Abstract
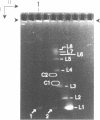
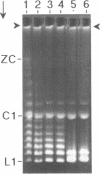
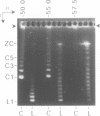
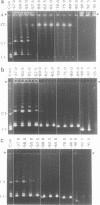
The 0.048502 megabase (Mb), primarily double-stranded DNA of bacteriophage lambda has single-stranded, complementary termini (cohesive ends) that undergo either spontaneous intramolecular joining to form open circular DNA or spontaneous intermolecular joining to form linear, end-to-end oligomeric DNAs (concatemers); concatemers also cyclize. In the present study, the effects of polyethylene glycol (PEG) on the cyclization and concatemerization of lambda DNA are determined at temperatures that, in the absence of PEG, favor dissociation of cohesive ends. Circular and linear lambda DNA, monomeric and concatemeric, are observed by use of pulsed field agarose gel (PFG) electrophoresis. During preparation of lambda DNA for these studies, hydrodynamic shear-induced, partial dissociation of joined cohesive ends is fortuitously observed. Although joined lambda cohesive ends progressively dissociate as their temperature is raised in the buffer used here (0.1 M NaCl, 0.01 M sodium phosphate, pH 7.4, 0.001 M EDTA), when PEG is added to this buffer, raising the temperature sometimes promotes joining of cohesive ends. Conditions for promotion of primarily either cyclization or concatemerization are described. Open circular DNAs as long as a 7-mer are produced and resolved. The concentration of PEG required to promote joining of cohesive ends decreases as the molecular weight of the PEG increases. The rate of cyclization is brought, the first time, to values that are high enough to be comparable to the rate observed in vivo. For double-stranded DNA bacteriophages that have a linear replicative form of DNA (bacteriophage T7, for example), a suppression, sometimes observed here, of cyclization mimics a suppression of cyclization previously observed in vivo. The PEG, temperature effects on DNA joining are explained by both the excluded volume of PEG random coils and an increase in this excluded volume that occurs when temperature increases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam R. E., Zimm B. H. Shear degradation of DNA. Nucleic Acids Res. 1977;4(5):1513–1537. doi: 10.1093/nar/4.5.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode V. C., Kaiser A. D. Changes in the structure and activity of lambda DNA in a superinfected immune bacterium. J Mol Biol. 1965 Dec;14(2):399–417. doi: 10.1016/s0022-2836(65)80190-5. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Smith C. L., Mathew M. K. Pulsed-field gel electrophoresis of very large DNA molecules. Annu Rev Biophys Biophys Chem. 1988;17:287–304. doi: 10.1146/annurev.bb.17.060188.001443. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Williams K. R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- Fuke M., Thomas C. A., Jr Isolation of open-circular DNA molecules by retention in agar gels. J Mol Biol. 1970 Sep 14;52(2):395–397. doi: 10.1016/0022-2836(70)90040-9. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Nakazawa M., Ishizaki Y., Hiraoka N., Obayashi A. Regulation of inter- and intramolecular ligation with T4 DNA ligase in the presence of polyethylene glycol. Nucleic Acids Res. 1986 Oct 10;14(19):7617–7631. doi: 10.1093/nar/14.19.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Nakazawa M., Ishizaki Y., Obayashi A. Influence of monovalent cations on the activity of T4 DNA ligase in the presence of polyethylene glycol. Nucleic Acids Res. 1985 May 10;13(9):3261–3271. doi: 10.1093/nar/13.9.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis T. C., Ring D. M., Daube S. S., von Hippel P. H. "Macromolecular crowding": thermodynamic consequences for protein-protein interactions within the T4 DNA replication complex. J Biol Chem. 1990 Sep 5;265(25):15160–15167. [PubMed] [Google Scholar]
- Laurent T. C., Preston B. N., Carlsson B. Conformational transitions of polynucleotides in polymer media. Eur J Biochem. 1974 Apr 1;43(2):231–235. doi: 10.1111/j.1432-1033.1974.tb03404.x. [DOI] [PubMed] [Google Scholar]
- Lerman L. S. A transition to a compact form of DNA in polymer solutions. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1886–1890. doi: 10.1073/pnas.68.8.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie D., Serwer P. A hybrid mode of rotating gel electrophoresis for separating linear and circular duplex DNA. Appl Theor Electrophor. 1989;1(3):169–173. [PubMed] [Google Scholar]
