Abstract
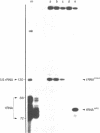
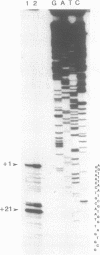
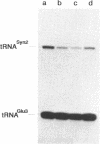
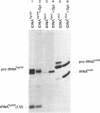
To monitor in vivo transcription and chromatin structure of yeast tRNA genes, we constructed a synthetic tRNA gene that can be used as a reporter. Constructs in which this synthetic tRNA gene is combined with different flanking regions can be integrated into the genome as single copies. The artificial tRNA gene is tagged by the insertion of an intron-like sequence that cannot be spliced out from the precursor and transcripts can thus be identified and quantitated. By several criteria, the artificial tRNA gene behaves like a resident tRNA gene. By measuring the accessibility towards DNaseI in chromatin, we found that the artificial tRNA gene exhibits the same characteristic pattern as resident tRNA genes. Three DNaseI-sensitive sites across the transcribed part of the gene and the immediate flanking regions reflect the formation of the stable transcription complex; positioned nucleosomes are observed in the upstream flanking region. We are confident that the system we have established will prove useful for studying regulatory aspects of tRNA gene expression as well as aspects of pre-tRNA processing and splicing.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi D. G., Margarit I., Tocchini-Valentini G. P. Structural alterations in mutant precursors of the yeast tRNALeu3 gene which behave as defective substrates for a highly purified splicing endoribonuclease. EMBO J. 1985 Dec 1;4(12):3289–3297. doi: 10.1002/j.1460-2075.1985.tb04079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt B., Feldmann H. Characterization of precursors to tRNA in yeast. FEBS Lett. 1973 Dec 1;37(2):129–133. doi: 10.1016/0014-5793(73)80441-7. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Chambers J., Raymond G. J., Kim D., Raymond K. C., Nelson C., Clark S., Johnson J. D. Splice junction mutations in a yeast tRNA gene which alter the rate and precision of processing. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):156–164. doi: 10.1016/0167-4781(90)90051-3. [DOI] [PubMed] [Google Scholar]
- DeLotto R., Schedl P. Internal promoter elements of transfer RNA genes are preferentially exposed in chromatin. J Mol Biol. 1984 Nov 15;179(4):607–628. doi: 10.1016/0022-2836(84)90158-x. [DOI] [PubMed] [Google Scholar]
- Eigel A., Feldmann H. Ty1 and delta elements occur adjacent to several tRNA genes in yeast. EMBO J. 1982;1(10):1245–1250. doi: 10.1002/j.1460-2075.1982.tb00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Coppo A., Fruscoloni P., Benedetti P., Di Segni G., Tocchini-Valentini G. P. Comparative mutational analysis of wild-type and stretched tRNA3(Leu) gene promoters. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8763–8767. doi: 10.1073/pnas.84.24.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fascher K. D., Schmitz J., Hörz W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae. EMBO J. 1990 Aug;9(8):2523–2528. doi: 10.1002/j.1460-2075.1990.tb07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gafner J., Robertis E. M., Philippsen P. Delta sequences in the 5' non-coding region of yeast tRNA genes. EMBO J. 1983;2(4):583–591. doi: 10.1002/j.1460-2075.1983.tb01467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Greer C. L., Söll D., Willis I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jan;7(1):76–84. doi: 10.1128/mcb.7.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Nelböck P., Pilz U., Feldmann H. Enhancer-like stimulation of yeast tRNA gene expression by a defined region of the Ty element micro-injected into Xenopus oocytes. Biol Chem Hoppe Seyler. 1986 Nov;367(11):1141–1146. doi: 10.1515/bchm3.1986.367.2.1141. [DOI] [PubMed] [Google Scholar]
- Hauber J., Stucka R., Krieg R., Feldmann H. Analysis of yeast chromosomal regions carrying members of the glutamate tRNA gene family: various transposable elements are associated with them. Nucleic Acids Res. 1988 Nov 25;16(22):10623–10634. doi: 10.1093/nar/16.22.10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5'-flanking region. Mol Cell Biol. 1989 Aug;9(8):3244–3252. doi: 10.1128/mcb.9.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Evans C. F., Engelke D. R. Comparison of tRNA gene transcription complexes formed in vitro and in nuclei. Mol Cell Biol. 1987 Sep;7(9):3212–3220. doi: 10.1128/mcb.7.9.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Raymond G. J., Clark S. D., Vranka J. A., Johnson J. D. Yeast tRNATrp genes with anticodons corresponding to UAA and UGA nonsense codons. Nucleic Acids Res. 1990 Jul 25;18(14):4215–4221. doi: 10.1093/nar/18.14.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey P. T., Sandmeyer S. B. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991 Mar 25;19(6):1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Irie T., Yoshida M., Takeishi K., Ukita T. The primary structure of yeast glutamic acid tRNA specific to the GAA codon. Biochim Biophys Acta. 1974 Oct 11;366(2):168–181. doi: 10.1016/0005-2787(74)90331-1. [DOI] [PubMed] [Google Scholar]
- Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae. Secondary and tertiary structures of the substrates. J Biol Chem. 1985 Mar 10;260(5):3108–3115. [PubMed] [Google Scholar]
- Lochmüller H., Stucka R., Feldmann H. A hot-spot for transposition of various Ty elements on chromosome V in Saccharomyces cerevisiae. Curr Genet. 1989 Oct;16(4):247–252. doi: 10.1007/BF00422110. [DOI] [PubMed] [Google Scholar]
- Mannhaupt G., Stucka R., Pilz U., Schwarzlose C., Feldmann H. Characterization of the prephenate dehydrogenase-encoding gene, TYR1, from Saccharomyces cerevisiae. Gene. 1989 Dec 28;85(2):303–311. doi: 10.1016/0378-1119(89)90422-8. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Nelböck P., Stucka R., Feldmann H. Different patterns of transposable elements in the vicinity of tRNA genes in yeast: a possible clue to transcriptional modulation. Biol Chem Hoppe Seyler. 1985 Nov;366(11):1041–1051. doi: 10.1515/bchm3.1985.366.2.1041. [DOI] [PubMed] [Google Scholar]
- O'Connor J. P., Peebles C. L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jan;11(1):425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri L., Philippsen P. Centromeric DNA from chromosome VI in Saccharomyces cerevisiae strains. EMBO J. 1982;1(12):1605–1611. doi: 10.1002/j.1460-2075.1982.tb01362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K. C., Raymond G. J., Johnson J. D. In vivo modulation of yeast tRNA gene expression by 5'-flanking sequences. EMBO J. 1985 Oct;4(10):2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. A synthetic substrate for tRNA splicing. Anal Biochem. 1987 Oct;166(1):90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988 Nov 18;55(4):719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucka R., Hauber J., Feldmann H. One member of the tRNA(Glu) gene family in yeast codes for a minor GAGtRNA(Glu) species and is associated with several short transposable elements. Curr Genet. 1987;12(5):323–328. doi: 10.1007/BF00405754. [DOI] [PubMed] [Google Scholar]
- Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990 Feb;6(2):52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- Warmington J. R., Green R. P., Newlon C. S., Oliver S. G. Polymorphisms on the right arm of yeast chromosome III associated with Ty transposition and recombination events. Nucleic Acids Res. 1987 Nov 11;15(21):8963–8982. doi: 10.1093/nar/15.21.8963. [DOI] [PMC free article] [PubMed] [Google Scholar]