Background: DOT1L is responsible for methylation of histone H3K79.
Results: DOT1L deficiency leads to senescence in lung cancer cells.
Conclusion: DOT1L is required for the proper proliferation of cancer cells.
Significance: The inhibition of DOT1L activity might act as a barrier to tumorigenesis.
Keywords: Cancer, Cell Cycle, Cell Proliferation, Histone Methylation, Senescence, DOT1L, H3K79
Abstract
Dot1-like protein (DOT1L) is an evolutionarily conserved histone methyltransferase that methylates lysine 79 of histone H3 (H3K79). Mammalian DOT1L participates in the regulation of transcription, development, erythropoiesis, differentiation, and proliferation of normal cells. However, the role of DOT1L in cancer cell proliferation has not been fully elucidated. DOT1L siRNA-transfected A549 or NCI-H1299 lung cancer cells displayed a nonproliferating multinucleated phenotype. DOT1L-deficient cells also showed abnormal mitotic spindle formation and centrosome number, suggesting that DOT1L deficiency leads to chromosomal missegregation. This chromosomal instability in DOT1L-deficient cells led to cell cycle arrest at the G1 phase and induced senescence as determined by enhanced activity of senescence-associated β-galactosidase activity. Meanwhile, overexpression of a catalytically active DOT1L, not an inactive mutant, restored DOT1L siRNA-induced phenotypes. Overall, these data imply that down-regulation of DOT1L-mediated H3K79 methylation disturbs proliferation of human cells. In addition, although H3K79 methylation is down-regulated in aged tissues, it is up-regulated in lung cancer cell lines and tumor tissues of lung cancer patients. Therefore, H3K79 methylation is a critical histone modification that regulates cell proliferation and would be a novel histone mark for aging and cancer.
Introduction
Histone modifications such as acetylation, methylation, phosphorylation, and ubiquitination have emerged as important mechanisms in the alteration of chromatin structure and the reprogramming of gene expression (1). Epigenetic changes consequently control disease development as well as normal physiological processes, and specific modifications have been used as epigenetic makers for several diseases including cancers (2, 3). Although most histone modifications occur at the histone tail, certain modifications such as the methylation of histone H3 lysine 79 (H3K79)3 occur within the globular domain (4). The enzyme responsible for mono-, di-, and trimethylation of H3K79 is a Dot1/KMT4-lacking the SET domain, which is a highly conserved motif among most histone methyltransferases (5, 6).
Dot1 was originally identified as a disruptor of telomeric silencing in Saccharomyces cerevisiae, and its orthologs are evolutionarily conserved from yeast to mammals (7, 8). Dot1 deletion or overexpression reduces silencing at the telomere, silent mating-type loci, and ribosomal DNA loci (4, 9). Beyond the repetitive regions, Dot1 is also known to regulate heterochromatin formation in the euchromatic region (10). The expression of the epithelial Na+ channel is also repressed by the Dot1a (murine DOT1 alternative splicing variant a)-AF9 complex (11, 12).
In contrast to transcriptional silencing, Dot1 and mammalian DOT1L (Dot1-like protein) participate in transcriptional activation. Genome-wide analysis reveals that H3K79 methylation is detected in actively transcribed genes (13–16). DOT1L is purified in a transcriptional elongation complex (17–20). Furthermore, during leukemogenesis, MLL fusion proteins such as MLL-AF4, MLL-AF10, and MLL-ENL recruit DOT1L to mediate methylation at H3K79 and actively transcribe hematopoietic genes (21, 22).
In addition to their role in transcription, Dot1 and DOT1L are involved in the control of cell cycle through H3K79 methylation. Budding yeast Dot1 has been shown to function in the regulation of the meiotic checkpoint (23) and the DNA damage checkpoint (24, 25). Trypanosoma brucei DOT1A and DOT1B, which mediate di- and trimethylation of H3K76, a homologue of H3K79 in other organisms, are required for normal proliferation and differentiation, respectively (26). Mammalian DOT1L is also known to control proliferation and differentiation in embryonic stem (ES) cells. DOT1L deficiency causes the accumulation of hyperploid cells and defects in differentiation (27). Moreover, it has been reported that Grappa, the fly ortholog of Dot1, and DOT1L regulate embryogenesis in Drosophila and mice, respectively (28–31).
Overall, DOT1L-mediated H3K79 methylation regulates normal cell cycle progression in yeast, trypanosome, and mouse. However, although the level of H3K79 methylation is dynamically changed depending on cell cycle phase (7), the effect of H3K79 methylation on the regulation of the cell cycle in human cells has not been deciphered. In addition, with the exception of hematomalignancy caused by MLL fusion proteins, it is not known whether methylation of H3K79 is able to control cell proliferation in other cancers. The present study suggests that the down-regulation of H3K79 methylation, which is highly modified in lung cancer cells, would be a novel approach to eliminating cancer cells via a senescence program.
EXPERIMENTAL PROCEDURES
Cell Culture and Synchronization
Non-small cell lung cancer (NSCLC) cells such as A549 and NCI-H1299 were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C in 5% (v/v) CO2. IMR-90 and WI-38 normal lung fibroblasts were maintained in Eagle's minimum essential medium with 10% fetal bovine serum. A549 cells were arrested at prometaphase by treatment with 0.2 μg/ml nocodazole, a microtubule depolymerizer for 16 h. By washing off the nocodazole with PBS, the cells were incubated in the medium to stimulate entry into the next cell cycle. G1/S synchronization was achieved by a double thymidine block. In brief, A549 cells were cultured in the presence of 2 mm thymidine for 19 h and then released to grow for 10 h. Cells were then treated for another 15 h with 2 mm thymidine, causing the cells to arrest at the G1/S boundary. The arrested cells were allowed to enter the S phase by washing off the thymidine with PBS.
Small Interfering RNA (siRNA) Transfection
Control and DOT1L siRNA were synthesized by Dharmacon, Inc. (Lafayette, CO) and ST Pharm. Co., Ltd. (Seoul, Korea). The siRNA duplexes were as follows: control siRNA sense strand, AUGUAUUGGCCUGUAUUAG; and DOT1L (NM_032482) siRNA sense strands 1, GCUAUGGAGAAUUACGUUU; 2, AAGAGUGGAGGGAGCGAAU; 3, UCACUAUGGCGUCGAGAAA; 4, GCAGAAUCGUGUCCUCGAA; 5, GCUGGAGCUGAGACUGAAG; and 6, GCAUGCAGAAUACACAUUG. Two DOT1L siRNA duplexes that target different regions were mixed to knock down DOT1L expression (a = 1 + 2; b = 3 + 4; c = 5 + 6). Transfection was performed with 20 nm siRNA using Lipofectamine RNAiMax according to the manufacturer's instructions (Invitrogen). All experiments were performed at 72 h after transfection.
