containing tandem repeats of the late strand of the circular viral genome. Late pre-mRNA processing involves the splicing of noncoding 'leader' exons to each other (removing genome-length introns), with the joining of the last leader to a coding 'body' exon. We have constructed a number of mutants blocked only in leader-leader splicing, or blocked in both leader-leader and leader-body splicing. We examined the accumulation of both nuclear and cytoplasmic late-strand RNAs in NIH3T3 cells. Consistent with our previous results, mutants lacking the 3' splice site of the late leader (leader-leader splicing blocked) showed a 10-20 fold defect in late RNA accumulation. Mutants which lacked the leader 5' splice site (leader-body splicing blocked) had a more profound defect, exhibiting virtually no late-strand cytoplasmic or nuclear RNA. This result was unexpected as a substantial proportion of wild type late cytoplasmic messages are unspliced. A mutant with no intron, but having functional 3' and 5' splice sites bordering the leader exon, is capable of producing large amounts of unspliced late mRNA. This demonstrates that an excisable intron is not a requirement for late mRNA accumulation. The accumulation of polyoma late mRNAs requires the presence of leader exons bordered by functional 3' and 5' splice sites, whether or not these sites are used during pre-mRNA processing.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acheson N. H. Kinetics and efficiency of polyadenylation of late polyomavirus nuclear RNA: generation of oligomeric polyadenylated RNAs and their processing into mRNA. Mol Cell Biol. 1984 Apr;4(4):722–729. doi: 10.1128/mcb.4.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H. Polyoma virus giant RNAs contain tandem repeats of the nucleotide sequence of the entire viral genome. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4754–4758. doi: 10.1073/pnas.75.10.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H. Transcription during productive infection with polyoma virus and Simian virus 40. Cell. 1976 May;8(1):1–12. doi: 10.1016/0092-8674(76)90179-3. [DOI] [PubMed] [Google Scholar]
- Adami G. R., Carmichael G. G. Polyomavirus late leader region serves an essential spacer function necessary for viability and late gene expression. J Virol. 1986 May;58(2):417–425. doi: 10.1128/jvi.58.2.417-425.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adami G. R., Carmichael G. G. The length but not the sequence of the polyoma virus late leader exon is important for both late RNA splicing and stability. Nucleic Acids Res. 1987 Mar 25;15(6):2593–2610. doi: 10.1093/nar/15.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adami G. R., Marlor C. W., Barrett N. L., Carmichael G. G. Leader-to-leader splicing is required for efficient production and accumulation of polyomavirus late mRNAs. J Virol. 1989 Jan;63(1):85–93. doi: 10.1128/jvi.63.1.85-93.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R. L., Allen J. M., Behringer R. R., Gelinas R. E., Palmiter R. D. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988 Feb;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988 Oct;8(10):4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill K. B., Carmichael G. G. Deletion analysis of the polyomavirus late promoter: evidence for both positive and negative elements in the absence of early proteins. J Virol. 1989 Sep;63(9):3634–3642. doi: 10.1128/jvi.63.9.3634-3642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill K. B., Roome A. J., Carmichael G. G. Replication-dependent transactivation of the polyomavirus late promoter. J Virol. 1990 Mar;64(3):992–1001. doi: 10.1128/jvi.64.3.992-1001.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J., Fromm M., Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987 Dec;1(10):1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987 Aug;7(8):2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund R., Mandel G., Carmichael G. G., Barncastle J. P., Dawe C. J., Benjamin T. L. Polyomavirus tumor induction in mice: influences of viral coding and noncoding sequences on tumor profiles. J Virol. 1987 Jul;61(7):2232–2239. doi: 10.1128/jvi.61.7.2232-2239.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser C. S., Simonsen C. C., Schilling J. W., Schimke R. T. Expression of abbreviated mouse dihydrofolate reductase genes in cultured hamster cells. