Abstract
Background
Single sample predictors (SSPs) and Subtype classification models (SCMs) are gene expression–based classifiers used to identify the four primary molecular subtypes of breast cancer (basal-like, HER2-enriched, luminal A, and luminal B). SSPs use hierarchical clustering, followed by nearest centroid classification, based on large sets of tumor-intrinsic genes. SCMs use a mixture of Gaussian distributions based on sets of genes with expression specifically correlated with three key breast cancer genes (estrogen receptor [ER], HER2, and aurora kinase A [AURKA]). The aim of this study was to compare the robustness, classification concordance, and prognostic value of these classifiers with those of a simplified three-gene SCM in a large compendium of microarray datasets.
Methods
Thirty-six publicly available breast cancer datasets (n = 5715) were subjected to molecular subtyping using five published classifiers (three SSPs and two SCMs) and SCMGENE, the new three-gene (ER, HER2, and AURKA) SCM. We used the prediction strength statistic to estimate robustness of the classification models, defined as the capacity of a classifier to assign the same tumors to the same subtypes independently of the dataset used to fit it. We used Cohen κ and Cramer V coefficients to assess concordance between the subtype classifiers and association with clinical variables, respectively. We used Kaplan–Meier survival curves and cross-validated partial likelihood to compare prognostic value of the resulting classifications. All statistical tests were two-sided.
Results
SCMs were statistically significantly more robust than SSPs, with SCMGENE being the most robust because of its simplicity. SCMGENE was statistically significantly concordant with published SCMs (κ = 0.65–0.70) and SSPs (κ = 0.34–0.59), statistically significantly associated with ER (V = 0.64), HER2 (V = 0.52) status, and histological grade (V = 0.55), and yielded similar strong prognostic value.
Conclusion
Our results suggest that adequate classification of the major and clinically relevant molecular subtypes of breast cancer can be robustly achieved with quantitative measurements of three key genes.
CONTEXTS AND CAVEATS
Prior knowledge
Single sample predictors (SSPs) are molecular classification models that use large sets of genes expressed in different tumors to classify different subtypes of breast cancer. Subtype classification models (SCMs) are based on groups of genes specifically correlated with three key breast cancer genes, estrogen receptor (ER), HER2, and aurora kinase A (AURKA). Both types of models use large numbers of genes. However, the robustness and prognostic value of these classifiers have not been compared with simplified models containing fewer genes.
Study design
A simplified SCM (SCMGENE) containing only ER, HER2, and AURKA was compared with three SSPs and two SCMs using data from 36 gene expression datasets in public databases. The models were compared with respect to concordance among themselves as well as association with clinical variables and disease-free survival.
Contribution
Among the SCMs, SCMGENE with only three genes was statistically more robust than SSPs and as robust and yielded similar prognostic value compared with the published SCMs that use large numbers of genes.
Implications
Adding more genes to a classification model may not improve the ability to discriminate among breast cancer subtypes. In addition, the complexity of multiple-gene classification models may limit their usefulness and translation into clinic.
Limitations
The datasets used were retrospectively accrued; therefore, the selection of patients may have resulted in unbalanced distribution of the different molecular subtypes. The gene expression datasets taken from public databases and websites were not renormalized. Software limitations precluded checking or correction for departure from proportional hazards assumptions.
From the Editors
Microarray-based expression studies have demonstrated that breast cancer is both a clinically diverse and molecularly heterogeneous disease comprising subtypes with distinct gene expression patterns that are associated with outcome (1–8). The relevance of these subtypes for basic and translational research has led to their use in prognostic assessments (3,9), prediction of therapeutic efficacy (10), and retrospective analysis of clinical trials (11). Independent of subtype analysis, several research groups have developed prognostic gene signatures by analyzing gene expression together with clinical outcome data [see (12) for a review]; Mammaprint (13,14), Oncotype Dx (15), and the Gene expression Grade Index (GGI) (16) are currently commercially available. Although the data and statistical methods used to develop these prognostic gene signatures differ from those used for breast cancer molecular subtyping, we and others reported a high rate of concordance between the predicted risk classifications and subtypes (1,8,17), shedding new light on the common biological processes relevant for predicting outcome in breast cancer.
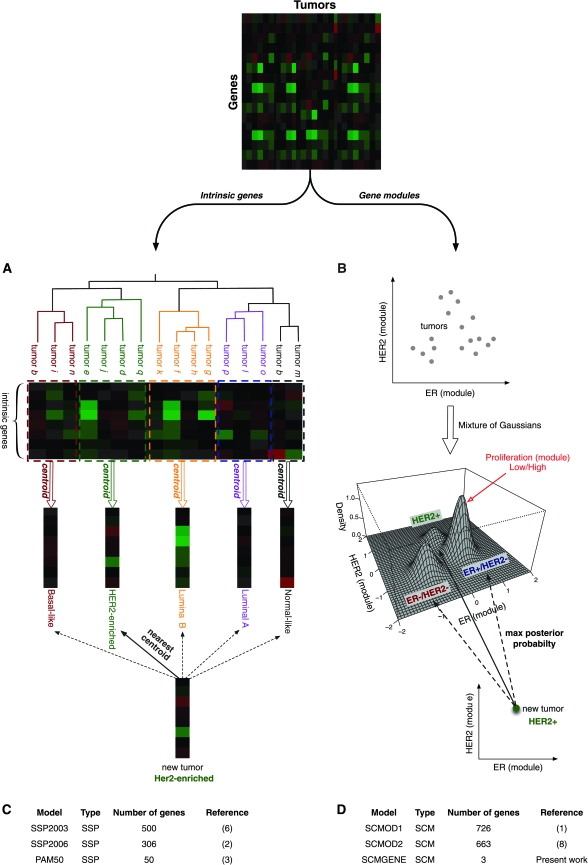
In their seminal work, Perou et al. (4) identified four breast tumor subtypes: the basal-like, HER2-enriched, the luminal (often differentiated into two or three subgroups), and the normal-like tumors. These molecular subtypes were identified by selecting a large set of “intrinsic” genes (those showing little variance within repeated samplings of the same tumor but with high variance across tumors) and then using hierarchical clustering to separate patients into transcriptionally distinct groups (4). However, because only samples in large retrospective studies could be classified by this method, the authors developed the Single Sample Predictor (SSP; Figure 1, A), which identifies the subtype of a single tumor using a nearest centroid classifier (2,3,6). This initial SSP [SSP2003; Figure 1, C; (6)] was further refined through iterations of the intrinsic gene list and the resulting two SSPs [SSP2006 and PAM50; Figure 1, C; (2,3)]. These SSPs have been applied to gene expression data generated from different cohorts of breast cancer patients and microarray technologies (2,17).
Figure 1.
Published classifiers for breast cancer molecular subtyping. Conceptual design of the two breast cancer molecular subtyping methods: A) the Single Sample Predictor (SSP) and B) the Subtype Classification Model (SCM). For SSP, the dimensionality of the data is first reduced by selecting intrinsic genes defined as those showing little variance in expression within repeated samplings of the same tumor but high variance across tumors. A hierarchical clustering of the tumors is performed to identify the main molecular subtypes and then a nearest centroid classifier is built by computing the average gene expression profiles for each subtype. A new tumor sample can be classified into one subtype based on its expression profile of intrinsic genes by computing the correlation with each of the centroids. For SCM, genes whose expression is specifically correlated with the estrogen receptor (ER), HER2, and aurora kinase A (AURKA) are first selected and summarized to quantify the activity of ER, HER2, and proliferation phenotypes, respectively. A mixture of three Gaussian distributions is then fitted on the data to represent the three main molecular subtypes of breast tumors (ER−/HER2−, HER2+, and ER+/HER2−), the proliferation module being used to discriminate between low and high proliferative ER+/HER2− tumors. A new tumor sample can therefore be classified into one subtype with respect to its maximum posterior probability to belong to each subtype. Panels (C) and (D) provide information about the published SSPs (SSP203, SSP2006, and PAM50 composed of 500, 306, and 50 genes, respectively) and SCMs (SCMOD1, SCMOD2, and SCMGENE, composed of 726, 663, and 3 genes, respectively).
However, all SSPs have severe limitations. Pusztai et al. (18) showed that small changes in the initial set of breast tumors can have a dramatic impact on the hierarchical clustering used in defining the initial subgroups for the SSPs, raising questions about the stability of the method (18,19). Kapp et al. (20) challenged the use of hundreds of intrinsic genes and showed that only genes related to estrogen receptor (ER) and HER2 phenotypes lead to a stable identification of three main subtypes: ER−/HER2− (basal-like tumors), HER2+ (HER2-enriched), and ER+/HER2− (combined luminal A and B tumors) (20). Weigelt et al. (21) reported that subtype classifications depend on the list of intrinsic genes because SSPs were only moderately concordant. Recently, Mackay et al. (22) highlighted the lack of interobserver agreement for manually identifying subtypes from dendrograms estimated by hierarchical clustering.