- Louie D., Serwer P. Blunt-ended ligation can be used to produce DNA ladders with rung spacing as large as 0.17 Mb. Nucleic Acids Res. 1990 May 25;18(10):3090–3090. doi: 10.1093/nar/18.10.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton A. P. The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol Cell Biochem. 1983;55(2):119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- Minton A. P., Wilf J. Effect of macromolecular crowding upon the structure and function of an enzyme: glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1981 Aug 18;20(17):4821–4826. doi: 10.1021/bi00520a003. [DOI] [PubMed] [Google Scholar]
- O'Neill R. R., Mitchell L. G., Merril C. R., Rasband W. S. Use of image analysis to quantitate changes in form of mitochondrial DNA after x-irradiation. Appl Theor Electrophor. 1989;1(3):163–167. [PubMed] [Google Scholar]
- Parsegian V. A., Rand R. P., Fuller N. L., Rau D. C. Osmotic stress for the direct measurement of intermolecular forces. Methods Enzymol. 1986;127:400–416. doi: 10.1016/0076-6879(86)27032-9. [DOI] [PubMed] [Google Scholar]
- Pheiffer B. H., Zimmerman S. B. Polymer-stimulated ligation: enhanced blunt- or cohesive-end ligation of DNA or deoxyribooligonucleotides by T4 DNA ligase in polymer solutions. Nucleic Acids Res. 1983 Nov 25;11(22):7853–7871. doi: 10.1093/nar/11.22.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson A. A theory for the displacement of proteins and viruses with polyethylene glycol. Prep Biochem. 1977;7(2):129–154. doi: 10.1080/00327487708061631. [DOI] [PubMed] [Google Scholar]
- Prouty M. S., Schechter A. N., Parsegian V. A. Chemical potential measurements of deoxyhemoglobin S polymerization. Determination of the phase diagram of an assembling protein. J Mol Biol. 1985 Aug 5;184(3):517–528. doi: 10.1016/0022-2836(85)90298-0. [DOI] [PubMed] [Google Scholar]
- Serwer P., Masker W. E., Allen J. L. Stability and in vitro DNA packaging of bacteriophages: effects of dextrans, sugars, and polyols. J Virol. 1983 Feb;45(2):665–671. doi: 10.1128/jvi.45.2.665-671.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P., Watson R. H., Hayes S. J. Multidimensional analysis of intracellular bacteriophage T7 DNA: effects of amber mutations in genes 3 and 19. J Virol. 1987 Nov;61(11):3499–3509. doi: 10.1128/jvi.61.11.3499-3509.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son M., Hayes S. J., Serwer P. Concatemerization and packaging of bacteriophage T7 DNA in vitro: determination of the concatemers' length and appearance kinetics by use of rotating gel electrophoresis. Virology. 1988 Jan;162(1):38–46. doi: 10.1016/0042-6822(88)90392-3. [DOI] [PubMed] [Google Scholar]
- Son M., Hayes S. J., Serwer P. Optimization of the in vitro packaging efficiency of bacteriophage T7 DNA: effects of neutral polymers. Gene. 1989 Oct 30;82(2):321–325. doi: 10.1016/0378-1119(89)90058-9. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. Cyclization of phage DNAs. Cold Spring Harb Symp Quant Biol. 1968;33:409–415. doi: 10.1101/sqb.1968.033.01.047. [DOI] [PubMed] [Google Scholar]
- Waterbury P. G., Lane M. J. Generation of lambda phage concatemers for use as pulsed field electrophoresis size markers. Nucleic Acids Res. 1987 May 11;15(9):3930–3930. doi: 10.1093/nar/15.9.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilf J., Minton A. P. Evidence for protein self-association induced by excluded volume. Myoglobin in the presence of globular proteins. Biochim Biophys Acta. 1981 Oct 28;670(3):316–322. doi: 10.1016/0005-2795(81)90103-3. [DOI] [PubMed] [Google Scholar]
- Zimm B. H., Reese H. R. The degradation of T7 DNA in converging flow. Nucleic Acids Res. 1990 Aug 11;18(15):4469–4470. doi: 10.1093/nar/18.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B., Harrison B. Macromolecular crowding accelerates the cohesion of DNA fragments with complementary termini. Nucleic Acids Res. 1985 Apr 11;13(7):2241–2249. doi: 10.1093/nar/13.7.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B., Harrison B. Macromolecular crowding increases binding of DNA polymerase to DNA: an adaptive effect. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1871–1875. doi: 10.1073/pnas.84.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. Macromolecular crowding allows blunt-end ligation by DNA ligases from rat liver or Escherichia coli. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5852–5856. doi: 10.1073/pnas.80.19.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]