Background: Although the role of p63 in skin development is well known, its action in regulating human adult epidermal homeostasis is not completely elucidated.
Results: p63 regulates keratinocyte proliferation via a MYC-regulated gene network and differentiation through a cell adhesion-related gene network.
Conclusion: The balance between these two networks may control epidermal homeostasis.
Significance: This is important to understand epithelial carcinogenesis.
Keywords: Differentiation, Keratinocytes, Myc, p63, Proliferation
Abstract
Although p63 and MYC are important in the control of epidermal homeostasis, the underlying molecular mechanisms governing keratinocyte proliferation or differentiation downstream of these two genes are not completely understood. By analyzing the transcriptional changes and phenotypic consequences of the loss of either p63 or MYC in human developmentally mature keratinocytes, we have characterized the networks acting downstream of these two genes to control epidermal homeostasis. We show that p63 is required to maintain growth and to commit to differentiation by two distinct mechanisms. Knockdown of p63 led to down-regulation of MYC via the Wnt/β-catenin and Notch signaling pathways and in turn reduced keratinocyte proliferation. We demonstrate that a p63-controlled keratinocyte cell fate network is essential to induce the onset of keratinocyte differentiation. This network contains several secreted proteins involved in cell migration/adhesion, including fibronectin 1 (FN1), interleukin-1β (IL1B), cysteine-rich protein 61 (CYR61), and jagged-1 (JAG1), that act downstream of p63 as key effectors to trigger differentiation. Our results characterized for the first time a connection between p63 and MYC and a cell adhesion-related network that controls differentiation. Furthermore, we show that the balance between the MYC-controlled cell cycle progression network and the p63-controlled cell adhesion-related network could dictate skin cell fate.
Introduction
The outer layer of human skin, the epidermis, is a self-renewing, stratified squamous epithelial tissue. In adult tissue, the maintenance of epidermal homeostasis depends on an exquisite regulation of the balance between keratinocyte proliferation and differentiation (1, 2). However, the specific molecular mechanisms governing each of these processes are not completely understood.
p63, a member of the p53 tumor suppressor gene family, is known as a key regulator of epidermis development and keratinocyte differentiation. Striking developmental defects have been discovered during embryonic development in p63-knock-out mice (3, 4). In addition, p63 is required for the maintenance of proliferative potential in epithelial stem cells (3, 5, 6), epithelial lineage commitment (4), differentiation of keratinocytes (7), and epithelial cell adhesion and survival (8). Dual roles of p63 in the initiation of epithelial stratification and maintenance of stem cell proliferative potential have been established (6). Because simultaneous p63 and p53 knockdown rescued the cell proliferation defect of p63 knockdown alone but failed to restore differentiation, the roles of p63 in proliferation and differentiation of developmentally mature keratinocytes appear to be distinct (7).
In vivo, as keratinocytes commit to differentiation, they detach from the basal layer and migrate outward into a spinous layer, accompanied by the expression of early differentiation markers keratin 1 (K1) and keratin 10 (K10) (9). The effects of p63 in keratinocyte differentiation have been investigated by gain and loss of function experiments. Ectopic expression of p63 induces the early marker K1 but suppresses the expression of late differentiation markers, such as loricrin and filagrin (10). Loss of p63 inhibited both stratification and differentiation in skin organotypic culture (7). However, little is known about key genes acting downstream of p63 to regulate the dynamic equilibrium of keratinocytes differentiation and proliferation as well as epidermal homeostasis (11).
Similar to p63, the MYC oncogene is predominantly expressed in the basal cell layers of epidermis and is absent in suprabasal layers (12, 13). MYC plays a vital role not only in keratinocyte proliferation (14–16) but also in accelerating the differentiation of epidermal stem cells (17–20). Specifically, MYC activation leads to epidermal stem cell proliferation, and the sustained elevated expression of MYC in turn stimulates the proliferating stem cells to enter the transit-amplifying compartment, thereby initiating terminal differentiation (21). Although p63 and MYC have been independently demonstrated to be key regulators in the dynamic equilibrium between proliferation and differentiation in epidermal stem cells, no connection between p63 and MYC has been reported so far. The genetic basis underlying the processes regulated by p63 and MYC in mature keratinocytes, as well as the nature of their downstream effectors, remains mostly unknown.
We hypothesized that the regulation of the equilibrium between proliferation and differentiation of keratinocytes could rely on gene networks acting downstream of p63 and MYC rather than on a single gene. To this end, we compared phenotypic outcomes and global gene expression profiles in differentiating human keratinocytes transiently depleted of either p63 or MYC. We demonstrate that p63 regulates the proliferation and commitment to differentiation of human mature keratinocytes by two independent mechanisms. p63 is necessary for the proper expression of MYC and in turn controls keratinocyte proliferation by regulating the expression of MYC via the Wnt/β-catenin and Notch signaling pathways. Moreover, p63 regulates human keratinocyte differentiation by controlling a network of genes implicated in cell migration and adhesion.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary human keratinocytes were isolated from human skin biopsy and cultured in KGM2 medium (Clonetics) on flasks coated with collagen type I (Falcon Biocoat) at 37 °C and 5% CO2. This study was approved by the ethical research committee “Comité de Protection des Personnes Sud-Est II” (CODECOH Number DC-2008-262). HaCaT is a non-tumorigenic, spontaneously transformed human keratinocyte cell line (22) generously provided by N. E. Fusenig (German Cancer Research Center, Heidelberg, Germany). HaCaT cells were grown at 37 °C in a humidified incubator with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum, 105 units/liter penicillin, 50 mg/liter streptomycin, and 2 mm GlutaMax (Invitrogen). HaCaT differentiation was induced by cultivating cells seeded in plastic flasks at 104 cells/cm2 as described previously (22).
Transient Transfection
siRNA transfection was used INTERFERin according to the manufacturer's instructions. All transfections were performed using 10 nm siRNA. siRNA against MYC (SI02662611), all p63 isoforms (SI00055118), FN1 (SI02664004), IL1B (SI00012166), MMP13 (SI03049277), CYR61 (SI02626421), JAG1 (SI02780134), and All Stars negative control siRNA (1027281) were obtained from Qiagen.
Cotransfection of siRNA with plasmids was done using an Amaxa Nucleofector II (Amaxa Biosystems) according to the manufacturer's recommendations. Plasmids containing the ORF of cleaved Notch1 (NICD)3 (Addgene plasmid 17623) (23) or Wnt-3 (Addgene plasmid 17993) (24) were obtained from Addgene.