To address these issues, we developed an alternative classification approach, the Subtype Classification Model (SCM; Figure 1, B). In contrast to SSPs, SCMs are based on a mixture of three Gaussian distributions in a two-dimensional space defined by the ER and HER2 gene modules, with a proliferation (aurora kinase A [AURKA]) module providing discrimination between low and high proliferative tumors (1,8). These modules are composed of genes whose expression is specifically correlated with their prototype gene—ER, HER2, or AURKA (1,8). Two versions of gene lists representing these modules have been published [Figure 1, D; (1,8)]. SCMs have been applied in datasets using different microarray platforms and normalization methods (1,8,9).
Although establishing breast cancer molecular subtypes has had a substantial impact on the way clinicians perceive the disease, we know surprisingly little about the reproducibility of the various classification algorithms (19) because of the intrinsic nature of subtype identification where the true classification remains unknown, rendering the validation of the corresponding classifiers difficult. Weigelt et al. (21) recently estimated the agreement of the three published SSPs in four public datasets and showed that these classifiers are only moderately concordant.
Here, we address the issue of reproducibility, comparing SSPs and SCMs to assess their robustness (defined as the capacity of a classifier to assign the same tumors to the same subtypes independently of the dataset used to fit it), their classification concordance, and their prognostic value. To do this, we first developed SCMGENE, a simplified version of the SCM using only the three key genes (ER, HER2, and AURKA) known to be the main discriminators of clinical and molecular breast cancer subtypes (1,8,20). We then used a large compendium of gene expression and clinical data from 5715 breast cancer patients to assess the relative performance of six subtype classifiers, the three published SSPs, two published SCMs, and the simplified version of SCM, namely SCMGENE.
Methods and Statistical Analysis
All analyses have been performed using R version 2.13.1 (http://www.r-project.org/). To ensure full reproducibility of our results, software and data are available at http://compbio.dfci.harvard.edu/pubs/sbtpaper/. Differences with P values less than .05 were considered statistically significant. All statistical tests were two-sided.
Gene Expression Data
Thirty-six gene expression datasets of expression profiles from 5715 tumors were retrieved from public databases or authors’ websites (Table 1); this includes 676 ER-positive (ER+ as defined by immunohistochemistry [IHC]) breast tumors from tamoxifen-treated patients (Table 1). We used normalized log2(intensity) for single-channel platforms and log2(ratio) in dual-channel platforms. Hybridization probes were mapped to Entrez GeneID as described in Shi et al. (58) using RefSeq and Entrez whenever possible; otherwise mapping was performed using IDconverter [http://idconverter.bioinfo.cnio.es/; (59)]. When multiple probes mapped to the same GeneID, we used the one with the highest variance in the dataset under study. To facilitate comparison between datasets, we applied a robust linear scaling to each gene or module score where expression quantiles 2.5% and 97.5% were set to −1 and +1, respectively. This procedure was particularly efficient in datasets with skewed populations of patients (such as those with different proportions of ER−/+ or HER2−/+ tumors), because only a few extreme cases (5%) were needed to perform the robust scaling, without relying on outliers. This scaling improved the consistency between classifiers in datasets using different microarray technologies and normalization procedures (60,61).
Table 1.
Compendium of microarray datasets of unique breast cancer patients*
| Dataset | Microarray technology | Survival data | Treatment | No. of patients | Number of probes | Source | Reference |
| EXPO | Affymetrix HGU | NA | NA | 353 | 54 675 | GEO: GSE2109 | (23) |
| VDX† | Affymetrix HGU | RFS, DMFS | Untreated | 344 | 22 283 | GEO: GSE2034/GSE5327 | (24,25) |
| NKI† | Agilent | RFS, DMFS,OS | Untreated, chemo | 337 | 24 481 | Rosetta Inpharmatics | (13,14) |
| UCSF† | In-house cDNA | DNFS, RFS, OS | Untreated, chemo, hormonal | 162 | 10 368 | Authors’ website | (26,27) |
| STNO2† | In-house cDNA | RFS, OS | Untreated, chemo, hormonal | 122 | 7787 | SMD | (6) |
| NCI† | In-house cDNA | RFS | Untreated, chemo, hormonal | 99 | 6878 | Authors’ website | (7) |
| MSK | Affymetrix HGU | DMFS | Heterogeneous | 99 | 22 283 | GEO: GSE2603 | (28) |
| UPP† | Affymetrix HGU | RFS | untreated, hormonal | 251 (190)‡ | 44 928 | GEO: GSE3494 | (29) |
| STK | Affymetrix HGU | RFS | untreated, chemo, hormonal | 159 | 44 928 | GEO: GSE1456 | (30) |
| UNT† | Affymetrix HGU | RFS, DMFS | untreated | 137 (92)‡ | 44 928 | GEO: GSE2990 | (16,31) |
| UNC4† | Agilent | RFS, OS | Heterogeneous | 337 | 17 779 | UNC DB | (32) |
| DUKE | Affymetrix HGU95 | OS | Heterogeneous | 171 | 12 625 | GEO: GSE3143 | (33) |
| CAL† | Affymetrix HGU | RFS, DMFS, OS | Chemo, hormonal | 118 | 22 283 | AE: E-TABM-158 | (34) |
| TRANSBIG† | Affymetrix HGU | RFS, DMFS, OS | Untreated | 198 | 22 283 | GEO: GSE7390 | (35) |
| DUKE2 | Affymetrix X3P | NA | Chemo | 160 | 61 359 | GEO: GSE6961 | (36) |
| MAINZ† | Affymetrix HGU | DMFS | Untreated | 200 | 22 283 | GEO: GSE11121 | (37) |
| LUND2 | Swegene | NA | Hormonal | 105 | 27 648 | GEO: GSE5325 | (38) |
| LUND | Swegene | NA | Heterogeneous | 143 | 26 824 | GEO: GSE5325 | (39) |
| FNCLCC | In-house cDNA | NA | Chemo | 150 | 9216 | GEO: GSE7017 | (40) |
| MDA4 | Affymetrix HGU | NA | Chemo | 129 (65)‡ | 22 283 | MDACC DB | (10,42) |
| EMC2† | Affymetrix HGU | DMFS | Chemo | 204 | 54 675 | GEO: GSE12276 | (43) |
| MUG | Operon | NA | Chemo | 152 | 35 788 | GEO: GSE10510 | (44) |
| NCCS | Affymetrix HGU | NA | NA | 183 | 22 283 | GEO: GSE5364 | (45) |
| MCCC | Illumina | NA | NA | 75 | 48 701 | GEO: GSE19177 | (46) |
| KOO† | Affymetrix HGU95 | NA | NA | 88 | 48 701 | Authors’ website | (47) |
| EORTC10994 | Affymetrix HGU | NA | Chemo | 49 | 22 283 | GEO: GSE1561 | (41) |
| HLP | Illumina | NA | Chemo | 53 | 48 701 | AE: E-TABM-543 | (48) |
| DFHCC† | Affymetrix HGU | DMFS | Heterogeneous | 115 | 54 675 | GEO: GSE19615 | (49) |
| DFHCC2 | Affymetrix HGU | NA | Chemo | 84 (75)‡ | 54 675 | GEO: GSE18864 | (51) |
| DFHCC3 | Affymetrix HGU | NA | Chemo | 40 (26)‡ | 54 675 | GEO: GSE3744 | (52) |
| DFHCC4† | Affymetrix HGU | NA | Untreated | 129 | 54 675 | GEO: GSE5460 | (53) |
| MAQC2 | Affymetrix HGU | NA | Chemo | 230 | 22 283 | GEO: GSE20194 | (54) |
| JBI | Affymetrix HGU | NA | NA | 92 | 54 675 | GEO: GSE20711 | (55) |
| Datasets of tamoxifen-treated patients only | |||||||
| TAM | Affymetrix HGU | DMFS, RFS | Hormonal | 345 (242)ठ| 44 928 | GEO: GSE6532/GSE9195 | (56) |
| MDA5 | Affymetrix HGU | DMFS | Hormonal | 298 | 22 283 | GEO: GSE17705 | (57) |
| VDX3 | Affymetrix HGU | DMFS | Hormonal | 136 | 22 283 | GEO: GSE12093 | (50) |
