Abstract
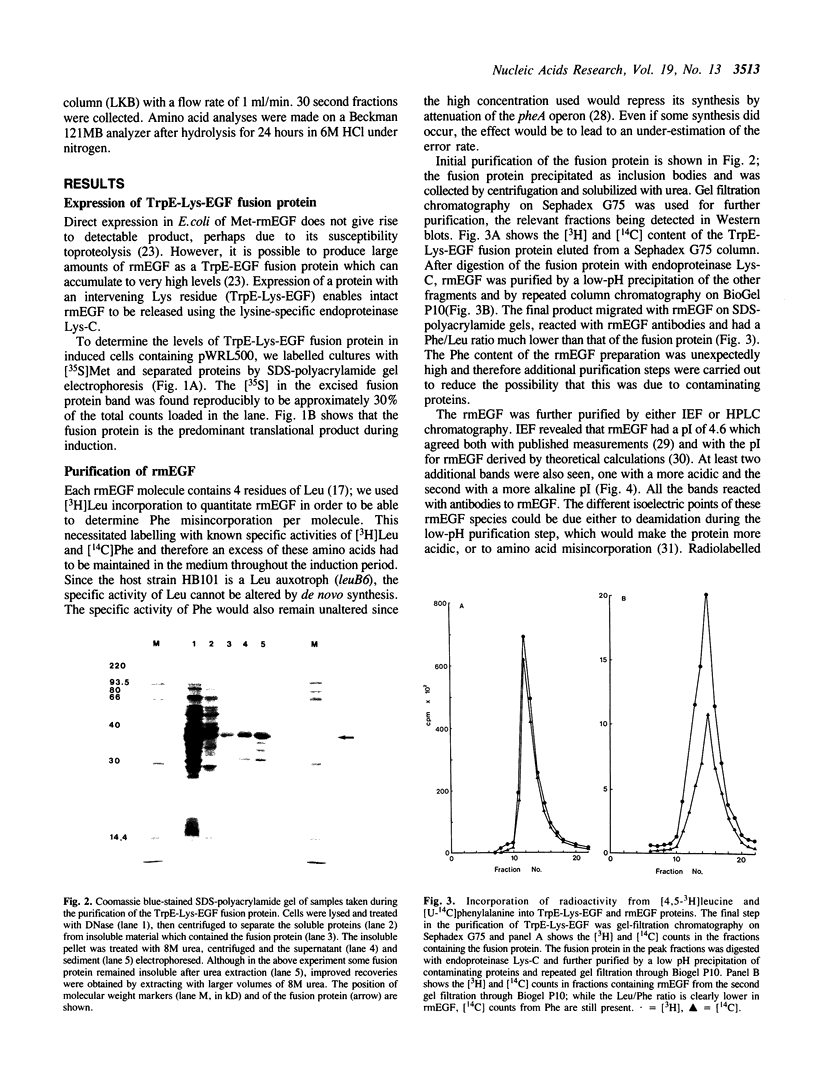
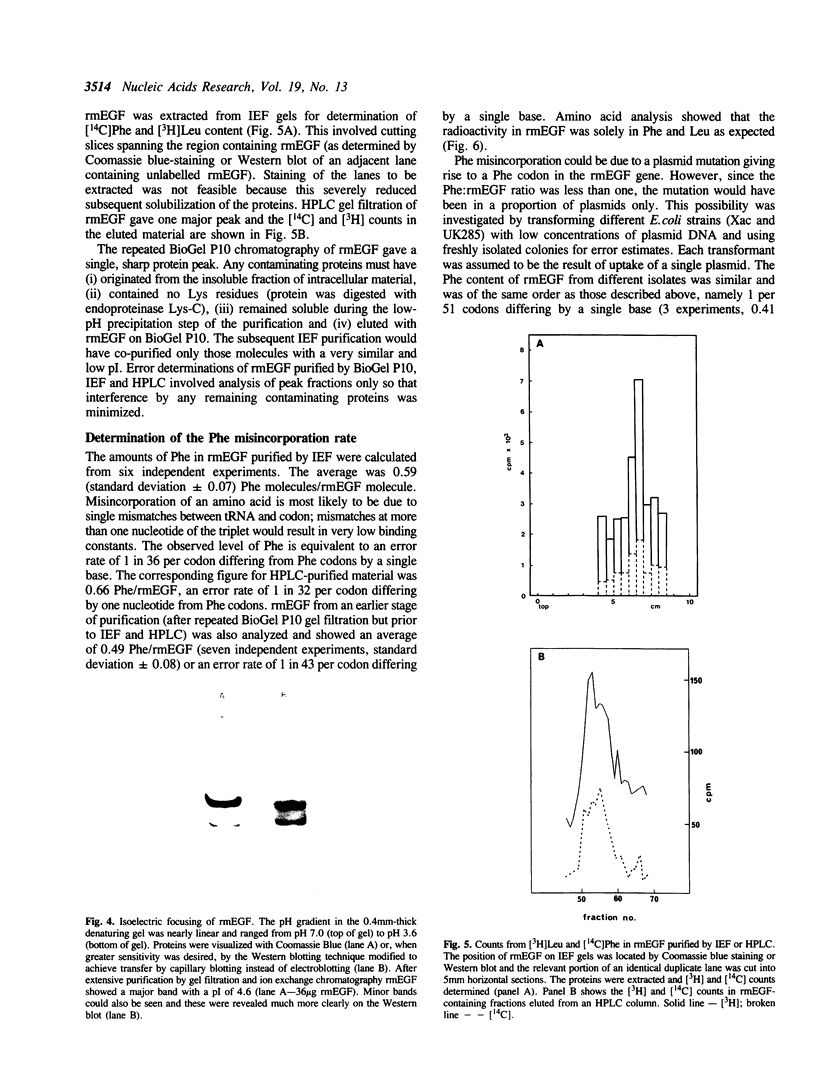
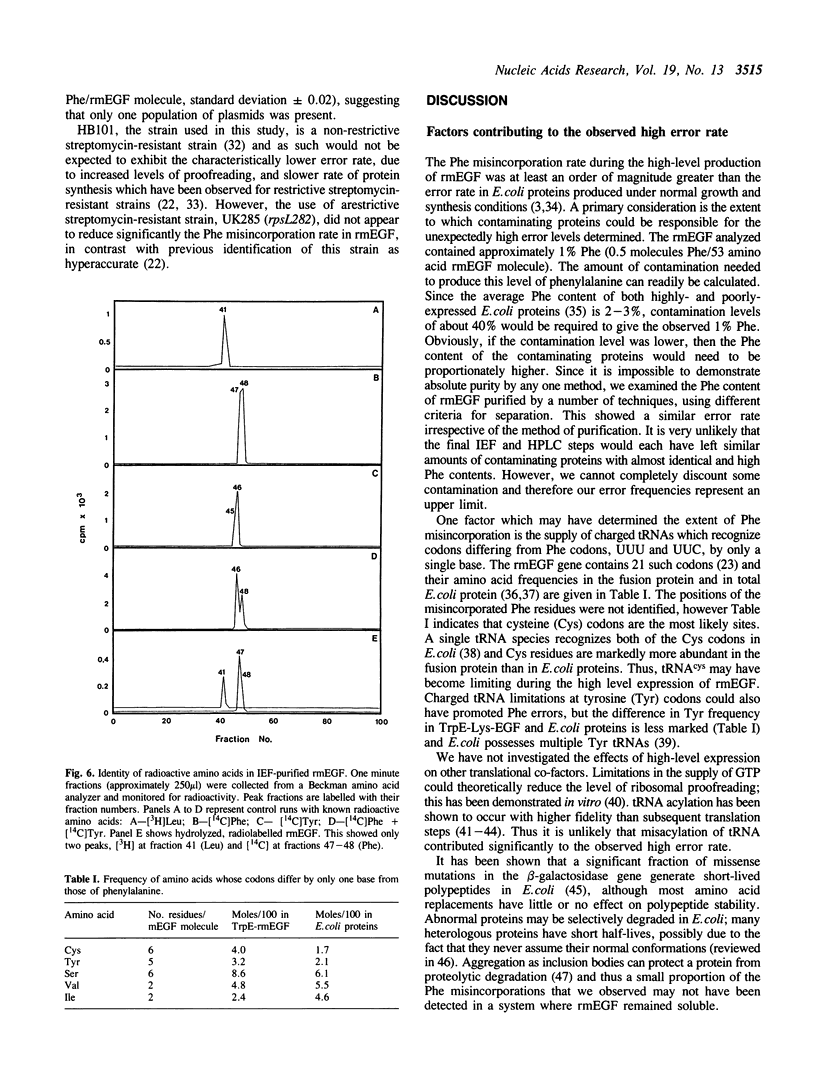
To determine whether the high-level expression of foreign proteins in Escherichia coli can lead to frequent translational errors, we analyzed amino acid misincorporation in mouse epidermal growth factor (mEGF) produced as a TrpE fusion protein. The mEGF DNA does not encode phenylalanine and determining the phenylalanine content of the purified protein will measure missense errors. Using this approach, we found an error frequency of about 1 in 40 for codons differing by a single base from those for phenylalanine. This is at least ten times higher than the error rate found for normal E. coli protein synthesis and may be due to limiting supply of charged tRNAs and GTP, brought about by the high-level production of the heterologous protein. The unexpectedly high error rate has implications for the clinical use of E. coli-derived therapeutic proteins.