Retrovirus Infection
The pMSCB-HA-DOT1L retroviral construct was a kind gift from Drs. Y. Zhang and Y. Okada (University of North Carolina). The siRNA-resistant construct containing silent mutations in the middle of the DOT1L siRNA 5 and 6 binding regions was created by site-directed mutagenesis. The siRNA-resistant DOT1L catalytic inactive mutant was then generated by the introduction of a missense mutation (G163R/S164C/G165R) in the S-adenosyl-l-methionine-binding domain (22). The retrovirus produced from BOSC23 packaging cells was infected in A549 cells in the presence of 8 μg/ml Polybrene.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
The mRNA levels of DOT1L and cell cycle-related genes were detected by RT-PCR. Total RNA was isolated using TRIzol reagent (Invitrogen), and cDNA was synthesized by PrimeScriptTM reverse transcriptase (Takara Bio Inc.). The primer sequences used for PCR are shown in supplemental Table S1. Synthesized cDNA was amplified, and the PCR product was then visualized on 1% agarose gel.
Western Blotting
Frozen lung tissues from Sprague-Dawley rats were kindly provided by the Aging Tissue Bank (Pusan National University, Korea). Tissue samples of lung cancer patients were obtained from the Korea Lung Tissue Bank, assigned and supported by the Korea Science and Engineering Foundation in the Ministry of Science and Technology. The utilization of the patient's specimens for this research was authorized by the Institutional Review Board of Kyung Hee University (KHU IRB 2010-003) and Korea University Guro Hospital (KU Guro Gene Bank 2010-005 and 2011-008). The frozen tissues were homogenized and lysed with a pestle in NETN buffer (100 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl, pH 8.0, 0.5% Nonidet P-40) containing 50 mm β-glycerophosphate, 10 mm NaF, and a protease inhibitor mixture (Calbiochem) for 10 min on ice. Normal lung fibroblasts and lung carcinoma cells were also lysed in the buffer described above. After centrifugation, the supernatant was saved as the crude cell extract, and the pellet was incubated with 0.2 n HC1 for 20 min on ice. After centrifugation, the HCl-soluble fraction was used as the chromatin fraction to detect the histones. The antibodies used for Western blotting were as follows: anti-H3K79me2 (Upstate 07-366), H3K79me3 (Abcam ab2621), GAPDH (Santa Cruz Biotechnology, Inc., sc-25778), β-actin (Cell Signaling 4970), cyclin D1 (Santa Cruz Biotechnology sc-8396), cyclin E (Santa Cruz Biotechnology sc-481), cyclin A (Santa Cruz Biotechnology sc-751), cyclin B (Santa Cruz Biotechnology sc-245), p21 (Calbiochem OP64), p18 (Santa Cruz Biotechnology sc-9965), Rb (Santa Cruz Biotechnology sc-50), phospho-Ser-807/811-Rb (pRb) (Cell Signaling 9308), and phospho-Thr-160-CDK2 (Cell Signaling 2561). Anti-histone H2AX phospho-Ser-139 (γH2AX) and anti-histone H3 phospho-Ser-10 (pH3) antibodies were generated as described previously (32).
Fluorescence-activated Cell Sorting (FACS)
Cells were fixed with 70% ethanol and then permeabilized with 0.25% Triton X-100. The cells were incubated with anti-histone H3 phospho-Ser-10 (pH3) antibody and then incubated with FITC-conjugated goat anti-rabbit antibody in PBS with 1% BSA. After staining, cells were treated with DNase-free RNase A, and the DNA content was determined by propidium iodide staining. The percentage of cells in each cell cycle phase and the pH3-positive cells were determined by FACS.
Immunocytochemical Fluorescent Staining
Cells grown on coverslips were fixed with cold methanol at −20 °C for 5 min and then rehydrated in PBS three times for 5 min. The cells were post-fixed with 3% paraformaldehyde at room temperature for 10 min followed by permeabilization with 0.5% Triton X-100 at room temperature for 5 min. The cells were incubated with rabbit polyclonal antibody against α-tubulin (Abcam ab18251) and mouse monoclonal antibody against γ-tubulin (Abcam ab11316) at 37 °C for 20 min. After washing with PBS, the cells were incubated with FITC-conjugated goat anti-rabbit IgG and rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) at 37 °C for 20 min. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Coverslips with stained cells were mounted with antifade solution.
5-Bromo-2′-deoxyuridine (BrdU) Pulse Labeling
Cells grown on coverslips were incubated with 30 μm BrdU (Sigma) for incorporation into the DNA for 30 min at 37 °C in a 5% (v/v) CO2 incubator. After fixation with 3% paraformaldehyde and permeabilization with 0.5% Triton X-100, the DNA was denatured by immersion with 2 n HCl for 10 min at 37 °C. After washing with PBS, the cells were incubated with mouse monoclonal anti-BrdU antibody (BD Pharmingen 555627). Cells were then incubated sequentially with rhodamine-conjugated goat anti-mouse IgG and DAPI.
Preparation of Metaphase Spread
Cells were treated with 25 ng/ml colcemid (Invitrogen) for 6 h at 37 °C in a 5% (v/v) CO2 incubator. The harvested cells were exposed to hypotonic shock with 0.075 m KCl at 37 °C for 30 min. After preparation of the metaphase spread, the cells were fixed by multiple changes of Carnoy's fixative (3:1 methanol:acetic acid). The fixed metaphase spread was dropped onto slides and then soaked in Giemsa staining solution. The number of chromosomes was scored under the microscope.
Senescence-associated β-Galactosidase Staining
The cells were washed with PBS and then fixed and stained at pH 6.0 using a senescence β-galactosidase (SA-β-gal) staining kit (Cell Signaling 9860).
RESULTS
H3K79 Methylation Is a Cell Cycle-regulated Histone Modification
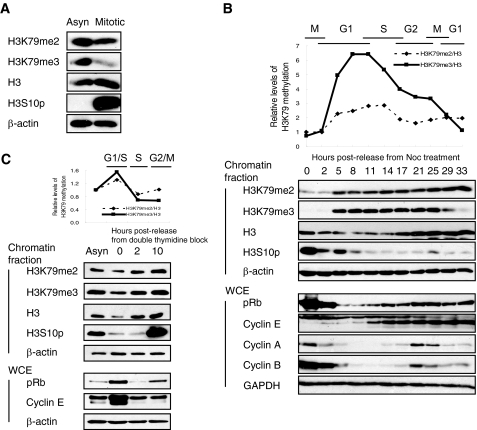
First, the level of H3K79 methylation was examined during cell cycle progression in A549 lung adenocarcinoma cells. Both H3K79me2 and H3K79me3 were decreased in the nocodazole-induced mitotic phase, implying that H3K79 methylation is a cell cycle-dependent modification (Fig. 1A). After cells were released from mitosis, the level of H3K79me2 and H3K79me3 began to increase in mid-G1 phase and peaked at the G1/S boundary (Fig. 1B). H3K79me3 levels then declined dramatically until cells reached the G1 phase of the next cell cycle. In addition, cells synchronized at the G1/S boundary using double thymidine block showed higher levels of H3K79 methylation (Fig. 1C). Overall, these findings suggest that H3K79 methylation plays a role in cell cycle regulation and is possibly required for the G1/S transition.