Microarray datasets of unique breast cancer patients (5715) used in this study were retrieved from authors’ websites, Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/), ArrayExpress (AE; http://www.ebi.ac.uk/arrayexpress/), Stanford Microarray Database (SMD; http://smd.stanford.edu/), MD Anderson Cancer Center Microarray database (MDACC DB; http://bioinformatics.mdanderson.org/pubdata.html), University of North Carolina database (UNC DB; https://genome.unc.edu/), and Rosetta Inpharmatics (http://www.rosettabio.com/). Each dataset was assigned a short acronym and an instance number if several datasets were published by the same institution or consortium. CAL = dataset of breast cancer patients from the University of California, San Francisco, and the California Pacific Medical Center (United States); DFHCC = Dana-Farber Harvard Cancer Center (United States); DUKE = Duke University Hospital (United States); EMC = Erasmus Medical Center (the Netherlands); EORTC10994 = Trial number 10994 from the European Organization for Research and Treatment of Cancer Breast Cancer (Europe); EXPO = expression project for oncology, large dataset of microarray data published by the International Genomics Consortium (United States); FNCLCC = Fédération Nationale des Centres de Lutte contre le Cancer (France); HLP = University Hospital La Paz (Spain); JBI = Jules Bordet Institute (Belgium); KOO = Koo Foundation Sun Yat-Sen Cancer Centre (Taiwan); LUND = Lund University Hospital (Sweden); MAINZ = Mainz hospital (Germany); MAQC = Microarray quality control consortium (United States); MCCC = Peter MacCallum Cancer Centre (Australia); MDA = MD Anderson Cancer Center (United States); MSK = Memorial Sloan-Kettering (United States); MUG = Medical University of Graz (Austria); NCCS = National Cancer Centre of Singapore (Singapore); NCI = National Cancer Institute (United States); NKI = National Kanker Instituut (the Netherlands); STK = Stockholm, Karolinska University Hospital (Sweden); STNO = Stanford/Norway (United States and Norway); TRANSBIG = dataset collected by the TransBIG consortium (Europe); UCSF = University of California, San Francisco (United States); UNC = University of North Carolina (United States); UNT = cohort of untreated breast cancer patients from the Oxford Radcliffe (United Kingdom) and Karolinska (Sweden) hospitals; UPP = Uppsala Hospital (Sweden); VDX = Veridex (the Netherlands). These datasets were generated with diverse microarray technologies developed either by Agilent (http://www.genomics.agilent.com), Affymetrix (HGU GeneChips, which include chips HG-U133A, HG-U133B and HG-U133PLUS2, and X3P GeneChip; http://www.affymetrix.com), Swegene (http://www.genomics.agilent.com), Operon (http://www.operon.com) or developed in-house (cDNA platforms). For most datasets, survival data (distant metastasis–free survival [DMFS], relapse-free survival [RFS], and overall survival [OS]) and information regarding the adjuvant treatment (untreated, chemo, hormonal, and heterogeneous standing for no treatment, chemotherapy, hormonal therapy, and heterogeneous combination of therapies, respectively) were available; otherwise missing information is referred to as not available (NA). Additional clinical characteristics are provided in Table 2. All untreated patients had surgery, and most of them had radiation therapy, although information is not available for all datasets.
Dataset containing untreated patients with node-negative breast tumor, as used in our survival analysis.
Duplicated patients were removed from the UNT, UPP, MDA4, DFHCC2, DFHCC3, and TAM datasets for the estimation of concordance and prognostic value.
Five tumors were removed because of negative or missing estrogen receptor status.
We also collected the publicly available clinical and demographic information for our compendium of datasets (Table 2).
Table 2.
Demographic and clinical characteristics of breast cancer patients in compendium of microarray datasets*
| Characteristics | All patients, %† | Untreated node-negative patients, %‡ |
| ER status | ||
| Negative | 25 | 30 |
| Positive | 58 | 65 |
| Missing | 17 | 5 |
| PGR status | ||
| Negative | 19 | 3 |
| Positive | 24 | 10 |
| Missing | 57 | 88 |
| HER2 status | ||
| Negative | 17 | 5 |
| Positive | 7 | 1 |
| Missing | 76 | 94 |
| Histological grade | ||
| Low | 10 | 14 |
| Intermediate | 25 | 32 |
| High | 30 | 36 |
| Missing | 36 | 18 |
| Tumor size, cm | ||
| ≤2 | 30 | 63 |
| >2 | 30 | 29 |
| Missing | 40 | 8 |
| Nodal status | ||
| Negative | 44 | 100 |
| Positive | 30 | 0 |
| Missing | 26 | 0 |
| Age at diagnosis, y | ||
| ≤50 | 28 | 42 |
| >50 | 43 | 44 |
| Missing | 29 | 14 |
Patients described in Table 1; ER = estrogen receptor; PGR = progesterone receptor.
Refers to the whole set of microarray data (n = 5715 patients).
Refers to the subset of patients having a node-negative tumor and who were not treated by systemic adjuvant therapy (n = 1260).
Subtype Classifiers
Classification gene lists were manually transcribed from original publications (Figure 1, C and D) and submitted to GeneSigDB (62). The SSP models were SSP2003 with 500 genes (6), SSP2006 with 306 genes (2), and PAM50 with 50 genes (3). The SCM models were SCMOD1 with 726 genes (1), SCMOD2 with 663 genes (8), and SCMGENE from this study. The SSP and SCM algorithms were implemented as described in the original publications and adapted for scaled data; the source code and documentation are available in the genefu R/Bioconductor package version 1.3.4 [http://www.bioconductor.org/packages/release/bioc/html/genefu.html; (63)]; the methodology underlying the SCMs and the corresponding R code are further detailed in Supplementary Methods, Parts 1 and 2, respectively (available online). Percent of genes used in the classifiers that are actually mapped in each dataset is reported in Supplementary Table 1 (available online). Throughout, we use the Perou et al. (4–6) subtype nomenclature (Figure 1, A): basal-like, HER2-enriched, and luminal A/B, which correspond, respectively, to the ER−/HER2−, HER2+, and ER+/HER2− low/high proliferation tumors of Sotiriou et al. (1,8) (Figure 1, B).
In this study, we developed SCMGENE by selecting the genes ER, HER2, and AURKA, which have been used as prototypes in the ER, HER2 signaling, and proliferation gene modules published in Desmedt et al. (1) and Wirapati et al. (8). We used the Affymetrix probesets published in Desmedt et al. (1), that is, 205225_at, 216836_s_at, and 208079_s_at, representing ER, HER2, and AURKA, respectively (see “scmgene” object in the genefu package).
Robustness of Subtype Classifiers.
To assess robustness, we used the “prediction strength” statistic (64) (Supplementary Methods Part 1, available online), as implemented in the genefu package. First, using each classifier, all samples were assigned “true” subtype labels for that classifier in each dataset separately. The data were then split into training and test sets; the classification model fitted on the training set was applied to the test sets (“predicted” labels) and compared with the true classifications. The prediction strength quantifies the similarity between the true and predicted classifications in each dataset. Values range from 0 (low similarity) to 1 (high similarity), and a prediction strength of at least 0.8 indicates a robust classification (64). Statistical comparison of classifier robustness was performed using the two-sided Wilcoxon signed rank test (65); P values were two-sided and Holm corrected for multiple testing (66).
Because there is no clear consensus about the number of breast cancer molecular subtypes, we analyzed robustness of classifiers for assignment to either three or four subtype groups (20,21). Robustness of SSPs was computed for three to five subtypes by selecting the main clusters, which contain at least five tumors, as defined by the dendrogram built using hierarchical clustering (correlation distance and average linkage). Robustness of SCMs was computed for three subtypes by fitting a mixture of three Gaussian distributions (equal variance and shape, see Supplementary Methods Part 1, available online) and for four subtypes by further estimating a cutoff for proliferation to discriminate between low and high proliferative tumors. Note that, in contrast to SSPs, SCMs are limited to the identification of three and four subtypes by construction [(1,8); see Supplementary Methods Part 1, available online].
Prognostic Gene Signatures
To assess the concordance of subtype classifications with published prognostic gene signatures, we implemented the original algorithms of the MammaPrint (MAMMAPRINT) (14), the Oncotype DX (ONCOTYPE) (15) gene signatures, and the Gene expression Grade Index (GGI) (16), in our compendium of microarray datasets, similarly to Fan et al. (17). The corresponding source code and documentation are available in the genefu package. The resulting risk predictions were labeled low, intermediate, and high risk to reflect the prognosis of the patients. Percent of genes in the signatures that are mapped in each dataset are reported in Supplementary Table 1 (available online).