Full text
PDF
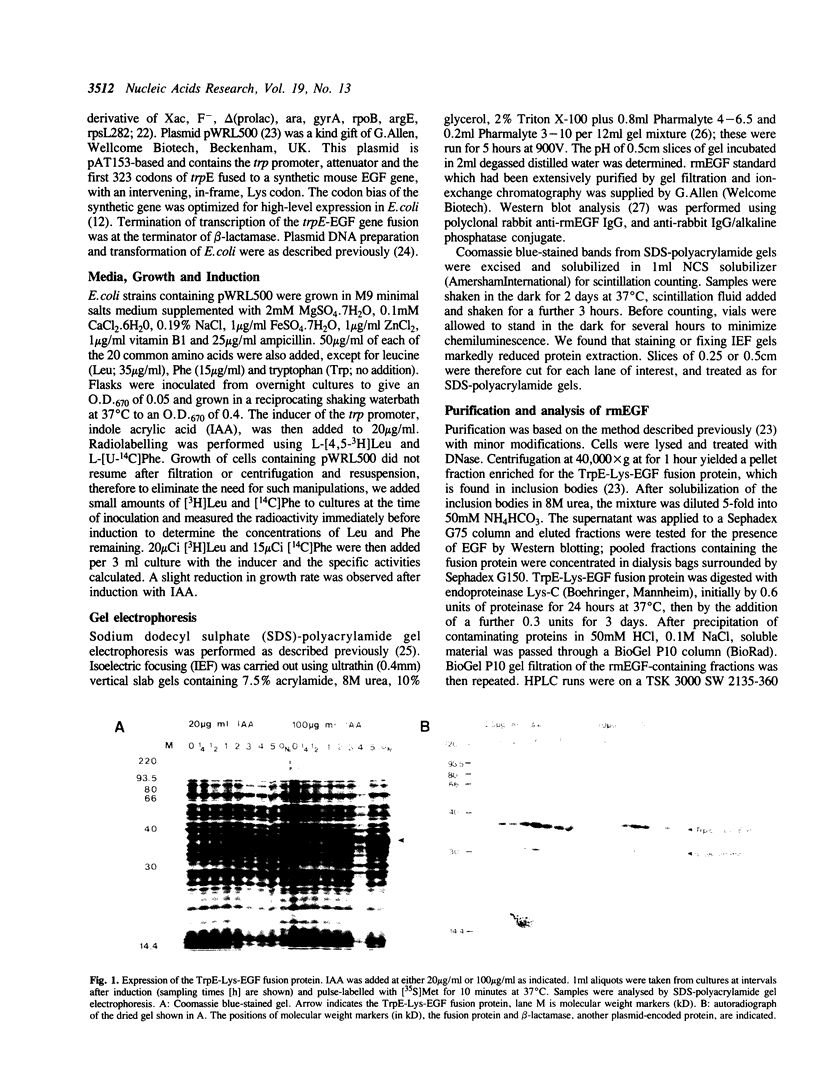

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen G., Paynter C. A., Winther M. D. Production of epidermal growth factor in Escherichia coli from a synthetic gene. J Cell Sci Suppl. 1985;3:29–38. doi: 10.1242/jcs.1985.supplement_3.4. [DOI] [PubMed] [Google Scholar]
- Bohman K., Ruusala T., Jelenc P. C., Kurland C. G. Kinetic impairment of restrictive streptomycin-resistant ribosomes. Mol Gen Genet. 1984;198(2):90–99. doi: 10.1007/BF00328706. [DOI] [PubMed] [Google Scholar]
- Borg-Olivier S. A., Tarlinton D., Brown K. D. Defective regulation of the phenylalanine biosynthetic operon in mutants of the phenylalanyl-tRNA synthetase operon. J Bacteriol. 1987 May;169(5):1949–1953. doi: 10.1128/jb.169.5.1949-1953.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouadloun F., Donner D., Kurland C. G. Codon-specific missense errors in vivo. EMBO J. 1983;2(8):1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Breckenridge L., Gorini L. Genetic analysis of streptomycin resistance in Escherichia coli. Genetics. 1970 May;65(1):9–25. doi: 10.1093/genetics/65.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. S., Kwoh D. Y., Kwoh T. J., Soltvedt B. C., Zipser D. Stabilization of a degradable protein by its overexpression in Escherichia coli. Gene. 1981 Jun-Jul;14(1-2):121–130. doi: 10.1016/0378-1119(81)90154-2. [DOI] [PubMed] [Google Scholar]
- Cohen S., Taylor J. M. Epidermal growth factor: chemical and biological characterization. Recent Prog Horm Res. 1974;30(0):533–550. doi: 10.1016/b978-0-12-571130-2.50017-1. [DOI] [PubMed] [Google Scholar]
- Cupples C. G., Miller J. H. Effects of amino acid substitutions at the active site in Escherichia coli beta-galactosidase. Genetics. 1988 Nov;120(3):637–644. doi: 10.1093/genetics/120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N., Gallant J. An estimate of the global error frequency in translation. Mol Gen Genet. 1982;188(2):169–172. doi: 10.1007/BF00332670. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Dingwall C. An editing mechanism for the methionyl-tRNA synthetase in the selection of amino acids in protein synthesis. Biochemistry. 1979 Apr 3;18(7):1250–1256. doi: 10.1021/bi00574a021. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Dingwall C. Cysteinyl-tRNA synthetase from Escherichia coli does not need an editing mechanism to reject serine and alanine. High binding energy of small groups in specific molecular interactions. Biochemistry. 1979 Apr 3;18(7):1245–1249. doi: 10.1021/bi00574a020. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Dingwall C. Establishing the misacylation/deacylation of the tRNA pathway for the editing mechanism of prokaryotic and eukaryotic valyl-tRNA synthetases. Biochemistry. 