FIGURE 1.
Di- and trimethyl H3K79 are regulated during cell cycle progression. Asynchronous (Asyn) A549 cells were arrested at the mitotic phase with nocodazole (Noc) (A) and released to enter the next cell cycle (B). C, A549 cells were arrested at the G1/S boundary by double thymidine block and then released to S phase. B and C, the cell cycle phase was determined by the level of pH3, pRb, and cyclins including cyclins E, A, and B. The level of H3K79 methylation, normalized to total H3, is shown during cell cycle progression in the upper panel. WCE, whole cell extract.
DOT1L Deficiency Inhibits Cell Proliferation
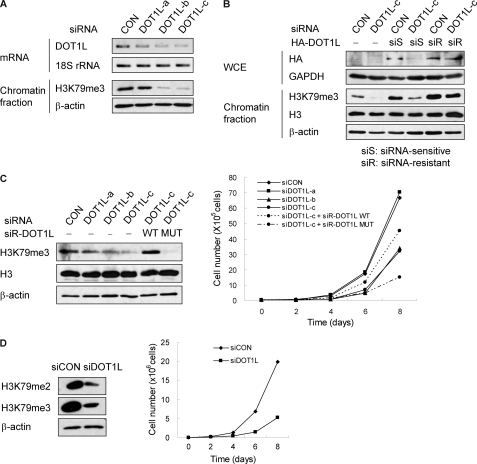
The mechanisms underlying the regulation of cell cycle progression by H3K79 methylation in human cells are unknown. To investigate the role of DOT1L-mediated H3K79 methylation in cell proliferation, A549 or NCI-H1299 human lung cancer cells were depleted of DOT1L by siRNA. The knockdown efficiency of several siRNA combinations was verified by RT-PCR for DOT1L and by Western blotting for H3K79me3.
siRNA-c resulted in the most significant reduction of DOT1L and H3K79me3 (Fig. 2A), although this reduction was rescued by overexpression of siRNA-c-resistant DOT1L but not by siRNA-c-sensitive DOT1L (Fig. 2B). In addition, the transduction of catalytically active (WT) and inactive (MUT) siRNA-resistant DOT1L was compared in DOT1L siRNA-transfected cells to determine whether DOT1L siRNA-induced phenotypes are indeed influenced by the activity of histone methyltransferase. DOT1L-deficient A549 cells showed a substantial reduction in cell proliferation, whereas siRNA-resistant DOT1L-WT-reconstituted cells, but not DOT1L-MUT, were resistant to slow proliferation (Fig. 2C). DOT1L deficiency resulted in similar growth retardation in NCI-H1299 cells (Fig. 2D). Overall, these results indicate that the methyltransferase activity of DOT1L is required to maintain the normal rate of cell proliferation.
FIGURE 2.
DOT1L-deficient cells show slow proliferation. A–C, A549 cells were transfected with control (siCON) or DOT1L siRNA (siDOT1L) in the absence or presence of siRNA-resistant DOT1L. A, after 72 h of transfection of different DOT1L siRNAs, the levels of DOT1L and di- and trimethyl H3K79 in the chromatin fraction were determined by RT-PCR and Western blotting, respectively. B, siRNA-c-sensitive (S) or -resistant (R) HA-DOT1L was reconstituted in endogenous DOT1L-deficient cells. WCE, whole cell extract. C, DOT1L siRNA-c was transfected after infection of the catalytically active (WT) and inactive (MUT) siRNA-c-resistant DOT1L. After 48 h of transfection, the total cell number was counted every other day using a hemocytometer. D, NCI-H1299 cells were transfected with control or DOT1L siRNA, and the total cell number was counted.
DOT1L Deficiency Leads to Abnormal Cell Cycle Progression in Lung Cancer Cells
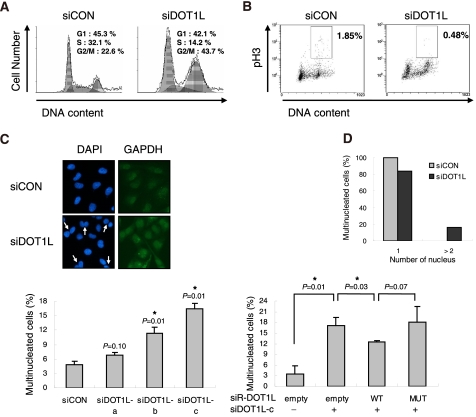
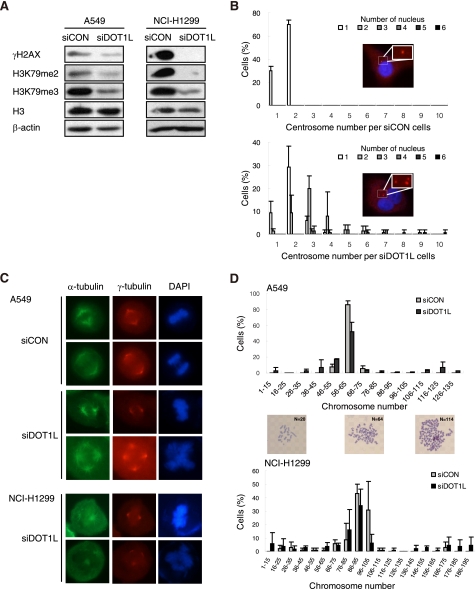
To examine how H3K79 hypomethylation inhibits cell proliferation rates, cell cycle distribution was determined by FACS. DOT1L-deficient A549 cells showed a significant accumulation of cells with a 4N DNA content and an apparent reduction in the S-phase population (Fig. 3A). To determine the identity of cells with a 4N DNA content as a G2/M-arrested population, the level of pH3 (histone H3 phospho-Ser-10), which is up-regulated at the late G2 and M phases, was evaluated. DOT1L siRNA-transfected A549 cells displayed a dramatic reduction in pH3 staining, suggesting that the accumulation of 4N DNA-containing cells induced by DOT1L deficiency was not due to G2/M arrest (Fig. 3B). Instead, immunofluorescent staining revealed that a multinucleated cell population with more than two nuclei was significantly enhanced in DOT1L-deficient A549 cells; this condition was restored by DOT1L-WT but not by catalytically inactive MUT (Fig. 3C). DOT1L-deficient NCI-H1299 cells also showed multinucleation (Fig. 3D). These results indicate that the increase in 4N DNA content, shown in Fig. 3A, results from multinucleation induced by the absence of DOT1L methyltransferase activity. Overall, the results demonstrate that DOT1L-mediated methylation of H3K79 is required for faithful cell cycle progression.
FIGURE 3.
DOT1L deficiency results in a multinucleated phenotype. A—C, A549 cells were transfected with control (siCON) or DOT1L siRNA (siDOT1L) in the absence or presence of siRNA-resistant DOT1L WT or MUT. Cell cycle distribution (A) and pH3 (B) were determined by FACS in DOT1L-deficient cells. C, the nucleus of the cells and the cell boundary were visualized by staining with DAPI and GAPDH, respectively. Multinucleated cells were counted. All bars represent mean ± S.D. (n ≥ 3). *, statistically significant differences (p < 0.05). D, multinucleated cells were counted in NCI-H1299 cells after transfection with DOT1L siRNA.