Concordance
We used color bars to represent the concordance of subtype classifications and prognostic gene signatures; in this representation, subtypes and risk groups are represented by unique colors. Tumors were ordered according to the probabilities estimated by a subtype classifier, such as SCMGENE or PAM50. To quantitatively assess concordance of subtype classifications and prognostic gene signatures, we used Cohen Kappa coefficient (κ) (67), as implemented in the R package epibasix version 1.1 (http://cran.r-project.org/web/packages/epibasix/); κ ranges from 0 to 1, with 0 indicating no relation and 1 indicating a perfect concordance. Typically qualitative descriptions are associated with intervals [κ ≤ 0.20, slight agreement; 0.20 < κ ≤ 0.40, fair agreement; 0.40 < κ ≤ 0.60, moderate agreement, 0.60 < κ ≤ 0.80, substantial agreement; and 0.80 < κ ≤ 0.99, almost perfect agreement, as described in Weigelt et al. (22)]. To assess the association between subtype classifications and clinical parameters, we used Cramer V statistic (68) as implemented in the R package vcd version 1.2-11 (http://cran.r-project.org/web/packages/vcd/). For comparison, we used the same intervals and descriptions used with κ. Statistical significance of the concordance and association was calculated using the χ2 test.
Survival Analysis
Subtypes were considered a categorical variable, with no assumption made on order across subtypes. Risk predictions as computed by the prognostic gene signatures were considered ordered categorical variables [low, intermediate, and high-risk groups as defined in the original publications, which are (14) for MAMMAPRINT, (15) for ONCOTYPE, and (16) for GGI, respectively]. Disease-free survival curves (distant metastasis–free survival whenever available, relapse-free survival otherwise) were estimated using the Kaplan–Meier estimator and compared using the two-sided log-rank test as implemented in the R package survival version 2.36-9 (http://cran.r-project.org/web/packages/survival/). To statistically compare the prognostic value of competitive risk prediction models, such as subtype classifiers or published gene signatures, we used a two-sided Wilcoxon signed rank test comparing 10-fold cross-validated partial likelihood (CVPL) (69) as implemented in the R/Bioconductor survcomp package (70), version 1.3.6 [http://www.bioconductor.org/packages/release/bioc/html/survcomp.html; (70)]; the lower the estimate, the better the prognostic value.
Results
In breast cancer classification, there have been two general methodological approaches to developing subtype classifiers (Figure 1). SSPs use hierarchical clustering to identify the main breast cancer subtypes from gene expression data, and a nearest centroid classifier is subsequently built to enable subtyping of a single tumor sample (Figure 1, A). Three versions of the SSPs and their corresponding centroids have been published so far and implemented in our study, SSP2003 (6), SSP2006 (2), and PAM50 [(3); Figure 1, C]. SCMs represent an alternative approach based on a mixture of three Gaussians to represent the main breast cancer molecular subtypes, which are the basal-like, HER2-enriched, and luminal tumors (Figure 1, B); the median of the AURKA module score within the luminal tumors was used to discriminate between the low and high proliferative luminal A and B tumors (31). Two versions of the SCMs have been published recently, SCMOD1 (1) and SCMOD2 (8) (Figure 1, D). Given the statistical and clinical challenges in implementing multigene classifiers, we wanted to explore whether we could simplify SCM-based classification to the smallest possible number of genes. Therefore, we developed SCMGENE, an SCM-based classifier reduced to its simplest form, which uses only the expression of the three key and most representative genes of breast cancer biology, ER, HER2, and AURKA. SCM-based classifiers were trained in the largest gene expression dataset, EXPO [EXpression Project for Oncology; dataset consisting of 353 primary breast tumors collected by the International Genomic Consortium, http://www.intgen.org/expo/; see Table 1].
Robustness of Classifiers
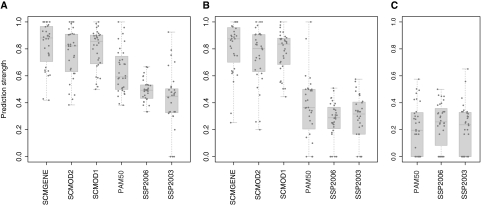
Validating subtype classification is difficult because the true subtypes are unknown. Tibshirani and Walter (64) developed a new statistic, called the prediction strength, to assess the robustness of a classifier, defined as the capacity of a classification model to assign the same tumors to the same subtypes independently of the dataset used to fit the model. The rationale is that if a classification model is strongly dependent on the training dataset, then it is likely to be unreliable (see “Methods and Statistical Analysis” and Supplementary Methods Part 3, available online). Prediction strength ranges from 0 to 1, and a value of at least 0.8 is characteristic of a robust classifier. We compared the robustness of the classification algorithms underlying the three SSPs and the three SCMs (Figure 1, C and D, respectively) in our large compendium of breast cancer datasets (Table 1). To do this, we fitted each model on a training set (EXPO, Table 1) and estimated their prediction strength on the remaining 32 independent (test) datasets.
Because there is no clear consensus as to the number of breast cancer subtypes (20,21), we analyzed robustness for all six classifiers for identifying both three and four subtypes. For the SSPs, we also considered five subtypes. SCMs are limited to three or four by construction (see Supplementary Methods, Part 1, available online). SCMs yielded median prediction strength (≥0.8) for three and four subtypes (Figure 2, A and B, respectively, Supplementary Tables 2 and 3, available online). SSPs yielded lower prediction strength for three subtypes (median prediction strength, 0.45–0.59), and their robustness dramatically decreased with increasing number of subtypes (Figure 2, Supplementary Tables 2–4, available online). SCMs were statistically significantly more robust than SSPs for the identification of three and four subtypes (two-sided Wilcoxon signed rank tests, Holm's corrected P < .005; median differences, confidence intervals, and P values, Supplementary Table 5, available online), although we observed only a trend to significance for the higher robustness of SCMOD2 compared with PAM50 for three subtypes [median prediction strength of 0.82 vs 0.59 for SCMOD2 and PAM50, respectively; two-sided Wilcoxon signed rank test, median difference = −0.13, 95% confidence interval = −0.03 to 0.26; Holm's corrected P = .078]. SCMGENE yielded the best median prediction strength among SCMs, although its superiority over SCMOD2 and SCMOD1 was not statistically significant.
Figure 2.
Robustness of classification into three, four, and five breast cancer molecular subtypes with respect to the models. To assess the robustness of the six subtype classification models, prediction strength is calculated in each dataset separately for the classification into three (A), four (B), and five (C) subtypes. PAM50 = single sample predictor (3); SCMGENE = three-gene subtype classification model; SCMOD1 = subtype classification model 1 (1); SCMOD2 = subtype classification model 2 (8); SSP2003 = single sample predictor (6); SSP2006 = single sample predictor (2).
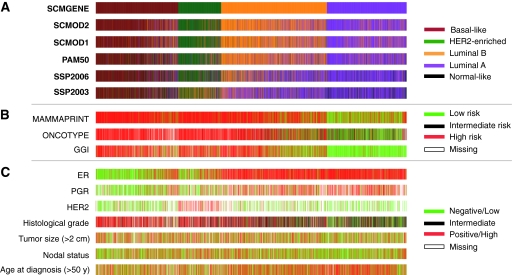
Concordance of Subtype Classifications
We used the published SSPs and SCMs (Figure 1, C and D) to assign molecular subtypes to each of the 5715 breast tumor samples in our compendium of datasets (Tables 1 and 2). As reported previously (71–73), luminal A/B tumors were the most frequently observed subtype (56%–63%), followed by the basal-like (19%–27%) and HER2-enriched (13%–15%) subtypes. SSPs identified a small percentage of normal-like tumors (11%, 8%, and 5% for SSP2003, SSP2006, and PAM50, respectively). Note that the two oldest SSPs, SSP2003 and SSP2006, identified substantially different percentages of luminal A (43% and 41%, respectively) and luminal B (15%) tumors compared with PAM50 and the SCMs (26%–31% and 31%–36% of luminal A and B tumors, respectively).