Cell Proliferation and Cell Death Assays
12 h post-transfection, HaCaT cells were cultured in serum-free medium for 24 h. After a 40-h culture in complete medium, cell growth was performed with the ViaLight Plus kit (Cambrex) according to the manufacturer's protocol. For EdU staining, HaCaT cells were pulse-labeled for 24 h with 10 μm EdU (Invitrogen) 24 h after transfection by siRNA. Cells were subsequently fixed on glass coverslips with paraformaldehyde immediately after the EdU pulse labeling. The Click-it EdU reaction was performed according to the manufacturer's instructions (Invitrogen) to label and detect EdU with the Alexa Fluor azide 555 dye. EdU staining was analyzed by BD LSRII (BD Biosciences). Student's t test was computed for statistical analysis. We used the fluorescence-based method LIVE/DEAD viability/cytotoxicity kit (Invitrogen) to evaluate cell death. HaCaT cells transfected by siRNA at 48 h after transfection were used for staining. Live cells (i.e. cells with esterase activity and intact membranes) were labeled with calcein AM, a green fluorescent product, and dead cells were labeled with ethidium homodimer-1, a red fluorescent nucleic acid stain. Cell staining was visualized with an Olympus BX61 straight microscope controlled with Metamorph software (Molecular Devices, Downington, PA).
Microarray Experiments and Genetic Network Analysis
For transcriptome analysis, microarrays with 25,342 spotted human oligonucleotides were used (Human-25K, Réseau National des Génopoles, France) (25). RNA was isolated, amplified, labeled, and hybridized following a published protocol (25). For each condition, three independent biological replicates were obtained, and the knockdown of corresponding genes was verified before RNA extraction (supplemental Fig. S2). Each biological replicate was hybridized to the arrays in four replicates using a dye swap strategy. Slides were scanned with an Agilent G2565AA Microarray Scanner (Agilent Technologies). Data analyses, including intensity-dependent Lowess normalization of raw data and differential analysis, were performed with GeneSpring 7.0 software (Silicon Genetics). Differentially expressed genes were identified using analysis of variance (p value corrected with Benjamini and Hoehberg false discovery rate), and only genes with a -fold change of ≥1.2 and p < 0.01 were used for further analysis.
Gene pathway and genetic network analyses were performed by Ingenuity Pathway Analysis (IPA) (Ingenuity Systems). Ontologies attached to each gene were used to classify altered genes according to main biological themes. The following criteria enter into generating the networks in the Ingenuity Pathways Analysis application. 1) We have designated the molecules of interest in the analysis parameters before running the analysis. Molecules that (a) meet the cut-off and/or filter criteria and (b) interact with other molecules in Ingenuity's Knowledge Base are identified as focus molecules (also called “network eligible molecules”). Focus molecules serve as the “seeds,” or focal points, for generating networks. 2) Networks are preferentially enriched for focus molecules with the most extensive interactions and for which interactions are specific with the other molecules in the network (rather than molecules that are promiscuous, those that interact with a broad selection of molecules throughout the Ingenuity knowledge base). 3) Additional non-focus molecules from the data set and from the Ingenuity knowledge base are then recruited and added to the growing networks. 4) Networks are scored for the likelihood of finding the focus molecule(s) in that given network. The higher the score, the lower the probability that you would find the focus molecules(s) you see in a given network by random chance. 5) In the current version of the application, there is a cut-off of 35 molecules/network to keep networks to a usable size.
mRNA Expression Analysis
RNA was extracted with an RNeasy minikit (Qiagen). For real-time quantitative PCR, 2 μg of RNA was reverse-transcribed in a total volume of 20 μl with the SuperScript II RNase H reverse transcriptase system (Invitrogen) and random primers according the manufacturer's instructions. Reverse transcription reactions were diluted to 500 μl of water, and 5 μl of the diluted cDNA was used for each quantitative PCR. Quantitative PCR was carried out with a Platinum quantitative PCR SuperMix-UDG Kit (Invitrogen) using an ABI 7500 fast real-time PCR system (Applied Biosystems). All experiments were run in triplicate, and the results were normalized to 18 S rRNA expression. Primer sequences are listed in supplemental Table S1.
Immunoblotting
Total cellular protein was extracted using radioimmune precipitation assay buffer supplemented with protease inhibitor mixture (Roche Applied Science) and 1 mm sodium orthovanadate. After quantification with a BCA protein assay kit (Pierce), 20 μg of protein from each sample was run on a NuPAGE BisTris gel (Invitrogen) and then transferred to a PVDF membrane. The membranes were blocked for 1 h at room temperature in 5% nonfat dry milk, TBST and incubated overnight at 4 °C with primary antibody and then for 1 h at room temperature with HRP-conjugated secondary antibody. Detection was performed with an ECL kit (Pierce). Primary antibodies used were as follows: antibodies against TP63 (4A4, sc-8431, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), MYC (N-262, sc-764, Santa Cruz Biotechnology, Inc.), cytokeratin 1 (PRB-149P, Covance), cytokeratin 10 (MMS-159S, Covance), total β-catenin (6B3, 9582, Cell Signaling), cleaved Notch1 (NICD, Val-1744, 2421, Cell Signaling), and β-actin (A3854, Sigma).
Cell Cycle Analysis
Cell cycle phase was determined by analyzing total DNA content using flow cytometry. HaCaT cells were synchronized by culturing in serum-free DMEM 24 h before transfection. Cells were transfected using INTERFERin with 10 nm siRNA. 48 h post-transfection, cells were collected, fixed, and stained with a cell cycle phase determination kit (Cayman Chemical). Flow cytometry analysis was performed in a MoFlo cell sorter (DakoCytomation).
Luciferase Reporter Assay
HaCaT cells were cotransfected with plasmid and siRNA in 24-well plates using an Amaxa Nucleofector II (Amaxa Biosystems) according to the manufacturer's recommendations. The human MYC promoter cloned into a luciferase plasmid was described previously (26). A TK-Renilla reporter was used as an internal normalization control. The ratio of firefly luciferase plasmid to Renilla luciferase was 10:1 in each nucleofection (2 μg:200 ng). At 48 h post-transfection, luciferase activity was measured using a Dual-Luciferase reporter assay system (Promega). The luciferase reporter constructions were obtained as follows: pDel-1∼pDel-4 reporter (Addgene plasmid 16601, 16602, 16603, 16604) (26), TCF-4/LEF reporter plasmid from SuperArray Bioscience, YY-1 reporter plasmid from Panomics, and JUN and FOS reporter plasmid from H. van Dam (Leiden University, Netherlands).