Cell Proliferation Is Inhibited in DOT1L-deficient Cells
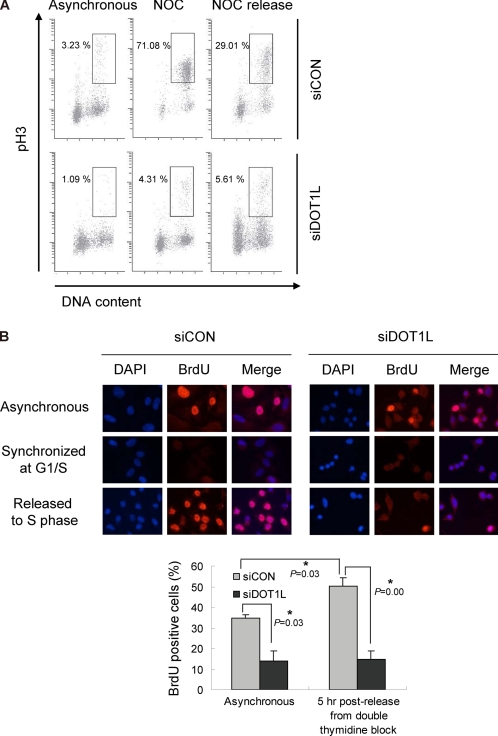
Based on the finding that DOT1L deficiency results in multinucleation, it was of interest to check the proliferative status of DOT1L-deficient cells by immunostaining against a mitotic marker, pH3, and DNA-incorporated BrdU. As observed in Fig. 3B, DOT1L siRNA resulted in decreased pH3 staining in asynchronous cells (Fig. 4A). Although nocodazole-arrested control cells were able to enter proliferating status after nocodazole withdrawal, DOT1L-deficient cells did not respond to nocodazole-induced arrest (Fig. 4A), suggesting that these cells are nonproliferating cells. To confirm that DOT1L-deficient cells did not actively proliferate, BrdU pulse labeling was used to determine the number of cells undergoing DNA replication in S phase. DOT1L-deficient cells showed a dramatic decrease in the uptake of BrdU in unsynchronized cultures, indicating reduced proliferation in the absence of DOT1L (Fig. 4B). These results are consistent with the low number of cells in S phase observed in Fig. 3A. To clarify the inhibition of proliferation induced by depletion of DOT1L, cells were arrested at the G1/S boundary by double thymidine block and then released to enter S phase. However, the depletion of DOT1L showed few BrdU-positive cells even after release from double thymidine block, whereas control siRNA-treated cells progressed normally into S phase (Fig. 4B). These results support the finding that DOT1L-deficient cells are nonproliferating cells.
FIGURE 4.
DOT1L-deficient cells are nonproliferating. A549 cells were transfected with control (siCON) or DOT1L siRNA (siDOT1L). A, cells were exposed to nocodazole (Noc) and then released to the next cell cycle. pH3 was stained and analyzed by FACS. B, siRNA-transfected cells were arrested by double thymidine block followed by their release to progress to the S phase. After BrdU labeling, BrdU-positive cells were detected at each time point by immunofluorescent staining. All bars represent mean ± S.D. (n = 3). *, statistically significant differences (p < 0.05).
DOT1L Deficiency Results in Chromosomal Instability
The next question was as to why the DOT1L-deficient cells showed nonproliferating aneuploidy. The abnormal proliferation might, in part, result from genomic damage or chromosomal missegregation. In comparison with control siRNA-treated cells, the level of γH2AX, a marker of DNA strand break, did not increase in DOT1L-deficient A549 and NCI-H1299 cells, indicating that DOT1L deficiency is not associated with genomic damage (Fig. 5A). As aneuploidy could also be due to abnormal mitotic division, the mitotic spindle and centrosome were examined by staining against α- and γ-tubulin, respectively. The number of cells with more than three centrosomes was significantly higher in DOT1L-depleted A549 cells than in control cells (Fig. 5B). Therefore, the centrosome aberrations might have induced asymmetrical mitotic division. In addition, multipolar spindles were observed in DOT1L-deficient cells, but the mitotic spindle was formed abnormally, which possibly led to aneuploidy with more than 4N DNA content (Fig. 5C). A549 is a hypotriploid human cell line in which the majority of cells contain 64–67 chromosomes. As expected, DOT1L-deficient cells showed a doubling of the chromosome number. However, 12% of the cells had less than 46 chromosomes (Fig. 5D), suggesting that DOT1L deficiency induced asymmetrical cell division. DOT1L-deficient NCI-H1299 cells also confer abnormalities of spindle formation or chromosome number (Fig. 5, C and D). These results indicate that DOT1L is required to prevent perturbed mitosis and enable proper chromosome segregation.
FIGURE 5.
DOT1L-deficient cells show an abnormal formation of centrosome and mitotic spindle. A549 or NCI-H1299 cells were transfected with control (siCON) or DOT1L siRNA (siDOT1L). A, the level of γH2AX in the chromatin fraction was determined by Western blotting. B, the centrosome (red) and nucleus (blue) were stained with γ-tubulin, and DAPI, respectively. The centrosome number per cell was counted in A549 cells. C, α-tubulin and γ-tubulin were stained to visualize the mitotic spindle and spindle pole body, respectively. D, the colcemid-treated metaphase spread was prepared to count the number of chromosomes. All bars represent mean ± S.D. (n = 3).