We then assessed the concordance between classifiers, quantitatively (using Cohen’s Kappa coefficient, κ, and calculating agreement between classifiers, as measured by the proportion of identical classifications), qualitatively [slight, fair, moderate, substantial, and almost perfect concordance based on ranges of κ (22,74), see “Methods and Statistical Analysis”; Table 3], and graphically using color bars (Figure 3). All models proved to be statistically significantly concordant (fair concordance, κ > 0.34, Holm's corrected P < .001 with 49%–86% agreement). SSPs were globally less concordant with each other than SCMs (fair to moderate concordance for SSPs: κ = 0.45–0.58 vs substantial to almost perfect concordance for SCMs: κ = 0.65–0.81; agreement, SSPs: 58%–68% vs SCMs: 75%–86%). However, the most recently developed SSP, that is PAM50, had moderate to substantial concordance with the SCMs (κ = 0.59–0.68, 70%–76% agreement). SCMGENE was statistically significantly concordant with published SCMs (κ = 0.65–0.70) and SSPs (κ = 0.34–0.59), statistically significantly associated with ER (V = 0.64), HER2 (V = 0.52) status, and histological grade (V = 0.55).
Table 3.
Concordance between classifiers and association with clinical parameters*
| Model | SCMGENE | SCMOD2 | SCMOD1 | PAM50 | SSP2006 | SSP2003 |
| Concordance between pairs of subtype classifications, Cohen’s κ (95% CI), %† | ||||||
| SCMOD2 | 0.70 (0.69 to 0.72), 78 | |||||
| SCMOD1 | 0.65 (0.64 to 0.67), 75 | 0.81 (0.80 to 0.82), 86 | ||||
| PAM50 | 0.59 (0.58 to 0.61] 70 | 0.68 (0.67 to 0.70), 76 | 0.67 (0.65 to 0.68), 76 | |||
| SSP2006 | 0.47 (0.45 to 0.48), 59 | 0.51 (0.50 to 0.53), 63 | 0.50 (0.49 to 0.52), 62 | 0.58 (0.56 to 0.59), 68 | ||
| SSP2003 | 0.34 (0.32 to 0.36), 49 | 0.38 (0.37 to 0.40), 53 | 0.36 (0.34 to 0.38), 51 | 0.45 (0.43 to 0.47), 58 | 0.55 (0.53 to 0.56), 67 | |
| Concordance between subtype classifications and risk predictions, Cohen's κ (95% CI), %‡ | ||||||
| MAMMAPRINT | 0.49 (0.46 to 0.51), 81 | 0.56 (0.53 to 0.58), 82 | 0.55 (0.52 to 0.57), 82 | 0.51 (0.49 to 0.54), 79 | 0.44 (0.42 to 0.46), 72 | 0.38 (0.36 to 0.40), 68 |
| ONCOTYPE | 0.61 (0.59 to 0.64), 83 | 0.62 (0.59 to 0.64), 83 | 0.56 (0.53 to 0.58), 81 | 0.61 (0.58 to 0.63), 82 | 0.60 (0.57 to 0.62), 80 | 0.56 (0.54 to 0.58), 77 |
| GGI | 0.58 (0.56 to 0.61), 80 | 0.67 (0.65 to 0.69), 84 | 0.61 (0.59 to 0.64), 81 | 0.73 (0.70 to 0.75), 87 | 0.70 (0.68 to 0.72), 85 | 0.58 (0.55 to 0.60), 79 |
| Association between subtype classifications and clinical parameters, Cramer's V (95% CI)§ | ||||||
| ER | 0.64 (0.61 to 0.67) | 0.71 (0.68 to 0.74) | 0.69 (0.66 to 0.72) | 0.71 (0.68 to 0.74) | 0.69 (0.66 to 0.72) | 0.69 (0.66 to 0.72) |
| PGR | 0.46 (0.42 to 0.5) | 0.54 (0.5 to 0.58) | 0.54 (0.5 to 0.58) | 0.54 (0.50 to 0.58) | 0.53 (0.49 to 0.57) | 0.52 (0.48 to 0.56) |
| HER2 | 0.52 (0.47 to 0.57) | 0.49 (0.44 to 0.54) | 0.48 (0.42 to 0.53) | 0.41 (0.36 to 0.47) | 0.39 (0.33 to 0.44) | 0.34 (0.29 to 0.39) |
| Histological grade‖ | 0.55 (0.51 to 0.59) | 0.58 (0.54 to 0.62) | 0.58 (0.54 to 0.62) | 0.59 (0.54 to 0.63) | 0.54 (0.5 to 0.58) | 0.51 (0.47 to 0.55) |
| Tumor size (>2 cm) | 0.10 (0.065 to 0.13) | 0.15 (0.11 to 0.18) | 0.18 (0.14 to 0.21) | 0.16 (0.12 to 0.19) | 0.13 (0.089 to 0.16) | 0.14 (0.11 to 0.17) |
| Nodal status | 0.07 (0.04 to 0.09) | 0.08 (0.06 to 0.1) | 0.08 (0.06 to 0.10) | 0.09 (0.07 to 0.11) | 0.08 (0.05 to 0.09) | 0.10 (0.07 to 0.11) |
| Age at diagnosis (>50 y) | 0.13 (0.10 to 0.16) | 0.09 (0.05 to 0.11) | 0.12 (0.09 to 0.15) | 0.12 (0.09 to 0.15) | 0.12 (0.08 to 0.14) | 0.14 (0.10 to 0.17) |
Concordance between subtype classifiers and prognostic gene signatures was estimated using Cohen's κ statistics. Association between subtype classifiers and clinical parameters was estimated using Cramer's V statistic. The statistical significance was calculated using a χ2 test. ER = estrogen receptor; GGI = prognostic gene signature (16); MAMMAPRINT = prognostic gene signature (14); PGR = progesterone receptor; ONCOTYPE = prognostic gene signature (15); PAM50 = single sample predictor (3); SCMGENE = three-gene subtype classification model; SCMOD2 = subtype classification model (8); SCMOD1 = subtype classification model (1); SSP2006 = single sample predictor (2); SSP2003: single sample predictor (6).
Cohen's κ statistic (point estimate and 95% confidence interval in parentheses; all Ps < .001, two-sided) estimating the concordance between each pair of subtype classifications as computed by the models under study (note that the SCMs do not identify normal-like tumors). In addition to the κ statistic, the agreement between classifiers, as estimated by the percentage (%) of identical subtype classifications, is given.
Cohen's κ statistic (point estimate and 95% confidence interval in parentheses; all Ps < .001, two-sided) estimating the concordance between the subtype classifications (where basal-like, HER2-enriched, and luminal B are considered high risk and luminal A and normal-like tumors are considered low risk) and the risk predictions as computed by the prognostic gene signatures (where the intermediate- and low-risk group, as defined by applying the published algorithm of the Oncotype DX prognostic model—ONCOTYPE—are combined). In addition to the κ statistic, the agreement between classifiers, as estimated by the percentage (%) of identical subtype classifications is given.
Cramer's V statistic (point estimate and 95% confidence interval in parentheses; all Ps < .001, two-sided) estimating the association between subtype classifications and widely used clinical parameters.
Intermediate histological grade was not considered for estimating the concordance with subtype classifications because they are present in all the molecular subtypes.
Figure 3.
Concordance of classifiers for breast cancer molecular subtyping. A) Colored bars illustrate the molecular subtypes as computed by each of the six classifiers applied to the compendium of 5715 breast tumors. SCMGENE, the three-gene subtype classification model, was used as the reference, that is, the patients (tumors) were unambiguously ordered using the maximum posterior probabilities estimated by SCMGENE. B) The corresponding risk predicted by the prognostic gene signatures. C) Clinical parameters: estrogen receptor (ER) and progesterone receptor (PGR) status defined by immunohistochemistry (IHC); HER2 status defined by IHC or fluorescent in situ hybridization (FISH); histological grade assessed separately in each dataset; and age at diagnosis (> 50 y) and tumor size (> 2 cm) are binary variables. GGI = prognostic gene signature (16); MAMMAPRINT = prognostic gene signature (14); ONCOTYPE = prognostic gene signature (15); PAM50 = single sample predictor (3); SCMOD1 = subtype classification model 1 (1); SCMOD2 = subtype classification model 2 (8); SSP2003 = single sample predictor (6); SSP2006 = single sample predictor (2).
For overall concordance (Figure 3), SCMGENE was used as the reference classifier (Supplementary Figure 1, available online, shows similar results for PAM50 as the reference). The basal-like subtype was the most consistently assigned subtype by all classifiers (basal-like vs the rest, median κ = 0.78). The HER2-enriched and luminal A subtypes were moderately concordant between different classifiers (median κ = 0.55). In contrast, the majority of the luminal B and normal-like tumors were classified differently depending on the classifier (median κ = 0.38 and 0.41, respectively).