RESULTS
siRNA-mediated Depletion of either p63 or MYC Decreased Mature Keratinocyte Proliferation, but Only Loss of p63 Inhibited Differentiation
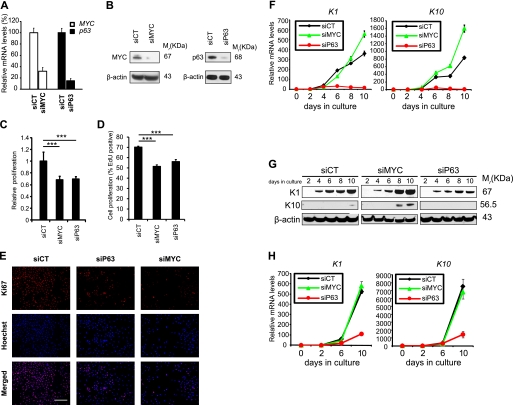
To investigate the specific roles of MYC and p63 in human keratinocyte differentiation, we knocked down each gene in HaCaT cells using specific siRNA and analyzed the phenotypic consequences. Because of the existence of six different isoforms of p63, we used an siRNA targeting the conserved DNA binding domain in all genes to achieve ablation of all p63 isoforms. We first verified that the expression of either MYC or p63 in siRNA-transfected keratinocytes was specifically knocked down at both the transcript (Fig. 1A) and protein levels (Fig. 1B) 48 h post-transfection. Compared with cells treated with control siRNA, we observed a 70 and 90% down-regulation of MYC and p63 expression, respectively. It is noteworthy that a significant loss of both genes was still observable after 10 days of culture (supplemental Fig. S1). Two days after siRNA transfection, we observed a reduced ATP content (Fig. 1C), whereas both EdU (Fig. 1D) and Ki67 staining (Fig. 1E) significantly decreased, both in p63- and MYC-knockdown HaCaT cells. Together, these data demonstrate a defect in keratinocyte proliferation upon ablation of either p63 or MYC. We next monitored the capacity of these cells to differentiate in vitro during 10 days of culture in the appropriate medium. As we focused on the commitment to differentiation, we chose to monitor the expression of two early differentiation markers, K1 and K10, rather than later ones, such as involucrin or filagrin. Indeed, K1 and K10 are markers of the basal-spinous layer transition in epidermis. Reduction of p63 levels in keratinocytes significantly inhibited the expression of K1 and K10 at the transcript level (Fig. 1F). In contrast, cells lacking MYC still expressed high levels of both markers, suggesting that these cells were still able to differentiate and did so even faster than the control (Fig. 1F). On the protein level, K1 was continuously induced except in cells lacking p63, which exhibited up-regulation of K1 until day 8, followed by a weak decrease in expression (Fig. 1G). Expression of the K10 protein started late in all conditions but became significant in MYC-depleted cells after 8 days of culture (Fig. 1G). In addition, we observed that loss of p63 in cultures of primary human keratinocytes inhibited expression of both K1 and K10 compared with control or MYC-depleted cells (Fig. 1H). These results demonstrate that the ablation of either MYC or p63 significantly reduced human mature keratinocyte proliferation, whereas only the siRNA-mediated loss of p63 inhibited differentiation.
FIGURE 1.
Influence of knockdown of MYC or p63 on keratinocyte differentiation. A and B, p63 and MYC expression was measured by qRT-PCR (A) and Western blot (B) after introduction of siRNA oligonucleotide duplexes. siCT is a non-targeting siRNA control, siP63 is an siRNA duplex against all p63 isoforms, and siMYC is an siRNA targeting the MYC oncogene. mRNA expression levels were normalized to siCT (A). Samples used in A and B were extracted 48 h after siRNA transfection. C, cell growth was inspected by Vialight. Error bars, S.D. of six biological replicates; Student's t test was used for statistical analysis. ***, p < 0.001. D, cell proliferation was assessed by EdU incorporation measured by flow cytometry. Three independent experiments were used, and Student's t test was used for statistical analysis. ***, p < 0.001. E, Ki67 staining of HaCaT cells 48 h post-transfection by siRNA. Red, Ki67; Blue, Hoechst. F and G, time course of qRT-PCR (F) and Western blot (G) analysis of human cytokeratin 1 (K1) and cytokeratin 10 (K10) expression in HaCaT cells transfected with siCT, siMYC, or siP63. mRNA expression levels were normalized to day 0. β-Actin was used as a loading control in the Western blot. H, time course of qRT-PCR analysis of K1 and K10 mRNA levels in primary human keratinocytes (PHK), transfected with siCT, siMYC, or siP63. mRNA expression levels were normalized to day 0. Error bars in all qRT-PCR graphs represent S.D. of triplicate samples from one representative experiment.
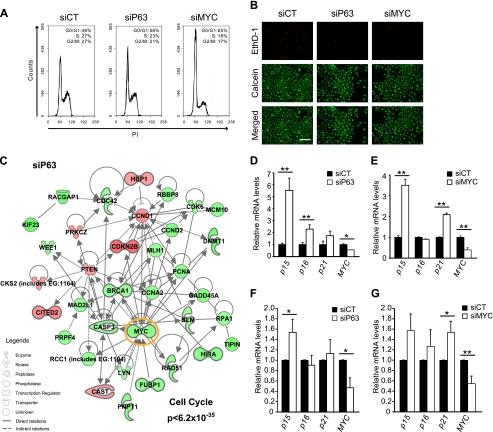
Knockdown of either MYC or p63 Leads to Cell Cycle Arrest
To determine the potential origin of the proliferation defect in keratinocytes lacking either p63 or MYC, we analyzed cell death and the cell cycle. Knockdown of either p63 or MYC triggered a significant arrest in G0/G1 (Fig. 2A) without affecting cell death (Fig. 2B). To identify the molecular mechanisms controlling this cell cycle arrest, we analyzed the gene expression profiles of keratinocytes depleted of either MYC or p63 after transfection (supplemental Table S2). For these experiments, we have particularly insisted on robustness of the data. We performed three independent siRNA transfections and three independent RNA extractions (supplemental Fig. S2), along with several technical replicates, to generate three independent expression profiles that we then averaged for either MYC- or p63-depleted keratinocytes. This transcriptome analysis showed that a small network of genes strongly associated with cell cycle regulation (p < 6.2 × 10−35) was significantly down-regulated in keratinocytes lacking either MYC or p63 (supplemental Fig. S3A and Fig. 2C). The majority of genes in this network were similarly regulated in both p63-depleted and MYC-depleted cells, except CSK2, GADD45A, and CCND2 (supplemental Fig. S3A). Analysis by qRT-PCR of the major cell cycle inhibitors confirmed that p15 and p21 were both significantly up-regulated in response to the knockdown of either MYC or p63 in HaCaT cells (Fig. 2, D and E). Interestingly, the same trend was observed in primary keratinocytes (Fig. 2, F and G). We also observed that MYC, a major hub (highly connected node) in this cell cycle-controlling network, was down-regulated in both HaCaT and primary keratinocyte cells lacking p63 (Fig. 2, C, D, and F). This result suggests that p63 is necessary for the proper expression of MYC in human keratinocytes.
FIGURE 2.
Shared cell cycle arrest mechanisms in cells treated with siP63 or siMYC. A, cell cycle phase determination analysis performed by flow cytometry. HaCaT cells were treated with siCT (left), siP63 (middle), or siMYC (right). PI, propidium iodide. B, Live/Dead staining of siRNA-transfected HaCaT cells. Red, EthD-1; green, calcein AM. C, genetic networks regulating the cell cycle were generated by IPA. The lists of genes used in IPA were obtained from transcriptome analysis of HaCaT cells treated with siP63 for 48 h. All cell cycle-related genes in siP63 were extracted from the IPA database with a very significant p value (<6.2 × 10−35) to generate this network (see “Experimental Procedures”). Nodes (genes or proteins) in the networks are shown by different shapes (biological functions) and colors (red indicates up-regulated, and green represents down-regulated). Edges are represented as solid or dashed lines to indicate direct and indirect interactions, respectively. D and E, qRT-PCR analysis of cyclin-dependent kinase inhibitors (such as p15, p16, and p21) and MYC in siP63 (D) or in siMYC (E) HaCaT keratinocytes. mRNA expression levels were normalized to siCT, and error bars in qRT-PCR graphs show S.D. of triplicate samples from one representative experiment. Student's t test was used for statistical analysis. *, p < 0.05; **, p < 0.01. F and G, identical to D and E in human primary keratinocytes lacking either p63 (F) or MYC (G).