DOT1L Deficiency Arrests Cells at the G1 Phase of the Cell Cycle by Affecting Cell Cycle Regulators
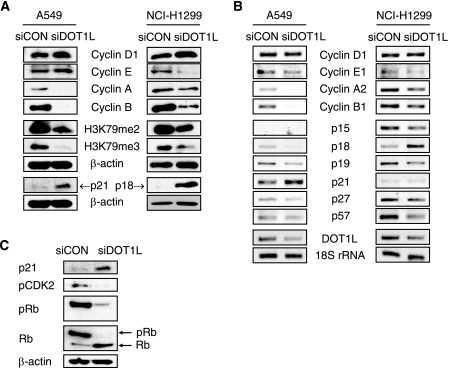
The above results imply that chromosomal instability caused by DOT1L depletion could lead to an inhibition of cell proliferation. The low level of pH3 and BrdU positivity in DOT1L siRNA-treated cells (Fig. 4) suggests that DOT1L deficiency could result in an arrest at the G1 phase of the cell cycle. To assess whether DOT1L depletion led to G1 arrest, the levels of each of the cyclin proteins were determined by Western blotting. The levels of cyclin A and cyclin B, which peak at G2 and M, respectively, were reduced in DOT1L-deficient A549 cells, whereas the levels of cyclin D1 and cyclin E, which rise in early and late G1 phases, respectively, were slightly induced (Fig. 6A). This result implies that cells treated with DOT1L siRNA were arrested at the G1 phase. NCI-H1299 cells also underwent possible G1 arrest as determined by the up-regulation of cyclin D1 and the down-regulation of cyclins E, A, and B (Fig. 6A). As expected, with cyclin D1 being the exception, the mRNA levels of cyclins E1, A2, and B1 were down-regulated by DOT1L siRNA in A549 and NCI-H1299 cells (Fig. 6B), confirming that DOT1L-deficient cells stop proliferating prior to S phase progression. Overall, although the mechanism by which down-regulation of H3K79 methylation leads to G1 arrest remains to be determined, the results suggest that aneuploid cells are arrested at the G1 phase of the cell cycle. Next, to investigate which cyclin-dependent kinase inhibitor (CKI) mediates possible G1 arrest, the mRNA levels of INK4 (p16INK4a, p15INK4b, p18INK4c, and p19INK4d) or CIP/KIP (p21CIP/WAF1, p27KIP1, and p57KIP2) family members were determined. A549 is a p15- and p16-null cell line, whereas NCI-H1299 cells are p53-null and p16-deficient. In A549 cells, DOT1L deficiency up-regulated p21 expression, whereas it down-regulated other CKIs (Fig. 6B and supplemental Fig. S1). On the other hand, p18 was up-regulated by DOT1L deficiency in NCI-H1299 cells, which have a low level of p21 due to the absence of p53 transcriptional factor (Fig. 6B). This indicates that p18 possibly compensates for the role of p21 in G1 arrest in p21-defective NCI-H1299 cells. The enhanced expression of p21 and p18 was confirmed by Western blotting in A549 and H1299 cells, respectively (Fig. 6A). Finally, the transcriptional up-regulation of p21 in DOT1L-deficient A549 cells induced the hypophosphorylation of CDK2 and Rb, which inhibited the progression from G1 to S phase (Fig. 6C). Overall, these results suggest that DOT1L deficiency up-regulates the transcriptional expression of CKIs and then arrest cells at the G1 phase.
FIGURE 6.
DOT1L deficiency leads to the dysregulation of cell cycle regulators and G1 cell cycle arrest. A549 or NCI-H1299 cells were transfected with control (siCON) or DOT1L siRNA (siDOT1L). A, protein levels of each cyclin and cyclin-dependent kinase inhibitor were detected by Western blotting. B, mRNA levels of each cyclin and cyclin-dependent kinase inhibitor were determined by RT-PCR. C, protein levels of p21, pCDK2, and pRb were measured in DOT1L-deficient A549 cells by Western blotting.
DOT1L Deficiency Induces Premature Senescence, and a Low Level of H3K79 Methylation Is a Possible Marker of Senescence
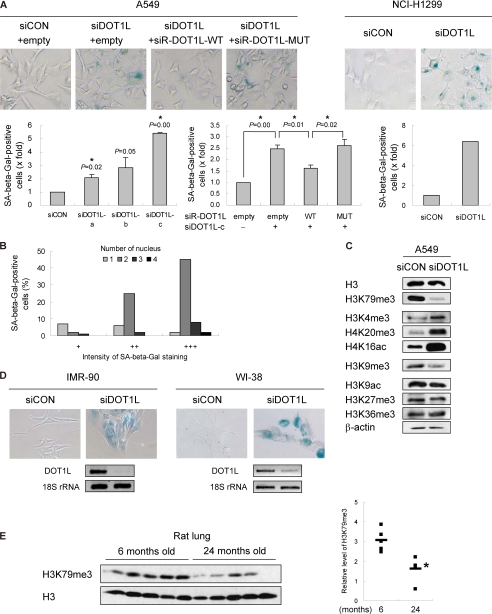
It was of interest to determine whether the nonproliferating status of DOT1L-deficient cells is reversible or irreversible. If DOT1L depletion leads to irreversible cell cycle arrest, this would be an alternative way to inhibit the growth of cancer cells. Irreversible G1 arrest refers to senescence, which is classified into replicative senescence and stress-induced premature senescence (33). Senescent cells display certain changes in morphology, biochemical factors, and chromatin (34). A significant biochemical change in senescent cells is an enhanced activity of SA-β-gal due to the expanded lysosomal compartment. DOT1L-deficient A549 and NCI-H1299 cells showed an enhanced SA-β-gal positivity (Fig. 7A). Even 8 days after siRNA transfection, SA-β-gal staining was still detected in DOT1L-deficient cells (data not shown), suggesting that senescence induced by DOT1L depletion is irreversible. The senescence phenotype was rescued by wild-type DOT1L overexpression, whereas the catalytically inactive DOT1L mutant failed to overcome senescence (Fig. 7A). In addition, cells having more than two nuclei displayed a higher intensity of SA-β-gal staining than mononuclear cells, suggesting that multinucleated cells are apparently senescent (Fig. 7B). Overall, these results indicate that the polyploid cells induced by the depletion of DOT1L constitute a senescence-like phenotype. To define a relationship between histone marks and senescence, other histone modifications were examined in DOT1L-depleted cells (Fig. 7C). Although no significant changes were found in H3K9ac, H3K27me3, and H3K36me3, some changes were observed in specific senescence-related histone residues. DOT1L depletion enhanced the level of H3K4me3, in which demethylase JHDM1B has been shown to inhibit senescence (35). H4K20me3, a substantially increased mark in aged rat liver and Hutchinson-Gilford progeria syndrome (36, 37), was increased by DOT1L deficiency. H4K16ac, which increases with age in yeast and is enhanced upon oncogene-induced senescence (38, 39), was also significantly induced. By contrast, DOT1L-deficient cells showed a decreased level of H3K9me3, which is globally undetectable in Hutchinson-Gilford progeria syndrome and is low in old skin fibroblast (37, 40). Overall, changes in senescence-related histone marks support DOT1L deficiency-induced senescence. However, although low levels of H3K79 methylation may affect histone cross-talk, it needs to be investigated further whether modifications at these residues are a cause or consequence of senescence.
FIGURE 7.
Hypomethylation of H3K79 is associated with senescence. A and B, A549 or NCI-H1299 cells were transfected with control (siCON) or DOT1L siRNA (siDOT1L) in the absence or presence of siRNA-resistant DOT1L. SA-β-gal-positive cells were detected by staining. All bars represent mean ± S.D. (n ≥ 3). *, statistically significant differences (p < 0.05). B, the percentage of SA-β-gal-positive DOT1L-deficient A549 cells was determined by the staining intensity depending on the number of nuclei. C, histone modifications in DOT1L siRNA-transfected A549 cells were examined by indicated antibodies. D, the level of DOT1L in control or DOT1L siRNA-transfected IMR-90 and WI-38 cells was determined by RT-PCR. E, the chromatin fractions isolated from lung tissues of young and old Sprague-Dawley rats were used to detect the levels of H3K79me3 and H3. The relative levels of H3K79 methylation in young and old tissues are shown as a scatter plot in the right panel. The means for each group are shown as a bar. *, statistically significant differences (p < 0.05).