We then computed risk predictions using the published algorithms of three prognostic gene signatures, MammaPrint (14) (MAMMAPRINT), Oncotype Dx (15) (ONCOTYPE), and Gene expression Grade Index (16) (GGI), and assessed the concordance between these risk classifications and the subtype classifications (Table 3, Figure 3, and Supplementary Figure 1, available online). Note that, similarly to Fan et al. (17), we did not use the commercially available assay, but we relied instead on the published microarray data to compute the risk classifications. Consistent with Fan et al. (17), basal-like, HER2-enriched, and luminal B tumors were almost all classified as high risk by the prognostic gene signatures, whereas luminal A and normal-like tumors were mostly classified as low risk (moderate to substantial concordance; Table 3) except for MAMMAPRINT, which may yield only fair concordance because of a small proportion of low-risk patients (approximately half of luminal A tumors are still predicted to be high risk; see Figure 3 and Supplementary Figure 1, available online).
We assessed the association between the subtype classifications and clinical parameters (Table 3, Figure 3, Supplementary Figure 1, available online). As expected, the majority of basal-like and luminal tumors were ER− and ER+, respectively (moderate to substantial concordance, Cramer’s V = 0.64–0.71). In contrast, the concordance with progesterone receptor was only moderate (V = 0.46–0.54). Most tumors defined as HER2 enriched are HER2 overexpressed by IHC or amplified by fluorescent in situ hybridization (fair to moderate concordance, V = 0.34–0.52), with the strongest concordance provided by the SCMs (V = 0.48–0.52). Tumors from the basal-like, HER2-enriched, and luminal B subtypes were mostly histological grade 3 tumors (moderate concordance, V = 0.51–0.58).
It is worth noting that no association was observed between the subtype classifications and the tumor size, nodal status, and age at diagnosis, suggesting that these features are independent of the molecular subtype (Table 3 and Figure 3, C).
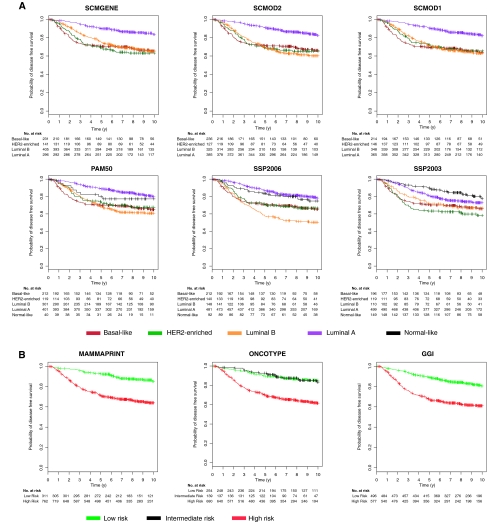
Survival of Untreated Early Breast Cancer Patients With Respect to Subtypes
Using data from the 1260 untreated patients with node-negative tumors (Tables 1 and 2), we analyzed the prognostic value of the six subtype classifiers and the published gene signatures. The survival curves were statistically significantly different between subtypes for all six classifiers (Figure 4, A; log-rank test, P < .001) and between the risk groups defined by prognostic gene signatures (Figure 4, B; log-rank test, P < .001). These results confirm the substantial prognostic value of molecular subtyping on early breast cancers without potentially confounding treatment effects. Although the survival curves from the SCMs, including SCMGENE, were virtually identical, SSPs exhibited some discrepancies for luminal B and normal-like tumors. We observed that a small group of 148 patients with luminal B tumors have the worst survival according to SSP2006, a result inconsistent with the other five classifiers (Figure 4, A). The survival curves for patients with tumors classified as normal-like vary depending on which SSP is used; the prognosis of patients with normal-like and luminal A tumors was similar according to SSP2006 (probability of survival at 5 years for patients with normal-like vs luminal A tumors, P = .84 and .87, respectively); normal-like was better for SSP2003 (P = .90 and .80 for normal-like vs luminal A, respectively) but slightly worse for PAM50 (P = .80 and .89 for normal-like vs luminal A, respectively).
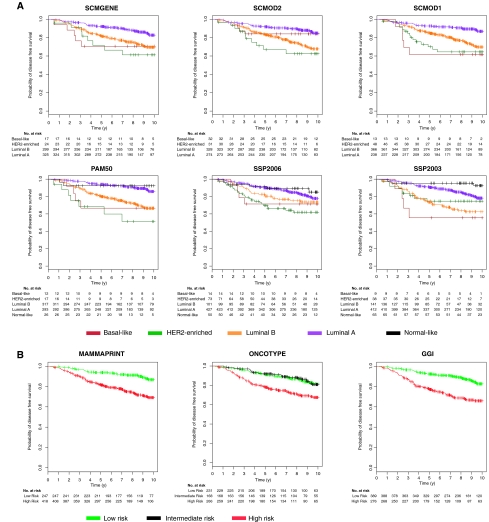
Figure 4.
Survival curves of untreated patients with respect to the subtype and risk classifications. A) Kaplan–Meier disease-free survival curves censored at 10 years for the subtypes identified by the six classifiers. B) The risk groups identified by the three prognostic gene signatures in the cohort of 1260 untreated patients with node-negative tumors (survival data were missing for 187 untreated patients). The statistically significant prognostic value of the subtype classifiers and published gene signatures was confirmed in this cohort (log-rank P < .001, two-sided). GGI = prognostic gene signature (16); MAMMAPRINT = prognostic gene signature (14); ONCOTYPE = prognostic gene signature (15); PAM50 = single sample predictor (3); SCMGENE = three-gene subtype classification model; SCMOD1 = subtype classification model 1 (1); SCMOD2 = subtype classification model 2 (8); SSP2003 = single sample predictor (6); SSP2006 = single sample predictor (2).
Given the good survival of the intermediate-risk group identified by ONCOTYPE (probability of survival at 5 years for patients predicted as intermediate and low risk: P = .93 and .90, respectively; Figure 4, B), we decided to merge it with the low-risk group as in Fan et al. (17). The luminal A subtype defined by the classifiers exhibited a survival similar to the low-risk group defined by the prognostic gene signatures (probability of disease-free survival at 5 years, SCMs: P = .90–.91; SSPs: P = .81–.90; and gene signatures: P = .89–.92; Figure 4). To confirm the similarity of the survival curves, we statistically compared the prognostic value of the subtype classifiers and gene signatures to test whether one classification was better than another. We used a 10-fold CVPL (see “Methods and Statistical Analysis”) to assess the prognostic value of each classification (Supplementary Table 6, available online), and we observed that all subtype classifiers yielded statistically similar prognostic value (CVPL = 1.651–1.667, two-sided Wilcoxon signed rank test, Holm's corrected P ≥ .05; Supplementary Table 7, available online). The gene signatures yielded better prognostic value (CVPL = 1.648, 1.648, and 1.651 for MAMMAPRINT, ONCOTYPE, and GGI, respectively), but they did not statistically significantly outperform the subtype classifiers (two-sided Wilcoxon signed rank test, Holm's corrected P ≥ .05; Supplementary Table 7, available online), except for SSP2003, which appeared to yield statistically significantly worse prognostic value than GGI (two-sided Wilcoxon signed rank test, Holm's corrected P = .035; Supplementary Table 7, available online). These results suggest that none of the subtype classifiers statistically significantly outperform the others and that we lack evidence to claim superiority of the published gene signatures for prognosis. We also showed in a series of 676 tamoxifen-treated patients with ER+ tumors as defined by locally reviewed IHC that tumors identified by SCMGENE and the other subtype classifiers as discordant (either basal-like or HER2-enriched subtype) had a poorer survival (probability of survival at 5 years for patients with either luminal A or B tumors compared with those with discordant ER status: P = .86–.88 and P = .63–.76, respectively) suggesting that these patients did not benefit from tamoxifen therapy (Figure 5).
Figure 5.
Survival curves of tamoxifen-treated patients with respect to the subtype and risk classifications. A) Kaplan–Meier disease-free survival curves censored at 10 years for the subtypes identified by the six classifiers. B) The risk groups identified by the three prognostic gene signatures in the cohort of 676 tamoxifen-treated patients with estrogen receptor–positive (ER+) tumors as defined by locally reviewed immunohistochemistry (survival data were missing for 11 patients). Despite their ER+ status, some tumors were classified as either basal-like or HER2-enriched subtypes by the six subtype classifiers, and the corresponding patients consistently exhibited poor survival. The statistically significant prognostic value of the subtype classifiers and published gene signatures were confirmed in this cohort (log-rank P < .001, two-sided). GGI = prognostic gene signature (16); MAMMAPRINT = prognostic gene signature (14); ONCOTYPE = prognostic gene signature (15); PAM50 = single sample predictor (3); SCMGENE = three-gene subtype classification model; SCMOD1 = subtype classification model 1 (1); SCMOD2 = subtype classification model 2 (8); SSP2003 = single sample predictor (6); SSP2006 = single sample predictor (2).