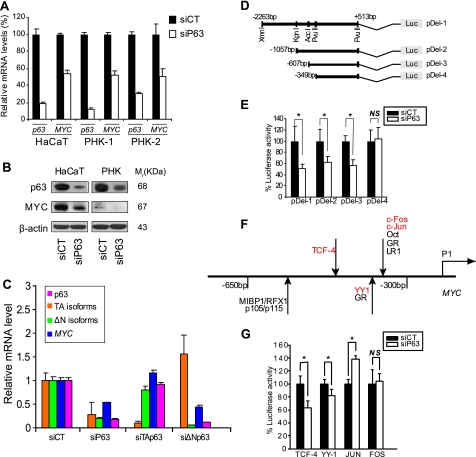
p63 Is Necessary for Proper Expression of MYC
We first confirmed the down-regulation of MYC expression in cells lacking p63 at the transcript level (Fig. 3A) and at the protein level (Fig. 3B), both in HaCaT cells and in primary human keratinocyte cultures from two different donors. Data mining in the Gene Expression Omnibus (GEO) confirmed that other groups have measured down-regulation of MYC in p63-deficient epithelial cell lines (supplemental Fig. S4) (27). Using specific siRNA targeting either ΔNp63 or TAp63, we were able to demonstrate that only the knockdown of ΔNp63 isoforms triggered down-regulation of MYC expression (Fig. 3C). This was consistent with the fact that ΔNp63 is the isoform predominantly expressed in adult keratinocytes. Conversely, overexpression of either ΔNp63 or TAp63 in HaCaT cells had no effect on MYC expression (supplemental Fig. S5). Together, these results demonstrated that p63 is necessary for the proper expression of MYC.
FIGURE 3.
Knockdown of p63 down-regulates MYC. qRT-PCR (A) and Western blot (B) analysis of MYC expression when p63 was knocked down by siRNA targeting all p63 isoforms in HaCaT cells or primary human keratinocytes (PHK). C, MYC expression in ΔNp63-, TAp63-, or p63-knockdown cells. 48 h post-transfection, MYC expression was measured by qRT-PCR. 18 S rRNA was an endogenous control to normalize the results. D, structure of the MYC promoter region cloned into the luciferase reporter plasmids. The heavy horizontal lines represent the promoter sequences in each reporter plasmid. E, HaCaT cells were cotransfected with siCT or siP63 and the MYC promoter luciferase reporter constructs. Luciferase activities (means ± S.D.) were measured 48 h post-transfection. Error bars, S.D. of five biological replicates. Firefly luciferase luminescence was normalized to an internal control Renilla luciferase. F, schematic representation of the MYC promoter region between −300 and −650 bp (which corresponds to the differences between pDel-3 and pDel-4). Known TF binding sites are indicated by arrows. G, HaCaT cells were cotransfected with control siRNA or all-p63 siRNA and TCF-4, YY-1, JUN, or FOS luciferase reporter plasmids (TFs marked in red in G). Detailed luminescence measurements and analysis can be found under “Experimental Procedures.” Error bars, S.D. of replicates. The Mann-Whitney test was used for statistical analysis in E and G. *, p < 0.05.
We next investigated the molecular mechanisms underlying the p63 knockdown-triggered down-regulation of MYC expression. Using luciferase reporter constructs fused to truncated MYC promoter regions (Fig. 3D), we characterized the sequence upstream of MYC and identified a putative p63-controlled region. As demonstrated in Fig. 3E, this region extended from −349 to −607 bp upstream of the transcription start site. A close-up view of this region revealed the absence of the p63 consensus binding site, in agreement with our previous published results, which showed that MYC was not a direct target of p63 in human keratinocytes (28, 29). These results suggested that the expression of MYC is under the indirect control of p63. The region upstream of the MYC gene also contains several other binding sites for transcription factors (TFs), including TCF-4 (TF responding to the Wnt/β-catenin signaling pathway), YY-1 (TF responding to the Notch signaling pathway), c-FOS, and c-JUN (AP1 TFs) (Fig. 3F). Because these TFs are known to play important roles in the control of keratinocyte proliferation and differentiation (30–32), we investigated whether or not they are involved in the p63 knockdown-triggered down-regulation of MYC promoter activity. In keratinocytes lacking p63, we observed a moderate inhibition of both TCF4 or YY1-dependent luciferase activities but no change or a slight activation in JUN- and FOS-driven luciferase expression (Fig. 3G). These results suggest that TCF4 and YY1, the TFs responding to the Wnt/β-catenin and Notch signaling pathways, respectively, were partially responsible for the p63-dependent down-regulation of MYC expression.
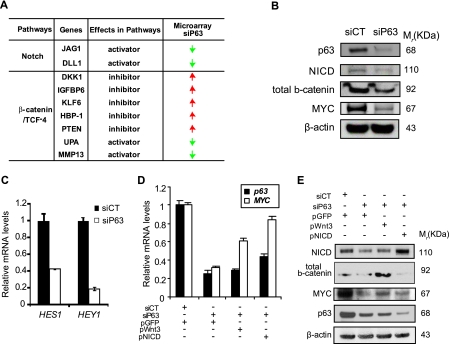
p63 Knockdown-triggered Down-regulation of MYC Expression Is Mediated by Notch and Wnt/β-Catenin Signaling Pathways
To determine whether the signaling cascades leading to the activation of YY1 and TCF4 are affected by p63 levels, we scrutinized the expression profiles of keratinocytes lacking either p63 or MYC. Interestingly, we found that the Wnt/β-catenin and Notch signaling cascades were indeed potently inhibited in keratinocytes lacking p63. JAG1 and DLL1, ligands of the Notch signaling pathway, were both down-regulated in p63-knockdown cells (Fig. 4A). In addition, several inhibitors of the Wnt/β-catenin pathway were up-regulated, whereas several activators were down-regulated, probably resulting in the inhibition of this signaling pathway (Fig. 4A). At the protein level, the ablation of p63 resulted in the down-regulation of both cleaved Notch1 (NICD) and total β-catenin (Fig. 4B). Taken together, these results suggest that both pathways were turned down in p63-knockdown cells. Inhibition of the Notch signaling pathway in the presence of p63-targeting siRNA was also indicated by the reduced expression of two reporter genes of the Notch-signaling cascade, HES1 and HEY1 (Fig. 4C). To further confirm the potential involvement of the Wnt and Notch pathways in down-regulation of MYC expression, we restored downstream signaling by transfecting HaCaT cells with either WNT3 or NICD expression vectors, together with p63-targeting siRNA or control siRNA. We observed that the overexpression of either WNT3 or NCID induced the up-regulation of MYC expression both at the transcript (Fig. 4D) and protein levels (Fig. 4E). This would partially restore keratinocyte proliferation. However, overexpression of those two genes was not sufficient to restore proper differentiation of keratinocytes lacking p63 (data not shown). These results suggest that the molecular role of p63 in differentiation of human mature keratinocytes is probably different from the ones that dictate their proliferation.