Senescence is also accelerated by DOT1L depletion in IMR-90 and WI-38 normal lung fibroblasts (Fig. 7D). The above results suggest that methylation of H3K79 could play a role in a senescence program. Finally, to determine whether the methylation level of H3K79 is dependent on organismal age, lung tissues isolated from Sprague-Dawley rats ranging in age from 6 to 24 months were used to detect H3K79me3. A low level of H3K79 methylation was detected in aged rats, strongly suggesting H3K79 methylation as an epigenetic mark related to senescence (Fig. 7E). Although the correlation between levels of H3K79me3 and DOT1L protein could not be validated, because of antibody unavailability, the levels of DOT1L mRNA were not different between young and aged tissues (supplemental Fig. S2). Taken together, these results suggest that a low level of H3K79 methylation accelerates senescence.
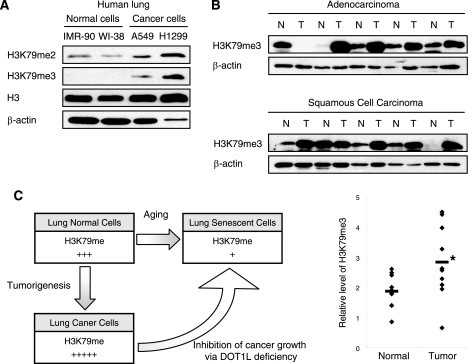
DOT1L-mediated H3K79 Methylation Is Up-regulated in Lung Cancer Cells
Given that DOT1L deficiency induces premature senescence in cancer cells, the down-regulation of H3K79 methylation may be a tumor suppressor mechanism. It implies that up-regulation of H3K79 methylation is a prerequisite for tumor cell proliferation. The levels of di- and trimethylation of H3K79 were compared between normal lung fibroblasts, including IMR-90 and WI-38, and lung carcinoma cells, including A549 and NCI-H1299. Lung cancer cells showed higher levels of both H3K79me2 and H3K79me3 than normal lung fibroblasts (Fig. 8A). The level of H3K79me3 was significantly higher in tumor lung tissues than in normal lung tissues isolated from lung cancer patients with adenocarcinoma or squamous cell carcinoma (Fig. 8B). However, the level of DOT1L mRNA in tumor lung tissues was not significantly different from that of normal lung tissues, implying that the increase in the level of H3K79 methylation in tumors is regulated by DOT1L protein and not mRNA (supplemental Fig. S3). Overall, these results suggest that the up-regulation of methylated H3K79 in cancer cells may be associated with tumorigenesis.
FIGURE 8.
DOT1L-mediated H3K79 methylation is higher in lung cancer. A, the chromatin fraction from normal lung fibroblasts and lung carcinoma cell lines was isolated to determine the level of H3K79 methylation. B, normal (N) and tumor (T) tissues of lung cancer patients were used to determine the protein levels of H3K79me3 and β-actin. The relative levels of H3K79 methylation in normal and tumor tissues are shown as a scatter plot in the right panel. The means for each group are shown as a bar. *, statistically significant differences (p < 0.05). C, diagram showing that DOT1L-mediated H3K79 methylation controls cancer and aging.
DISCUSSION
The present study demonstrates that DOT1L-mediated H3K79 methylation is required for coordinated cell cycle progression in cancer cells. DOT1L deficiency caused multinucleation, abnormal mitotic spindle formation, and increased centrosome number. Finally, DOT1L-deficient cells underwent irreversible G1 arrest, which is defined as premature senescence. Based on evidence that methylation of H3K79 decreases during the normal aging process and is higher in cancer cells than in normal cells, methylated H3K79 would be a novel histone mark for aging and cancer (Fig. 8C).
Although DOT1L is responsible for mono-, di-, and trimethylation of H3K79, several aspects of the different roles of each methylation status have been suggested. H3K79me2 is distributed diffusely in nucleus, whereas H3K79me3 is localized to pericentric heterochromatin in oocytes (41), suggesting the role of H3K79me2 and H3K79me3 as a transcriptionally active marker and a repressive marker, respectively. Furthermore, whereas H3K79me3 decorates only open the reading frame (ORF), H3K79me2 is enriched in the intergenic region as well as the ORF in S. cerevisiae (42). In addition, the level of H3K79me2 is lowest in the G1 phase and peaks at the G2/M transition, whereas the level of H3K79me3 is constant during the cell cycle in S. cerevisiae and T. brucei (26, 42). By contrast, the present study, as well as the previous report (7), shows that in higher eukaryotes, H3K79me2 is maintained at the highest level in late G1 phase, decreases in S phase, and is low in the G2/M phase. Although the pattern of H3K79 methylation during the cell cycle differs between lower and higher eukaryotes, these results suggest that H3K79me2 and H3K79me3 act as differential marks for the regulation of transcription and cell cycling.
The cell cycle progression controlled by DOT1L could be explained by H3K79 methylation-mediated expression of several cell cycle-regulatory genes. H3K79me2 is enriched in the open reading frames and promoters of G1/S-regulated genes in yeast (42). In addition, a subset of genes related to cell growth and cell cycle has also been reported to be up- or down-regulated in DOT1L-deficient ES cells (27). As in the differential transcriptome pattern in DOT1L-deleted yeast and ES cells, the cancer cells treated with DOT1L siRNA might undergo transcriptional changes in the genes related to cell cycle regulation. It is therefore important to identify the specific genes that are regulated transcriptionally in a DOT1L-dependent manner, leading to senescence in DOT1L-deficient cancer cells. We have demonstrated that CKIs, which are known tumor suppressors, were up-regulated by DOT1L depletion. Although p21-induced senescence is a well known pathway (43), p18-dependent senescence is yet to be defined (44). Interestingly, DOT1L depletion is reported to lead to G1 cell cycle arrest via up-regulation of p16 in MLL-AF9-transformed cells (45). Therefore, the mechanism by which the hypomethylation of H3K79 induces the expression of specific CKIs in a certain context remains to be elucidated.
In addition to cell cycle-regulatory genes, the other tumor suppressor genes are also regulated by DOT1L in cancer cells. DOT1L is required for the reactivation of tumor suppressor genes such as HOXA9 and RASSF1A when their CpG islands are unmethylated (46). In contrast to the DOT1L-dependent tumor suppression suggested by Jacinto et al. (46), the present data suggest that DOT1L down-regulation would be an alternative method of tumor suppression mediated by the induction of CKIs. Although the design of cancer therapeutics based on DOT1L regulation would be complex because of the specificity of genes that are targeted transcriptionally by DOT1L, our study suggests that epigenetic modulation of H3K79 methylation might be useful in controlling tumor growth.
