Abstract
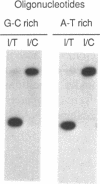
Double-stranded synthetic oligonucleotides with a centrally located dIMP residue in a 5'-32P-labeled strand were employed as substrates for hypoxanthine-DNA glycosylase. The enzyme activity was monitored by the generation of a piperidine-sensitive site in the labeled oligonucleotide. The enzyme was purified approximately 5000-fold from calf thymus. The purified enzyme removed efficiently a hypoxanthine base residue from an I.T base pair, but 15-20 times more slowly from an I.C base pair. Similar results were obtained with oligonucleotides in which the deoxyinosine residue was placed in different surrounding nucleotide sequences. The enzyme had no detectable activity on mismatched G.T, A.G or A.C base pairs. The data indicate that hypoxanthine-DNA glycosylase participates in the repair of deaminated adenine residues in DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Au K. G., Clark S., Miller J. H., Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Lederer F., Gouyette A., Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990 Mar 5;265(7):3916–3922. [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Brynolf K., Eliasson R., Reichard P. Formation of Okazaki fragments in polyoma DNA synthesis caused by misincorporation of uracil. Cell. 1978 Mar;13(3):573–580. doi: 10.1016/0092-8674(78)90330-6. [DOI] [PubMed] [Google Scholar]
- Dehazya P., Sirover M. A. Regulation of hypoxanthine DNA glycosylase in normal human and Bloom's syndrome fibroblasts. Cancer Res. 1986 Aug;46(8):3756–3761. [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980 Oct 9;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Frederico L. A., Kunkel T. A., Shaw B. R. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990 Mar 13;29(10):2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- Harosh I., Sperling J. Hypoxanthine-DNA glycosylase from Escherichia coli. Partial purification and properties. J Biol Chem. 1988 Mar 5;263(7):3328–3334. [PubMed] [Google Scholar]
- Hill-Perkins M., Jones M. D., Karran P. Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat Res. 1986 Sep;162(2):153–163. doi: 10.1016/0027-5107(86)90081-3. [DOI] [PubMed] [Google Scholar]
- Kammen H. O., Spengler S. J. The biosynthesis of inosinic acid in transfer RNA. Biochim Biophys Acta. 1970 Aug 8;213(2):352–364. doi: 10.1016/0005-2787(70)90043-2. [DOI] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T. Enzymatic excision of free hypoxanthine from polydeoxynucleotides and DNA containing deoxyinosine monophosphate residues. J Biol Chem. 1978 Sep 10;253(17):5877–5879. [PubMed] [Google Scholar]
- Karran P., Lindahl T. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry. 1980 Dec 23;19(26):6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Myrnes B., Guddal P. H., Krokan H. Metabolism of dITP in HeLa cell extracts, incorporation into DNA by isolated nuclei and release of hypoxanthine from DNA by a hypoxanthine-DNA glycosylase activity. Nucleic Acids Res. 1982 Jun 25;10(12):3693–3701. doi: 10.1093/nar/10.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka E., Matsuki S., Ikehara M., Takahashi Y., Matsubara K. An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J Biol Chem. 1985 Mar 10;260(5):2605–2608. [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990 Sep-Nov;236(2-3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Manlapaz-Ramos P., Lundquist R., Olivera B. M. Formation of Okazaki pieces at the Escherichia coli replication fork in vitro. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):231–237. doi: 10.1101/sqb.1979.043.01.028. [DOI] [PubMed] [Google Scholar]
- Tomkinson A. E., Lasko D. D., Daly G., Lindahl T. Mammalian DNA ligases. Catalytic domain and size of DNA ligase I. J Biol Chem. 1990 Jul 25;265(21):12611–12617. [PubMed] [Google Scholar]
- Tye B. K., Chien J., Lehman I. R., Duncan B. K., Warner H. R. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1978 Jan;75(1):233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner H. R., Duncan B. K. In vivo synthesis and properties of uracil-containing DNA. Nature. 1978 Mar 2;272(5648):32–34. doi: 10.1038/272032a0. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]