FIGURE 4.
The Wnt/β-catenin and Notch pathways are responsible for the MYC down-regulation in p63 knockdown keratinocytes. A, list of modulators of the Notch and β-catenin pathways that were differentially expressed according to siP63 transcriptome analysis. Green arrows, decrease in expression; red arrows, increase compared with siCT-treated cells. B, Western blots of cleaved NICD, total β-catenin, and MYC in siP63 as compared with siCT. C, qRT-PCR analysis of expression of HES1 and HEY1, which are two transcriptional target genes of the Notch signaling pathway. The values represent the mean ± S.D. of three replicate samples from one representative experiment. D and E, MYC expression study at mRNA level (D) and at protein level (E) after activation of the β-catenin or Notch pathway via overexpression of Wnt3 or NICD, respectively. A plasmid expressing GFP was used as a transfection control. Co-transfection of siRNA and plasmids was done by nucleofection using Amaxa (see details under “Experimental Procedures”). Error bars in all qRT-PCR graphs represent the S.D. of triplicates from one representative experiment.
Cell Migration/Adhesion-related Network Acts Downstream of p63 to Induce Onset of Keratinocyte Differentiation
Our results demonstrated that MYC is down-regulated in cells lacking p63 (Figs. 2 and 3), thus functionally corresponding, at least partially, to a siRNA-mediated knockdown of MYC. However, these cells exhibited completely opposite differentiation outcomes (Fig. 1E). To investigate the molecular mechanisms enabling keratinocyte differentiation downstream of p63, we compared the expression profiles of p63-depleted and MYC-depleted cells.
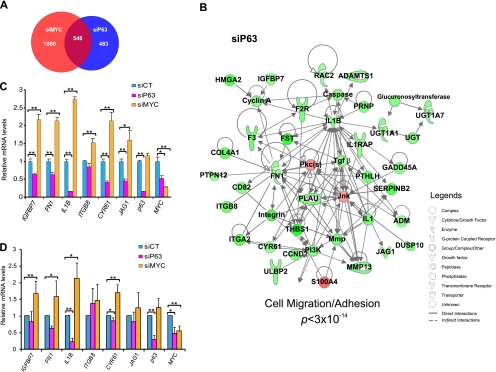
As demonstrated in Fig. 5A, 546 genes were common to both expression profiles. It is noteworthy that there were more genes common to both profiles than specific to the p63-depleted cells. This again suggests that part of the transcriptional response to p63 ablation in human keratinocytes was also due to the down-regulation of MYC. The down-regulated genes in keratinocytes lacking either MYC (supplemental Fig. S6A) or p63 (supplemental Fig. S6B) shared similar gene ontology terms (e.g. cell cycle, DNA replication, and DNA repair). Genes up-regulated in either MYC-depleted (supplemental Fig. S6C) or p63-depleted keratinocytes (supplemental Fig. S6D) also shared some gene ontology terms, such as cellular movement or cell death.
FIGURE 5.
Candidate keratinocyte differentiation effectors identified by comparative siMYC and siP63 transcriptome analysis. A, Venn diagram indicates distribution of differentially expressed genes in siMYC and siP63. 546 genes are common to siMYC and siP63. B, IPA with the list of oppositely expressed genes in the common part of A extracted a genetic network associated with a function of cell migration with a very significant p value (p < 3 × 10−14). The genes in this network are down-regulated in siP63 keratinocytes and up-regulated in siMYC keratinocytes (supplemental Fig. S6). This network was named the KFC network and contains candidate keratinocyte differentiation effectors acting downstream of p63. C and D, expression levels of genes identified in genetic networks validated by qRT-PCR in HaCaT cells (C) and primary keratinocytes (D). Student's t test was used for statistical analysis. *, p < 0.05; **, p < 0.01. Error bars, S.D.
Among the 546 genes common to both expression profiles, we found 71 genes that were antagonistically regulated (supplemental Table S3). We hypothesized that these antagonistically regulated genes could mechanistically explain, at least partially, the opposite differentiation outcomes between p63- and MYC-lacking keratinocytes. We used the Ingenuity knowledge base using IPA software to analyze the networks and functions associated with these 71 genes. Strikingly, a network of 41 nodes was extracted and significantly associated with a single function, cell migration/adhesion (p < 3 × 10−14). In cells lacking p63, this network was strongly down-regulated (Fig. 5B), whereas in MYC-depleted keratinocytes, this same network was up-regulated (supplemental Fig. S3B). We further validated the expression of some genes in the network by qRT-PCR in HaCaT cells (Fig. 5C) and in primary keratinocytes (Fig. 5D). As expected, the expression of these genes was up-regulated in MYC-depleted cells and down-regulated in keratinocytes lacking p63. These results suggest that this migration/adhesion-related gene network could contain potential effectors acting downstream of p63 to induce the onset of terminal differentiation in human keratinocytes.
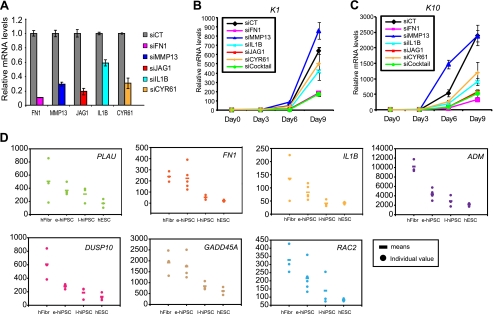
To validate our hypothesis, we used several functional approaches. First, we searched for known phenotypes associated with this 41-gene network in the Mouse Genome Informatics database. There are 19 knock-out mice with abnormal skin phenotypes reported in that database. Strikingly, 15 genes of these 19 KO mice were present in this network (Table 1). These data suggest that nearly 80% of all known skin dysfunction-related genes belong to the network we have characterized and functionally validate it. We also monitored in vitro differentiation of keratinocytes lacking different genes belonging to this migration/adhesion network: FN1, MMP13, JAG1, IL1B, and CYR61 (Fig. 6A). Except for MMP13, we observed a strong inhibition of differentiation in cells lacking any of these genes, as demonstrated by the delayed expression of K1 (Fig. 6B) and K10 (Fig. 6C) transcripts. It is noteworthy that although the ablation of IL1B was only partial (40%), it significantly inhibited differentiation. Furthermore, ablation of all of these genes together (siCocktail) strongly inhibited keratinocyte differentiation. Finally, if the genes belonging to this network promote commitment to differentiation, we postulated that their expression should be down-regulated in non-differentiated and/or pluripotent cells. We data-mined the NCBI Gene Expression Omnibus database, and interestingly, we found that seven hubs in this network, PLAU, FN1, IL1B, ADM, DUSP10, GADD45A, and RAC2, were significantly down-regulated in induced pluripotent stem cells (iPS) and are even part of the iPS transcriptomic signature (Fig. 6D) (33). Together, these results show that a p63-controlled migration/adhesion-related network plays a key role in the onset of human mature keratinocyte differentiation. As a consequence, we named this network the keratinocyte cell fate (KCF) network.