As with the phenotypes observed in DOT1L-deficient cancer cells in the present study, previous findings have also reported abnormal mitotic spindles with misalignment and tetraploidy in murine DOT1L-deficient ES cells. This abnormal mitotic regulation induced by DOT1L deficiency consequently results in mitotic arrest (27, 29). However, although DOT1L-deficient cancer cells showed similar phenotypes such as multipolar mitotic spindles and the resulting aneuploidy, these cells showed irreversible arrest at G1 phase rather than at the mitotic phase, suggesting that DOT1L-deficient cancer cells bypassed the spindle assembly checkpoint. One possible cause of aberrant microtubule attachment could be a loss of H3K79me2 at the centromeric or pericentric region. Although H3K79me2 is found in the transcriptionally active region (15, 47), it is also enriched in centromeric heterochromatin as well as in the telomeric region in embryonic stem cells (29). This implies that improper methylation at H3K79 results in a failure of kinetochore-microtuble attachment. Finally, chromosomal missegregation, associated with the cytokinesis failure, would result in aneuploidy.
The nonproliferating aneuploidy induced by DOT1L deficiency leads to cellular senescence, which is generally characterized by focal histone H3 lysine 9 trimethylation together with HP1 recruitment in senescence-associated heterochromatin foci (48, 49). However, senescence-associated heterochromatin foci and focal H3K9me3 were not observed in DOT1L-deficient A549 cancer cells (data not shown). Murine DOT1L-deficient ES cells have already been shown to possess a decreased level of H4K20me3 and H3K9me2, which are markers for heterochromatin, at the centromere and telomere, suggesting that DOT1L is required for heterochromatin formation (29). In addition, DOT1L-deficient mouse ES cells show telomere elongation. Therefore, the senescence induced by DOT1L deficiency is not due to heterochromatin formation and telomere shortening but possibly is caused by chromatin changes affecting the expression of the G1/S transition-regulatory genes such as p21 and p18 in lung cancer cells.
Another possible explanation for DOT1L-regulated senescence is that DOT1L does not directly regulate the expression of target genes but, rather, acts indirectly by mediating the cross-talk between different histone modifications. Cross-talk between histone modifications related to DOT1L-mediated H3K79 methylation has been reported in yeast and human. The di- and trimethylation of H3K79 is dependent on the monoubiquitination of H2BK120 and H2BK34 (50–52) and on the interaction between Dot1 and histone H4 in yeast (53, 54). However, the effect of H3K79 methylation on histone modification at other residues is still unknown. As shown in the present study, various histone modifications affected by DOT1L depletion could be involved in either transcriptional activation or repression. Although the intricate network of histone modifications that coordinate gene expression has been studied, the effect of DOT1L deficiency on gene regulation through cross-talk with other histone modifications remains to be determined in cancer cells.
Several epigenetic modifications are interconnected with aging and aging-related diseases (55). For instance, H4K20 methylation increases during aging, whereas it decreases in cancer (56). The present study also shows that H3K79 methylation is one of the histone modifications that change dynamically in aged and tumor tissues. However, in contrast to our finding that H3K79me3 was down-regulated in an aged lung tissue, it has been reported previously that H3K79me1 and H3K79me2 are up-regulated in the brain tissue of senescence-accelerated prone mice (57). The differences in the level of H3K79 methylation can be explained by the aging rate or tissue specificity. Further investigation is needed to decipher the mechanism by which DOT1L-mediated H3K79 methylation regulates aging and cancer.
Overall, the findings of the present study suggest a new model whereby DOT1L-mediated methylation of H3K79 is required for the survival of cancer cells. The irreversible arrest of cell proliferation called senescence, induced by DOT1L deficiency, acts as a barrier to tumorigenesis, possibly by reprogramming the cell cycle-regulatory genes and affecting the histone cross-talk (58). Therefore, the functional link identified between epigenetic modifications and cell cycle regulation contributes to the understanding of tumor development and treatment. However, because DOT1L inhibition also induces senescence in normal cells, DOT1L should be considered carefully as a cancer therapeutic target because of the potential side effects.
Supplementary Material
Acknowledgments
We thank Drs. Y. Zhang, Y. Okada, J. Ha, K. T. Lee, J. H. Park, H. J. You, E. Y. Moon, and E. Lee for providing several reagents.
This research was in part supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST) (2010-0009414 and 2008-314-E00054). This work was also supported by the NRF Grant funded by the Korea government (MEST) (2010-0001754 and 2011-0030722).

This article contains supplemental Figs. S1–S3 and Table S1.
- H3K79
- histone H3 lysine 79
- DOT1L
- Dot1-like protein
- MLL
- mixed lineage leukemia
- CKI
- cyclin-dependent kinase inhibitor
- SA-β-gal
- senescence-associated β-galactosidase
- WT
- wild type
- MUT
- mutant.
REFERENCES
- 1. Bhaumik S. R., Smith E., Shilatifard A. (2007) Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 2. Esteller M. (2008) Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 [DOI] [PubMed] [Google Scholar]
- 3. Egger G., Liang G., Aparicio A., Jones P. A. (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 [DOI] [PubMed] [Google Scholar]
- 4. Ng H. H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. (2002) Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16, 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Min J., Feng Q., Li Z., Zhang Y., Xu R. M. (2003) Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 112, 711–723 [DOI] [PubMed] [Google Scholar]
- 6. Sawada K., Yang Z., Horton J. R., Collins R. E., Zhang X., Cheng X. (2004) Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J. Biol. Chem. 279, 43296–43306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. (2002) Methylation of H3 lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 8. van Leeuwen F., Gafken P. R., Gottschling D. E. (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 [DOI] [PubMed] [Google Scholar]
- 9. Singer M. S., Kahana A., Wolf A. J., Meisinger L. L., Peterson S. E., Goggin C., Mahowald M., Gottschling D. E. (1998) Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150, 613–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verzijlbergen K. F., Faber A. W., Stulemeijer I. J., van Leeuwen F. (2009) Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae. BMC Mol. Biol. 10, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang W., Xia X., Reisenauer M. R., Hemenway C. S., Kone B. C. (2006) Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J. Biol. Chem. 281, 18059–18068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reisenauer M. R., Wang S. W., Xia Y., Zhang W. (2010) Dot1a contains three nuclear localization signals and regulates the epithelial Na+ channel (ENaC) at multiple levels. Am. J. Physiol. Renal Physiol. 299, F63–F76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ng H. H., Ciccone D. N., Morshead K. B., Oettinger M. A., Struhl K. (2003) Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. U.S.A. 100, 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schübeler D., MacAlpine D. M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D. E., O'Neill L. P., Turner B. M., Delrow J., Bell S. P., Groudine M. (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., Vakoc C. R. (2008) DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao P. F., Wang Z. F., Guo H. S., He N. Y., Lu Z. H. (2005) Combinational synthesis of oligonucleotides and assembly fabrication of oligonucleotide array. Colloids Surf. B Biointerfaces 40, 165–168 [DOI] [PubMed] [Google Scholar]