1979 Apr 3;18(7):1238–1245. doi: 10.1021/bi00574a019. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Dingwall C. Evidence for the double-sieve editing mechanism in protein synthesis. Steric exclusion of isoleucine by valyl-tRNA synthetases. Biochemistry. 1979 Jun 12;18(12):2627–2631. doi: 10.1021/bi00579a030. [DOI] [PubMed] [Google Scholar]
- Giulian G. G., Moss R. L., Greaser M. Analytical isoelectric focusing using a high-voltage vertical slab polyacrylamide gel system. Anal Biochem. 1984 Nov 1;142(2):421–436. doi: 10.1016/0003-2697(84)90486-x. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Zipser D. Deg phenotype of Escherichia coli lon mutants. J Bacteriol. 1978 Feb;133(2):844–851. doi: 10.1128/jb.133.2.844-851.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980 May 10;8(9):1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Heller K., Kadner R. J. Nucleotide sequence of the gene for the vitamin B12 receptor protein in the outer membrane of Escherichia coli. J Bacteriol. 1985 Mar;161(3):904–908. doi: 10.1128/jb.161.3.904-908.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981 Feb 15;146(1):1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Jelenc P. C., Kurland C. G. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3174–3178. doi: 10.1073/pnas.76.7.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston T. C., Borgia P. T., Parker J. Codon specificity of starvation induced misreading. Mol Gen Genet. 1984;195(3):459–465. doi: 10.1007/BF00341447. [DOI] [PubMed] [Google Scholar]
- Kirkwood T. B., Holliday R., Rosenberger R. F. Stability of the cellular translation process. Int Rev Cytol. 1984;92:93–132. doi: 10.1016/s0074-7696(08)61325-x. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Isaksson L. A. Involvement of ribosomal protein L7/L12 in control of translational accuracy. Proc Natl Acad Sci U S A. 1985 Feb;82(3):717–721. doi: 10.1073/pnas.82.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. R. Tinkering with enzymes: what are we learning? Science. 1987 Jun 5;236(4806):1252–1258. doi: 10.1126/science.3296192. [DOI] [PubMed] [Google Scholar]
- Kurland C. G., Ehrenberg M. Growth-optimizing accuracy of gene expression. Annu Rev Biophys Biophys Chem. 1987;16:291–317. doi: 10.1146/annurev.bb.16.060187.001451. [DOI] [PubMed] [Google Scholar]
- Kurland C. G. The role of guanine nucleotides in protein biosynthesis. Biophys J. 1978 Jun;22(3):373–392. doi: 10.1016/S0006-3495(78)85494-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langridge J. Mutation spectra and the neutrality of mutations. Aust J Biol Sci. 1974 Jun;27(3):309–319. doi: 10.1071/bi9740309. [DOI] [PubMed] [Google Scholar]