TABLE 1.
List of all knockout mice exhibiting abnormal skin phenotypes reported in the Mouse Genome Informatics database
Genes that are also present in the KCF network (Fig. 5B) and are antagonistically regulated in MYC- and p63-deficient keratinocytes are shown in boldface type. JNK and PI3K families figure in the network, and therefore no specific accession number could be given.
| Gene ID | GenBankTM accession number | Phenotype |
|---|---|---|
| CCND2 | NM_001759 | Abnormal external granule cell layer morphology |
| COL17A1 | NM_130778 | Abnormal epidermal layer morphology;skin lesions |
| DFNA5 | NM_004403 | Decreased cochlear outer hair cell number |
| DST | NM_001723 | Abnormal skin pigmentation |
| F2R | NM_001992 | Abnormal skin condition |
| F3 | NM_001993 | Abnormal skin condition |
| FN1 | NM_002026 | Skin lesions |
| FST | NM_006350 | Thickened epidermis |
| GADD45A | NM_001924 | Increased sensitivity to skin irradiation |
| HMGA2 | NM_003483 | Long hair |
| IL1RAP | NM_002182 | Abnormal skin condition/morphology |
| ITGA2 | NM_002203 | Abnormal skin condition |
| JAG1 | NM_000214 | Abnormal cochlear hair cell morphology |
| Jnk | Decreased hair follicle number | |
| KRT6A | NM_005554 | Abnormal coat/hair morphology |
| PI3K | Skin lesions | |
| PTHLH | NM_002820 | Thin epidermis |
| THBS1 | NM_003246 | Abnormal skin condition |
| PRNP | NM_000311 | Abnormal skin condition |
FIGURE 6.
An extracellular matrix genetic network is involved in keratinocyte differentiation. A, validations of siRNA knockdown targeting FN1, MMP13, JAG1, IL1B, or CYR61. Cells were transfected with 10 nm siRNA for 48 h, and then the gene expression levels were quantified by qRT-PCR. The effect of knockdown of each gene was compared with siCT. B and C, time course analysis of K1 (B) and K10 (C) expression were determined by qRT-PCR 9 days post-transfection of HaCaT cells with siRNA against FN1, MMP13, IL1B, JAG1, CYR61, or siCock (mixture of these five siRNAs). D, expression of oncogene hubs in human iPS. hFibr, human fibroblast; e-hiPSC, early human iPS; l-hiPSC, late human iPS; hESC, human embryonic stem cells. Each spot represents expression data from one biological sample. Error bars, S.D.
DISCUSSION
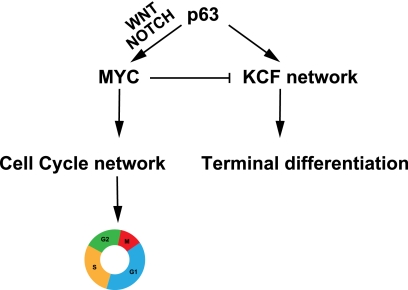
Our findings confirm that sustained expression of both p63 and MYC, two major regulators of epidermal homeostasis (5–7, 18, 21, 34, 35), is required to maintain growth and differentiation of human developmentally mature keratinocytes. However, we demonstrate that their respective roles are very different, as already suggested by some authors (36). We propose a model to illustrate the distinct mechanisms of action of p63 on human developmentally mature keratinocyte proliferation and differentiation (Fig. 7). p63 is required for the proper expression of MYC through the combined regulation of the Wnt/β-catenin and Notch signaling pathways, leading in turn to cell cycle regulation and cell proliferation. p63 also regulates a KCF network that contains several potential “differentiation effectors,” some located in the extracellular space. The up-regulation of the KCF network would promote the onset of terminal differentiation of keratinocytes (Fig. 7). In this study, we show that the siRNA-mediated loss of MYC triggers down-regulation of the “proliferation network” and up-regulation of the KCF network, promoting human keratinocyte differentiation; in contrast, p63 knockdown down-regulates both cell proliferation and mobility/adhesion-related networks, thus inhibiting differentiation. These results were observed both in the HaCaT cell line (mutated p53) and in normal human primary keratinocytes (wild-type p53), suggesting that the model we propose is independent of the p53 status of skin cells.
FIGURE 7.
Schematic representation of the mechanism of commitment of keratinocyte differentiation controlled by p63. p63 controls keratinocyte proliferation and differentiation independently via different genetic networks. p63 regulates the cell cycle in part by regulating MYC expression through both the Wnt/β-catenin and Notch pathways. To trigger the onset of differentiation, p63 controls the KCF network composed of keratinocyte differentiation effectors. Most of these effectors, such as FN1, IL1B, JAG1, and CYR61, are located in the extracellular matrix and are implicated in cell migration.
Because MYC is a transcriptional repressor of p15 and p21, two cyclin-dependent kinase inhibitors (37–39), and p63 was reported to repress p21 and p16 (40–42), it was not a surprise to observe reduced proliferation in cells lacking either gene. More surprising was the p63 knockdown-triggered down-regulation of MYC expression that we report here for the first time. This result suggests that although MYC is not a direct target of p63 (28, 29), the expression of this transcription factor is necessary for the proper expression of MYC both in HaCaT cell line and primary human keratinocyte culture. Our results establish that MYC expression is mediated, at least in part, by the Wnt/β-catenin and Notch signaling pathways. Interestingly, both pathways are important regulators of proliferation and differentiation in epidermal stem cell maintenance and wound healing (1, 43–46). The interplay between the Wnt/β-catenin and Notch pathways has been reported in epidermal homeostasis and differentiation as well (47, 48). We show that both pathways act in concert downstream of p63 to control the proper expression of MYC and, in turn, regulate the keratinocyte cell cycle.
Although the inhibition of proliferation of cells lacking p63 was partially MYC-dependent, the differentiation defects appeared to be independent of MYC. Indeed, the siRNA-mediated loss of MYC did not impair keratinocyte commitment to terminal differentiation. The differentiation was even slightly accelerated in MYC-depleted cells. By comparing expression profiles from mature keratinocytes lacking either MYC or p63, we found 546 common genes, of which 71 were antagonistically regulated. Strikingly, from that list of 71 genes, we extracted a gene network containing 41 genes that were significantly associated (p <3 × 10−14) with a single function, cell migration/adhesion. These results are consistent with recent reports showing that p63 functions as an inhibitor of cell migration (27) and that a p53 mutant forms a complex with p63 to antagonize its cell migration-inhibitory function, leading to TGFβ-dependent metastasis (49). Similarly, it was shown that p63 regulates a cell adhesion program, including integrins, in epithelial cells (8). Although we cannot exclude the possibility that other genes that are among the 546 common genes but are not found in our network might regulate early differentiation, we have clearly demonstrated that this p63-controlled cell migration/adhesion network contains several effectors acting downstream of p63 to trigger differentiation of mature keratinocytes.