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6522–6526. doi: 10.1073/pnas.79.21.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Roy P., Barkan A., Mertz J. E., Weissman S. M., Lebowitz P. Unspliced functional late 19S mRNAs containing intervening sequences are produced by a late leader mutant of simian virus 40. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1386–1390. doi: 10.1073/pnas.78.3.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M. K., Kainz M. S., Merrill G. F. Introns are inconsequential to efficient formation of cellular thymidine kinase mRNA in mouse L cells. Mol Cell Biol. 1987 Dec;7(12):4576–4581. doi: 10.1128/mcb.7.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Khoury G. Rescue of a splicing defective mutant by insertion of an heterologous intron. Nature. 1980 Aug 7;286(5773):634–637. doi: 10.1038/286634a0. [DOI] [PubMed] [Google Scholar]
- Gruss P., Lai C. J., Dhar R., Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D. H., Smith K. D., Boyer S. H., Leder P. SV40 recombinants carrying rabbit beta-globin gene coding sequences. Cell. 1979 Jul;17(3):725–735. doi: 10.1016/0092-8674(79)90279-4. [DOI] [PubMed] [Google Scholar]
- Hattori K., Angel P., Le Beau M. M., Karin M. Structure and chromosomal localization of the functional intronless human JUN protooncogene. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9148–9152. doi: 10.1073/pnas.85.23.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde-DeRuyscher R. P., Carmichael G. G. Polyomavirus late pre-mRNA processing: DNA replication-associated changes in leader exon multiplicity suggest a role for leader-to-leader splicing in the early-late switch. J Virol. 1990 Dec;64(12):5823–5832. doi: 10.1128/jvi.64.12.5823-5832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde-DeRuyscher R., Carmichael G. G. Polyomavirus early-late switch is not regulated at the level of transcription initiation and is associated with changes in RNA processing. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8993–8997. doi: 10.1073/pnas.85.23.8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J. Topography of the three late mRNA's of polyoma virus which encode the virion proteins. J Virol. 1980 Feb;33(2):637–651. doi: 10.1128/jvi.33.2.637-651.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedes L. H. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- Legon S., Flavell A. J., Cowie A., Kamen R. Amplification in the leader sequence of late polyoma virus mRNAs. Cell. 1979 Feb;16(2):373–388. doi: 10.1016/0092-8674(79)90013-8. [DOI] [PubMed] [Google Scholar]
- May P., Kress M., Lange M., May E. New genetic information expressed in SV40-transformed cells: characterization of the 55K proteins and evidence for unusual SV40 mRNAs. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):189–200. doi: 10.1101/sqb.1980.044.01.022. [DOI] [PubMed] [Google Scholar]
- Nagata S., Mantei N., Weissmann C. The structure of one of the eight or more distinct chromosomal genes for human interferon-alpha. Nature. 1980 Oct 2;287(5781):401–408. doi: 10.1038/287401a0. [DOI] [PubMed] [Google Scholar]
- Neuberger M. S., Williams G. T. The intron requirement for immunoglobulin gene expression is dependent upon the promoter. Nucleic Acids Res. 1988 Jul 25;16(14B):6713–6724. doi: 10.1093/nar/16.14.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu W. S., Mertz J. E. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J Virol. 1989 Oct;63(10):4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Treisman R. Characterisation of polyoma late mRNA leader sequences by molecular cloning and DNA sequence analysis. Nucleic Acids Res. 1980 Nov 11;8(21):4867–4888. doi: 10.1093/nar/8.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Kamen R. Structure of polyoma virus late nuclear RNA. J Mol Biol. 1981 May 25;148(3):273–301. doi: 10.1016/0022-2836(81)90539-8. [DOI] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- Tseng R. W., Acheson N. H. Use of a novel S1 nuclease RNA-mapping technique to measure efficiency of transcription termination on polyomavirus DNA. Mol Cell Biol. 1986 May;6(5):1624–1632. doi: 10.1128/mcb.6.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]