- Loftfield R. B., Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972 Aug;128(5):1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzara G. P., McClain W. H. Cysteine transfer RNA of Escherichia coli: nucleotide sequence and unusual metabolic properties of the 3' C-C-A terminus. J Mol Biol. 1977 Dec 25;117(4):1061–1079. doi: 10.1016/s0022-2836(77)80013-2. [DOI] [PubMed] [Google Scholar]
- Nene V., Glass R. E. Genetic studies on the beta subunit of Escherichia coli RNA polymerase. IV. Structure-function correlates. Mol Gen Genet. 1984;194(1-2):166–172. doi: 10.1007/BF00383512. [DOI] [PubMed] [Google Scholar]
- Nichols B. P., van Cleemput M., Yanofsky C. Nucleotide sequence of Escherichia coli trpE. Anthranilate synthetase component I contains no tryptophan residues. J Mol Biol. 1981 Feb 15;146(1):45–54. doi: 10.1016/0022-2836(81)90365-x. [DOI] [PubMed] [Google Scholar]
- Parker J., Johnston T. C., Borgia P. T., Holtz G., Remaut E., Fiers W. Codon usage and mistranslation. In vivo basal level misreading of the MS2 coat protein message. J Biol Chem. 1983 Aug 25;258(16):10007–10012. [PubMed] [Google Scholar]
- Parker J., Johnston T. C., Borgia P. T. Mistranslation in cells infected with the bacteriophage MS2: direct evidence of Lys for Asn substitution. Mol Gen Genet. 1980;180(2):275–281. doi: 10.1007/BF00425839. [DOI] [PubMed] [Google Scholar]
- Parker J., Precup J. Mistranslation during phenylalanine starvation. Mol Gen Genet. 1986 Jul;204(1):70–74. doi: 10.1007/BF00330189. [DOI] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruusala T., Andersson D., Ehrenberg M., Kurland C. G. Hyper-accurate ribosomes inhibit growth. EMBO J. 1984 Nov;3(11):2575–2580. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C. R., Jr, Inagami T., Cohen S. The primary structure of epidermal growth factor. J Biol Chem. 1972 Dec 10;247(23):7612–7621. [PubMed] [Google Scholar]
- Sillero A., Ribeiro J. M. Isoelectric points of proteins: theoretical determination. Anal Biochem. 1989 Jun;179(2):319–325. doi: 10.1016/0003-2697(89)90136-x. [DOI] [PubMed] [Google Scholar]
- Squires C., Konrad B., Kirschbaum J., Carbon J. Three adjacent transfer RNA genes in Escherichia coli. Proc Natl Acad Sci U S A. 1973 Feb;70(2):438–441. doi: 10.1073/pnas.70.2.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G., Ehrenberg M., Kurland C. G. Kinetic suppression of translational errors by (p)ppGpp. Mol Gen Genet. 1982;185(2):269–274. doi: 10.1007/BF00330797. [DOI] [PubMed] [Google Scholar]
- Wagner E. G., Kurland C. G. Translational accuracy enhanced in vitro by (p)ppGpp. Mol Gen Genet. 1980;180(1):139–145. doi: 10.1007/BF00267363. [DOI] [PubMed] [Google Scholar]