Some of the differentiation effectors we have identified (such as integrins, FN1, PLAU, JAG1, IL1, and CYR61) are also involved in cancer progression in inducible human tissue neoplasia (50). Integrin signaling plays an important role in epidermal adhesion, growth, and differentiation (51, 52). JAG1 is a ligand of the Notch signaling pathway and acts on keratinocyte differentiation (32, 53). JAG1 is also a transcriptional target of p63 (54). Interleukin-1 (IL1) is implicated in human epidermal keratinocyte proliferation (55) and even in the regeneration of epidermal tissue in vitro (56). IL1α is the active form of interleukin-1 in human epidermis. IL1β was considered non-functional in keratinocytes, but our data suggest that IL1β is necessary to induce keratinocyte differentiation.
Although most of these “differentiation effectors” do not seem to be direct transcriptional targets of p63, the control exerted by p63 on this network was dominant. Indeed, we were unable to rescue differentiation of p63-depleted keratinocytes by ectopic expression of NICD or JAG1 with the use of JAG1 or IL1β recombinant proteins or even with an acellular matrix obtained from normal fully differentiated keratinocytes (data not shown).
We report for the first time the role of this p63-regulated cell migration/adhesion network in the commitment of developmentally mature keratinocytes to differentiation. A normal expression of this network seems to be required to trigger differentiation, whereas its down-regulation prevents it. Furthermore, we believe that misregulation of this gene network may play a major role in tumorigenesis. Indeed, Khavari's group recently reported a core tumor progression signature network in keratinocytes, which contained 282 nodes and was involved in carcinogenesis (50). This core tumor progression signature network contained several oncogene hubs, and eight of the top 10 nodes were extracellular or cell surface proteins. It is noteworthy that four of these eight extracellular oncogene hubs, PLAU, CYR61, FN1, and IL1, also belong to the p63-regulated cell migration/adhesion network we describe in this study. Other oncogene hubs reported in the core tumor progression signature network (50), such as SERPINE1 and ITGA6, were also down-regulated in human keratinocytes depleted of p63 (supplemental Table S2).
In conclusion, the siRNA-mediated loss of MYC triggers down-regulation of the “proliferation network” and up-regulation of the “KCF migration/adhesion-related network,” promoting human keratinocyte differentiation; in contrast, p63 knockdown down-regulates both cell proliferation and KCF networks, thus inhibiting differentiation. We believe that the balance between levels of expression of both cell proliferation and KCF networks could dictate keratinocyte cell fate. Furthermore, we think that this network approach would reconcile much of the existing data on the regulation of the balance between proliferation and differentiation in skin.
Supplementary Material
Acknowledgment
We thank Jernej Murn for suggestions for the manuscript.

This article contains supplemental Tables S1–S3 and Figs. S1–S6.
All of the microarray data used in this study have been deposited in the NCBI Gene Expression Omnibus under accession number GSE17394.
- NICD
- Notch intracellular domain
- EdU
- 5-ethynyl-2′-deoxyuridine
- KCF
- keratinocyte cell fate
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- qRT-PCR
- quantitative RT-PCR
- TF
- transcription factor
- iPS
- induced pluripotent stem cell(s).
REFERENCES
- 1. Fuchs E. (2007) Scratching the surface of skin development. Nature 445, 834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fuchs E., Raghavan S. (2002) Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 3, 199–209 [DOI] [PubMed] [Google Scholar]
- 3. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) p63 is essential for regenerative proliferation in limb, craniofacial, and epithelial development. Nature 398, 714–718 [DOI] [PubMed] [Google Scholar]
- 4. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 [DOI] [PubMed] [Google Scholar]
- 5. Senoo M., Pinto F., Crum C. P., McKeon F. (2007) p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129, 523–536 [DOI] [PubMed] [Google Scholar]
- 6. Koster M. I., Kim S., Mills A. A., DeMayo F. J., Roop D. R. (2004) p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 18, 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Truong A. B., Kretz M., Ridky T. W., Kimmel R., Khavari P. A. (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 20, 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carroll D. K., Carroll J. S., Leong C. O., Cheng F., Brown M., Mills A. A., Brugge J. S., Ellisen L. W. (2006) p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8, 551–561 [DOI] [PubMed] [Google Scholar]
- 9. Dlugosz A. A., Yuspa S. H. (1993) Coordinate changes in gene expression which mark the spinous to granular cell transition in epidermis are regulated by protein kinase C. J. Cell Biol. 120, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ogawa E., Okuyama R., Egawa T., Nagoshi H., Obinata M., Tagami H., Ikawa S., Aiba S. (2008) p63/p51-induced onset of keratinocyte differentiation via the c-Jun N-terminal kinase pathway is counteracted by keratinocyte growth factor. J. Biol. Chem. 283, 34241–34249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fuchs E. (2009) Finding one's niche in the skin. Cell Stem Cell 4, 499–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osterland C. K., Wilkinson R. D., St Louis E. A. (1990) Expression of c-myc protein in skin and synovium in psoriasis and psoriatic arthritis. Clin. Exp. Rheumatol. 8, 145–150 [PubMed] [Google Scholar]
- 13. Bull J. J., Müller-Röver S., Patel S. V., Chronnell C. M., McKay I. A., Philpott M. P. (2001) Contrasting localization of c-Myc with other Myc superfamily transcription factors in the human hair follicle and during the hair growth cycle. J. Invest. Dermatol. 116, 617–622 [DOI] [PubMed] [Google Scholar]
- 14. Jensen K. B., Watt F. M. (2006) Single-cell expression profiling of human epidermal stem and transit-amplifying cells. Lrig1 is a regulator of stem cell quiescence. Proc. Natl. Acad. Sci. U.S.A. 103, 11958–11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hashiro M., Matsumoto K., Okumura H., Hashimoto K., Yoshikawa K. (1991) Growth inhibition of human keratinocytes by antisense c-myc oligomer is not coupled to induction of differentiation. Biochem. Biophys. Res. Commun. 174, 287–292 [DOI] [PubMed] [Google Scholar]
- 16. Pietenpol J. A., Holt J. T., Stein R. W., Moses H. L. (1990) Transforming growth factor β1 suppression of c-myc gene transcription. Role in inhibition of keratinocyte proliferation. Proc. Natl. Acad. Sci. U.S.A. 87, 3758–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arnold I., Watt F. M. (2001) c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 11, 558–568 [DOI] [PubMed] [Google Scholar]
- 18. Gandarillas A., Watt F. M. (1997) c-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 11, 2869–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gebhardt A., Frye M., Herold S., Benitah S. A., Braun K., Samans B., Watt F. M., Elsässer H. P., Eilers M. (2006) Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J. Cell Biol. 172, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waikel R. L., Kawachi Y., Waikel P. A., Wang X. J., Roop D. R. (2001) Deregulated expression of c-Myc depletes epidermal stem cells. Nat. Genet. 28, 165–168 [DOI] [PubMed] [Google Scholar]