- 17. Bitoun E., Oliver P. L., Davies K. E. (2007) The mixed lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 16, 92–106 [DOI] [PubMed] [Google Scholar]
- 18. Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., Shilatifard A. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 19. Mueller D., Bach C., Zeisig D., Garcia-Cuellar M. P., Monroe S., Sreekumar A., Zhou R., Nesvizhskii A., Chinnaiyan A., Hess J. L., Slany R. K. (2007) A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 110, 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park G., Gong Z., Chen J., Kim J. E. (2010) Characterization of the DOT1L network: implications of diverse roles for DOT1L. Protein J. 29, 213–223 [DOI] [PubMed] [Google Scholar]
- 21. Krivtsov A. V., Feng Z., Lemieux M. E., Faber J., Vempati S., Sinha A. U., Xia X., Jesneck J., Bracken A. P., Silverman L. B., Kutok J. L., Kung A. L., Armstrong S. A. (2008) H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14, 355–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V. M., Su L., Xu G., Zhang Y. (2005) hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178 [DOI] [PubMed] [Google Scholar]
- 23. San-Segundo P. A., Roeder G. S. (2000) Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell 11, 3601–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M. (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J. Biol. Chem. 280, 9879–9886 [DOI] [PubMed] [Google Scholar]
- 25. Wysocki R., Javaheri A., Allard S., Sha F., Côté J., Kron S. J. (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 25, 8430–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Janzen C. J., Hake S. B., Lowell J. E., Cross G. A. (2006) Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell 23, 497–507 [DOI] [PubMed] [Google Scholar]
- 27. Barry E. R., Krueger W., Jakuba C. M., Veilleux E., Ambrosi D. J., Nelson C. E., Rasmussen T. P. (2009) ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells 27, 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G. A., Kadam S., Zhai H., Valdez R., Gonzalo S., Zhang Y., Li E., Chen T. (2008) The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet. 4, e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mohan M., Herz H. M., Takahashi Y. H., Lin C., Lai K. C., Zhang Y., Washburn M. P., Florens L., Shilatifard A. (2010) Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev. 24, 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shanower G. A., Muller M., Blanton J. L., Honti V., Gyurkovics H., Schedl P. (2005) Characterization of the Grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169, 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim J. E., McAvoy S. A., Smith D. I., Chen J. (2005) Human TopBP1 ensures genome integrity during normal S phase. Mol. Cell. Biol. 25, 10907–10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mathon N. F., Lloyd A. C. (2001) Cell senescence and cancer. Nat. Rev. Cancer 1, 203–213 [DOI] [PubMed] [Google Scholar]
- 34. Campisi J., d'Adda di Fagagna F. (2007) Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740 [DOI] [PubMed] [Google Scholar]
- 35. Tzatsos A., Pfau R., Kampranis S. C., Tsichlis P. N. (2009) Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc. Natl. Acad. Sci. U.S.A. 106, 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sarg B., Koutzamani E., Helliger W., Rundquist I., Lindner H. H. (2002) Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J. Biol. Chem. 277, 39195–39201 [DOI] [PubMed] [Google Scholar]
- 37. Shumaker D. K., Dechat T., Kohlmaier A., Adam S. A., Bozovsky M. R., Erdos M. R., Eriksson M., Goldman A. E., Khuon S., Collins F. S., Jenuwein T., Goldman R. D. (2006) Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. U.S.A. 103, 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller K. M., Tjeertes J. V., Coates J., Legube G., Polo S. E., Britton S., Jackson S. P. (2010) Human HDAC1 and HDAC2 function in the DNA damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17, 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dang W., Steffen K. K., Perry R., Dorsey J. A., Johnson F. B., Shilatifard A., Kaeberlein M., Kennedy B. K., Berger S. L. (2009) Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459, 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scaffidi P., Misteli T. (2006) Lamin A-dependent nuclear defects in human aging. Science 312, 1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ooga M., Inoue A., Kageyama S., Akiyama T., Nagata M., Aoki F. (2008) Changes in H3K79 methylation during preimplantation development in mice. Biol. Reprod. 78, 413–424 [DOI] [PubMed] [Google Scholar]
- 42. Schulze J. M., Jackson J., Nakanishi S., Gardner J. M., Hentrich T., Haug J., Johnston M., Jaspersen S. L., Kobor M. S., Shilatifard A. (2009) Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell cycle regulation of H3K79 dimethylation. Mol. Cell 35, 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bringold F., Serrano M. (2000) Tumor suppressors and oncogenes in cellular senescence. Exp. Gerontol. 35, 317–329 [DOI] [PubMed] [Google Scholar]
- 44. Solomon D. A., Kim J. S., Jenkins S., Ressom H., Huang M., Coppa N., Mabanta L., Bigner D., Yan H., Jean W., Waldman T. (2008) Identification of p18 INK4c as a tumor suppressor gene in glioblastoma multiforme. Cancer Res. 68, 2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nguyen A. T., Taranova O., He J., Zhang Y. (2011) DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 117, 6912–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jacinto F. V., Ballestar E., Esteller M. (2009) Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene 28, 4212–4224 [DOI] [PubMed] [Google Scholar]
- 47. Sawado T., Halow J., Im H., Ragoczy T., Bresnick E. H., Bender M. A., Groudine M. (2008) H3 K79 dimethylation marks developmental activation of the beta-globin gene but is reduced upon LCR-mediated high-level transcription. Blood 112, 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hayflick L., Moorhead P. S. (1961) Exp. Cell Res. The serial cultivation of human diploid cell strains. 25, 585–621 [DOI] [PubMed] [Google Scholar]
- 49. Narita M., Nnez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- 50. Nakanishi S., Lee J. S., Gardner K. E., Gardner J. M., Takahashi Y. H., Chandrasekharan M. B., Sun Z. W., Osley M. A., Strahl B. D., Jaspersen S. L., Shilatifard A. (2009) Histone H2BK123 monoubiquitination is the critical determinant for H3K4 and H3K79 trimethylation by COMPASS and Dot1. J. Cell Biol. 186, 371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ng H. H., Xu R. M., Zhang Y., Struhl K. (2002) Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277, 34655–34657 [DOI] [PubMed] [Google Scholar]
- 52. Wu L., Zee B. M., Wang Y., Garcia B. A., Dou Y. (2011) The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in cross-talk with H3 K4 and K79 methylation. Mol. Cell 43, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Altaf M., Utley R. T., Lacoste N., Tan S., Briggs S. D., Côté J. (2007) Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28, 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fingerman I. M., Li H. C., Briggs S. D. (2007) A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 21, 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Calvanese V., Lara E., Kahn A., Fraga M. F. (2009) The role of epigenetics in aging and age-related diseases. Ageing Res. Rev. 8, 268–276 [DOI] [PubMed] [Google Scholar]
- 56. Fraga M. F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., Iyer N. G., Pérez-Rosado A., Calvo E., Lopez J. A., Cano A., Calasanz M. J., Colomer D., Piris M. A., Ahn N., Imhof A., Caldas C., Jenuwein T., Esteller M. (2005) Loss of acetylation at Lys-16 and trimethylation at Lys-20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37, 391–400 [DOI] [PubMed] [Google Scholar]
- 57. Wang C. M., Tsai S. N., Yew T. W., Kwan Y. W., Ngai S. M. (2010) Identification of histone methylation multiplicities patterns in the brain of senescence-accelerated prone mouse 8. Biogerontology 11, 87–102 [DOI] [PubMed] [Google Scholar]
- 58. Campisi J. (2005) Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120, 513–522 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.