Discussion
Despite the widespread recognition of the value of molecular subtyping, the complexity of the classification models, which use dozens to hundreds of genes, and uncertainty about their robustness and clinical relevance have been impediments to their general clinical use (18–21). Furthermore, quality assessment of molecular subtyping is complex because the truth is unknown. Using a collection of data from 5715 breast tumors, we analyzed five previously described classifiers (three SSPs and two SCMs) and compared these to SCMGENE, a simplified SCM-based classifier that uses only three genes that capture key biological processes in breast cancer namely ER signaling, HER2 signaling, and proliferation. We used the prediction strength statistic (64) to quantify robustness of subtype classifications, defined as the capacity of an algorithm to assign the same tumors to the same subtypes regardless of the gene expression data used to build the classifier. We found SCMs to be statistically significantly more robust than SSPs. Moreover, among the SCMs, SCMGENE, our simple three-gene model, was statistically as robust as the published SCMs, which use hundreds of genes.
Each classifier demonstrated fair to substantial concordance, underscoring the validity of the subtypes. Among the molecular subtypes, the basal-like subtype was consistently identified independently of the classifier used. In contrast, the luminal A, luminal B, and normal-like tumors were more difficult to classify, consistent with the recent study of Mackay et al. (21); the separation of the luminal group into A and B was not well supported by our analysis, probably because these subtypes are defined by expression of proliferation-related genes, which exhibit a continuum of expression levels (1,8,20,22). Like others (20,22), we did not find support for the normal-like subtype. It may be that this subtype is an artifact resulting from stromal contamination (22).
In the survival analysis of a large set of untreated node-negative breast cancer patients, we confirmed that all six classifiers had a statistically significant prognostic value (9,22). When assessing concordance with published prognostic gene signatures, we found that the vast majority of basal-like, HER2-enriched, and luminal B tumors were classified as high risk (8,17). Again, all the subtype classifiers and gene signatures yielded statistically similar prognostic value. Notably, we also showed that for a cohort of patients with ER+ tumors defined initially by IHC who were treated with adjuvant tamoxifen monotherapy, those patients with tumors identified by SCMGENE and the other subtype classifiers as basal like and HER2 enriched had a poorer survival, suggesting that these patients may not benefit from tamoxifen therapy. However, the clinical relevance in terms of response to therapy—for example, endocrine or anti-HER2—of those patients classified differently using IHC and gene expression remains unknown.
All subtype classifiers were statistically significantly associated with clinical variables widely used in management of breast cancer patients; the ER+ (IHC) tumors were particularly well identified by SCMOD2 and PAM50, whereas the HER2 amplified/overexpressed (FISH/IHC) tumors were highly concordant with the SCMGENE classification. However, we found no association with the subtype classifiers and tumor size, nodal status, and age at diagnosis. A large study involving central pathology measurement of traditional clinical parameters and gene expression profiling is needed to definitively draw conclusions about the complementarity or superiority of one technology over another; in addition, this would help determine the clinical relevance of the above concordance issues, that is, which method of subtype classification or central pathology using IHC would yield better predictive value for prescription of anti-HER2 or endocrine therapies. Ongoing prospective trials such as the MINDACT may facilitate such comparisons (75). Our data also suggest that accurate and reproducible measurements of ER, HER2, and proliferation can be used for molecular subtyping in breast cancer. This holds true for currently used methods of centrally reviewed IHC for ER, HER2, and Ki67, particularly for large clinical studies. Although IHC has well-known limitations in terms of intra-laboratory reproducibility and subjective and semiquantitative assessment of protein expression, IHC performed in a central laboratory undoubtedly provides significant additional prognostic value compared with local pathology. However, the good technical reproducibility and the quantitative nature of gene expression profiling (58) makes expression-based classification models promising candidates to complement the current IHC markers widely used in breast cancer. Our results also support the use of SCMGENE to provide molecular subtype classification for samples in large meta-analysis studies of gene expression profiling that involve data generated by different laboratories using diverse microarray technologies.
This study has several potential limitations. First, because our collection of breast cancer microarray data is composed of datasets that were retrospectively accrued, the selection of these patients may result in unbalanced distribution of the different molecular subtypes. Second, we used the normalized gene expression data as provided in public databases and authors’ websites; no attempts to renormalize the microarray data were made, although a robust scaling procedure ensured that the gene expressions were similarly distributed across datasets. Third, depending on the dataset, we did not annotate and map some probes used in the subtype classifiers because of the diversity of microarray platforms used in our compendium of datasets (Supplementary Table 3, available online). Fourth, the current implementation of the CVPL does not allow checking and correction for departure from the proportional hazards assumption. Finally, in contrast to SCMs, SSPs rely on hierarchical clustering, which makes automated identification of the main subtypes present in a specific dataset challenging (21); this may have affected their robustness estimations but also highlights the difficulties of using this type of classification method.
In conclusion, our study demonstrated that for breast cancer molecular subtyping, the simplest classification model, SCMGENE, which is based on the expression levels of three key genes and a simple Gaussian probabilistic model, was surprisingly concordant with the more complex published classifiers and yielded similar prognostic value. It also proved to be one of the most robust classifiers because it uses only ER, HER2, and AURKA gene expression, whereas the other classifiers rely on many more genes. The simplicity and robustness of the SCMGENE model provide an opportunity for wide application using a variety of expression data types. Moreover, our results suggest that, at present, for molecular subtyping of breast cancer, three genes provide adequate discrimination for clinical implementation; the clinical and biological relevance of the value of adding more genes remains to be demonstrated.
Funding
B.H.-K. was supported by a grant in aid from Fulbright Commission for Educational Exchange for postdoctoral research. B.H.-K. and J.Q. were supported by a grant from the National Library of Medicine of the US National Institutes of Health (R01 LM010129-01). A.C.C. and J.Q. were supported by a grant from the Claudia Adams Barr Program in Innovative Basic Cancer Research. C.S. was supported by the Belgian National Foundation for Research (FNRS), the MEDIC Foundation and the Breast Cancer Research Foundation (BCRF). C.D. was supported by the Belgian National Foundation for Research (FNRS), Belgium. S.L. was supported by the National Health and Medical Research Council of Australia (NHMRC) and the European Society of Medical Oncology (ESMO).
Supplementary Material
Footnotes
Supplementary File 1 (demo sbt.csv; available online) is a CSV file containing all the data necessary to easily reproduce the results of the article. The file contains the clinical information and the subtype classifications for all the datasets described in Table 1, with duplicated samples removed.
B. Haibe-Kains was responsible for the design and execution of the study and statistical analysis; B. Haibe-Kains, C. Desmedt, A. C. Culhane, S. Loi, G. Bontempi, and J. Quackenbush were responsible for data interpretation and article writing; J. Quackenbush and C. Sotiriou co-supervised the study. All authors read and approved the final article. J. Quackenbush and C. Sotiriou are co-last authors.
The funders did not have any involvement in the design of the study; the collection, analysis, and interpretation of the data; the writing of the article; or the decision to submit the article for publication. C. Sotiriou is named inventor on a patent application for the Gene expression Grade Index (GGI) used in this study. There are no other conflicts of interest.
We thank Mauro Delorenzi and Pratyaksha “Asa” Wirapati for their fruitful collaboration on the development of the Subtype Classification Model and Stefan Bentink for helpful discussion and advice. We also thank Sonal Jhaveri for her editorial assistance.