- 21. Watt F. M., Frye M., Benitah S. A. (2008) MYC in mammalian epidermis. How can an oncogene stimulate differentiation? Nat. Rev. Cancer 8, 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988) Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu X., Alder J. K., Chun J. H., Friedman A. D., Heimfeld S., Cheng L., Civin C. I. (2006) HES1 inhibits cycling of hematopoietic progenitor cells via DNA binding. Stem Cells 24, 876–888 [DOI] [PubMed] [Google Scholar]
- 24. Shimizu H., Julius M. A., Giarré M., Zheng Z., Brown A. M., Kitajewski J. (1997) Transformation by Wnt family proteins correlates with regulation of beta-catenin. Cell Growth Differ. 8, 1349–1358 [PubMed] [Google Scholar]
- 25. Le Brigand K., Russell R., Moreilhon C., Rouillard J. M., Jost B., Amiot F., Magnone V., Bole-Feysot C., Rostagno P., Virolle V., Defamie V., Dessen P., Williams G., Lyons P., Rios G., Mari B., Gulari E., Kastner P., Gidrol X., Freeman T. C., Barbry P. (2006) An open-access long oligonucleotide microarray resource for analysis of the human and mouse transcriptomes. Nucleic Acids Res. 34, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 27. Barbieri C. E., Tang L. J., Brown K. A., Pietenpol J. A. (2006) Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 66, 7589–7597 [DOI] [PubMed] [Google Scholar]
- 28. Viganò M. A., Lamartine J., Testoni B., Merico D., Alotto D., Castagnoli C., Robert A., Candi E., Melino G., Gidrol X., Mantovani R. (2006) New p63 targets in keratinocytes identified by a genome-wide approach. EMBO J. 25, 5105–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pozzi S., Zambelli F., Merico D., Pavesi G., Robert A., Maltère P., Gidrol X., Mantovani R., Vigano M. A. (2009) Transcriptional network of p63 in human keratinocytes. PLoS ONE 4, e5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mehic D., Bakiri L., Ghannadan M., Wagner E. F., Tschachler E. (2005) Fos and jun proteins are specifically expressed during differentiation of human keratinocytes. J. Invest. Dermatol. 124, 212–220 [DOI] [PubMed] [Google Scholar]
- 31. Slavik M. A., Allen-Hoffmann B. L., Liu B. Y., Alexander C. M. (2007) Wnt signaling induces differentiation of progenitor cells in organotypic keratinocyte cultures. BMC Dev. Biol. 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen B. C., Lefort K., Mandinova A., Antonini D., Devgan V., Della Gatta G., Koster M. I., Zhang Z., Wang J., Tommasi di Vignano A., Kitajewski J., Chiorino G., Roop D. R., Missero C., Dotto G. P. (2006) Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 20, 1028–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chin M. H., Mason M. J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J., Khvorostov I., Ott V., Grunstein M., Lavon N., Benvenisty N., Croce C. M., Clark A. T., Baxter T., Pyle A. D., Teitell M. A., Pelegrini M., Plath K., Lowry W. E. (2009) Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McKeon F. (2004) p63 and the epithelial stem cell. More than status quo? Genes Dev. 18, 465–469 [DOI] [PubMed] [Google Scholar]
- 35. Dai X., Segre J. A. (2004) Transcriptional control of epidermal specification and differentiation. Curr. Opin. Genet. Dev. 14, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Truong A. B., Khavari P. A. (2007) Control of keratinocyte proliferation and differentiation by p63. Cell Cycle 6, 295–299 [DOI] [PubMed] [Google Scholar]
- 37. Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massagué J., Hänel F., Eilers M. (2001) Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 3, 392–399 [DOI] [PubMed] [Google Scholar]
- 38. Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., Massagué J. (2001) TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 3, 400–408 [DOI] [PubMed] [Google Scholar]
- 39. Seoane J., Le H. V., Massagué J. (2002) Myc suppression of the p21Cip1 Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419, 729–734 [DOI] [PubMed] [Google Scholar]
- 40. Su X., Cho M. S., Gi Y. J., Ayanga B. A., Sherr C. J., Flores E. R. (2009) Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 28, 1904–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Westfall M. D., Mays D. J., Sniezek J. C., Pietenpol J. A. (2003) The ΔNp63α phosphoprotein binds the p21 and 14-3-3 σ promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell. Biol. 23, 2264–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Testoni B., Mantovani R. (2006) Mechanisms of transcriptional repression of cell cycle G2/M promoters by p63. Nucleic Acids Res. 34, 928–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blanpain C., Lowry W. E., Pasolli H. A., Fuchs E. (2006) Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, 3022–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rangarajan A., Talora C., Okuyama R., Nicolas M., Mammucari C., Oh H., Aster J. C., Krishna S., Metzger D., Chambon P., Miele L., Aguet M., Radtke F., Dotto G. P. (2001) Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Watt F. M., Estrach S., Ambler C. A. (2008) Epidermal Notch signaling. Differentiation, cancer, and adhesion. Curr. Opin. Cell Biol. 20, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ito M., Yang Z., Andl T., Cui C., Kim N., Millar S. E., Cotsarelis G. (2007) Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447, 316–320 [DOI] [PubMed] [Google Scholar]
- 47. Devgan V., Mammucari C., Millar S. E., Brisken C., Dotto G. P. (2005) p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 19, 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Estrach S., Ambler C. A., Lo Celso C., Hozumi K., Watt F. M. (2006) Jagged 1 is a β-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development 133, 4427–4438 [DOI] [PubMed] [Google Scholar]
- 49. Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M. B., Guzzardo V., Parenti A. R., Rosato A., Bicciato S., Balmain A., Piccolo S. (2009) A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 [DOI] [PubMed] [Google Scholar]
- 50. Reuter J. A., Ortiz-Urda S., Kretz M., Garcia J., Scholl F. A., Pasmooij A. M., Cassarino D., Chang H. Y., Khavari P. A. (2009) Modeling inducible human tissue neoplasia identifies an extracellular matrix interaction network involved in cancer progression. Cancer Cell 15, 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Müller E. J., Williamson L., Kolly C., Suter M. M. (2008) Outside-in signaling through integrins and cadherins. A central mechanism to control epidermal growth and differentiation? J. Invest. Dermatol. 128, 501–516 [DOI] [PubMed] [Google Scholar]
- 52. Watt F. M. (2002) Role of integrins in regulating epidermal adhesion, growth, and differentiation. EMBO J. 21, 3919–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nickoloff B. J., Qin J. Z., Chaturvedi V., Denning M. F., Bonish B., Miele L. (2002) Jagged-1-mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-κB and PPARγ. Cell Death Differ. 9, 842–855 [DOI] [PubMed] [Google Scholar]
- 54. Sasaki Y., Ishida S., Morimoto I., Yamashita T., Kojima T., Kihara C., Tanaka T., Imai K., Nakamura Y., Tokino T. (2002) The p53 family member genes are involved in the Notch signal pathway. J. Biol. Chem. 277, 719–724 [DOI] [PubMed] [Google Scholar]
- 55. Yano S., Banno T., Walsh R., Blumenberg M. (2008) Transcriptional responses of human epidermal keratinocytes to cytokine interleukin-1. J. Cell. Physiol. 214, 1–13 [DOI] [PubMed] [Google Scholar]
- 56. Maas-Szabowski N., Stärker A., Fusenig N. E. (2003) Epidermal tissue regeneration and stromal interaction in HaCaT cells is initiated by TGF-α. J. Cell Sci. 116, 2937–2948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.