References
- 1.Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14(16):5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 2.Hu Z, Fan C, Oh D, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96–107. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 5.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;1(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100(18):10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wirapati P, Sotiriou C, Kunkel S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10(4):R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haibe-Kains B, Desmedt C, Rothé F, et al. A fuzzy gene expression-based computational approach improves breast cancer prognostication. Genome Biol. 2010;11(2):R18. doi: 10.1186/gb-2010-11-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 11.Pusztai L, Broglio K, Andre F, et al. Effect of molecular disease subsets on disease-free survival in randomized adjuvant chemotherapy trials for estrogen receptor-positive breast cancer. J Clin Oncol. 2008;26(28):4679–4683. doi: 10.1200/JCO.2008.17.2544. [DOI] [PubMed] [Google Scholar]
- 12.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360(8):790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 13.Van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 14.Van t Veer LJ, Dai H, Van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 15.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 16.Sotiriou C, Wirapati P, Loi SM, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 17.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression–based predictors for breast cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 18.Pusztai L, Mazouni C, Anderson K, et al. Molecular classification of breast cancer: limitations and potential. Oncologist. 2006;11(8):868–877. doi: 10.1634/theoncologist.11-8-868. [DOI] [PubMed] [Google Scholar]
- 19.Andre F, Pusztai L. Molecular classification of breast cancer: implications for selection of adjuvant chemotherapy. Nat Clin Prac Oncol. 2006;3(11):621–632. doi: 10.1038/ncponc0636. [DOI] [PubMed] [Google Scholar]
- 20.Kapp A, Jeffrey S, Langerod A, et al. Discovery and validation of breast cancer subtypes. BMC Genomics. 2006;7(1):231. doi: 10.1186/1471-2164-7-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weigelt B, Mackay A, A’Hern R, et al. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 2010;11(4):339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 22.Mackay A, Weigelt B, Grigoriadis A, et al. Microarray-based class discovery for molecular classification of breast cancer: analysis of interobserver agreement. J Natl Cancer Inst. 2011;103(8):662–673. doi: 10.1093/jnci/djr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bittner M. Expression Project for Oncology (expO) http://www.intgen.org/expo/ [Google Scholar]
- 24.Minn AJ, Gupta GP, Padua D, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A. 2007;104(16):6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Klijn JGM, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 26.Korkola JE, Blaveri E, DeVries S, et al. Identification of a robust gene signature that predicts breast cancer outcome in independent data sets. BMC Cancer. 2007;7(1):61–73. doi: 10.1186/1471-2407-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korkola JE, DeVries S, Fridlyand J, et al. Differentiation of lobular versus ductal breast carcinomas by expression microarray analysis. Cancer Res. 2003;63(21):7167–7175. [PubMed] [Google Scholar]
- 28.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loi SM, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 32.Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 34.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Desmedt C, Piette F, Loi SM, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clinical Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 36.Bonnefoi H, Potti A, Delorenzi M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8(12):1071–1078. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt M, Bohm D, von Torne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68(13):5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 38.Saal LH, Johansson P, Holm K, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104(18):7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nimeus-Malmstrom E, Krogh M, Malmstrom P, et al. Gene expression profiling in primary breast cancer distinguishes patients developing local recurrence after breast-conservation surgery, with or without postoperative radiotherapy. Breast Cancer Res. 2008;10(2):R34. doi: 10.1186/bcr1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campone M, Campion L, Roché H, et al. Prediction of metastatic relapse in node-positive breast cancer: establishment of a clinicogenomic model after FEC100 adjuvant regimen. Breast Cancer Res Treat. 2008;109(3):491–501. doi: 10.1007/s10549-007-9673-x. [DOI] [PubMed] [Google Scholar]
- 41.Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24(29):4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 42.Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24(26):4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 43.Bos PD, Zhang XH-F, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calabro A, Beissbarth T, Kuner R, et al. Effects of infiltrating lymphocytes and estrogen receptor on gene expression and prognosis in breast cancer. Breast Cancer Res Treat. 2009;116(1):69–77. doi: 10.1007/s10549-008-0105-3. [DOI] [PubMed] [Google Scholar]
- 45.Yu K, Ganesan K, Tan LK, et al. A precisely regulated gene expression cassette potently modulates metastasis and survival in multiple solid cancers. PLoS Genet. 2008;4(7):e1000129. doi: 10.1371/journal.pgen.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waddell N, Arnold J, Cocciardi S, et al. Subtypes of familial breast tumours revealed by expression and copy number profiling. Breast Cancer Res Treat. 2009;123(3):661–677. doi: 10.1007/s10549-009-0653-1. [DOI] [PubMed] [Google Scholar]
- 47.Huang E, Cheng SH, Dressman H, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361(9369):1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 48.Natrajan R, Weigelt B, Mackay A, et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2009;121(3):575–589. doi: 10.1007/s10549-009-0501-3. [DOI] [PubMed] [Google Scholar]
- 49.Li Q, Eklund AC, Juul N, et al. Minimising immunohistochemical false negative ER classification using a complementary 23 gene expression signature of ER status. PloS One. 2010;5(12):e15031. doi: 10.1371/journal.pone.0015031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Sieuwerts A, McGreevy M, et al. The 76-gene signature defines high-risk patients that benefit from adjuvant tamoxifen therapy. Breast Cancer Res Treat. 2008;116(2):303–309. doi: 10.1007/s10549-008-0183-2. [DOI] [PubMed] [Google Scholar]
- 51.Silver DP, Richardson AL, Eklund AC, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28(7):1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 53.Lu X, Lu X, Wang ZC, et al. Predicting features of breast cancer with gene expression patterns. Breast Cancer Res Treat. 2008;108(2):191–201. doi: 10.1007/s10549-007-9596-6. [DOI] [PubMed] [Google Scholar]
- 54.Popovici V, Chen W, Gallas BG, et al. Effect of training-sample size and classification difficulty on the accuracy of genomic predictors. Breast Cancer Res. 2010;12(1):R5. doi: 10.1186/bcr2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dedeurwaerder S, Desmedt C, Calonne E, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011;3(12):726–741. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loi SM, Haibe-Kains B, Desmedt C, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9(1):239–250. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Symmans WF, Hatzis C, Sotiriou C, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J Clin Oncol. 2010;28(27):4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi L, Reid LH, Jones WD, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alibes A, Yankilevich P, Canada A, et al. IDconverter and IDClight: Conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8(1):9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perou CM, Parker JS, Prat A, et al. Clinical implementation of the intrinsic subtypes of breast cancer. Lancet Oncol. 2010;11(8):718–719. doi: 10.1016/S1470-2045(10)70176-5. [DOI] [PubMed] [Google Scholar]
- 61.Sorlie T, Borgan E, Myhre S, et al. The importance of gene-centring microarray data. Lancet Oncol. 2010;11(8):719–720. doi: 10.1016/S1470-2045(10)70174-1. [DOI] [PubMed] [Google Scholar]
- 62.Culhane AC, Schwarzl T, Sultana R, et al. GeneSigDB—a curated database of gene expression signatures. Nucleic Acids Res. 2010;38(Database issue):D716–D725. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haibe-Kains B, Schroder M, Bontempi G, et al. genefu R/Bioconductor package: Relevant Functions for Gene Expression Analysis, Especially in Breast Cancer. http://www.bioconductor.org/help/bioc-views/devel/bioc/html/genefu.html. Accessed June 1, 2011. [Google Scholar]
- 64.Tibshirani R, Walther G. Cluster validation by prediction strength. J Comput Graph Stat. 2005;14(3):511–528. [Google Scholar]
- 65.Wilcoxon F. Individual comparisons of grouped data by ranking methods. J Econ Entomol. 1946;39(6):80–83. doi: 10.1093/jee/39.2.269. [DOI] [PubMed] [Google Scholar]
- 66.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6(2):65–70. [Google Scholar]
- 67.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20(1):37–46. [Google Scholar]
- 68.Cramer H. Mathematical Methods of Statistics. Princeton, NJ: Princeton University Press; 1999. [Google Scholar]
- 69.Verweij PJ, Van Houwelingen HC. Cross-validation in survival analysis. Stat Med. 1993;12(24):2305–2314. doi: 10.1002/sim.4780122407. [DOI] [PubMed] [Google Scholar]
- 70.Schroder MS, Culhane AC, Quackenbush J, et al. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27(22):3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Calza S, Hall P, Auer G, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8(4):R34. doi: 10.1186/bcr1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conforti R, Boulet T, Tomasic G, et al. Breast cancer molecular subclassification and estrogen receptor expression to predict efficacy of adjuvant anthracyclines-based chemotherapy: a biomarker study from two randomized trials. Ann Oncol. 2007;18(9):1477–1483. doi: 10.1093/annonc/mdm209. [DOI] [PubMed] [Google Scholar]
- 73.Sihto H, Lundin J, Lehtimaki T, et al. Molecular subtypes of breast cancers detected in mammography screening and outside of screening. Clin Cancer Res. 2008;14(13):4103–4110. doi: 10.1158/1078-0432.CCR-07-5003. [DOI] [PubMed] [Google Scholar]
- 74.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 75.Cardoso F, Piccart-Gebhart M, Van’t Veer L, et al. The MINDACT trial: the first prospective clinical validation of a genomic tool. Molecular Oncol. 2007;1(3):246–251. doi: 10.1016/j.molonc.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.