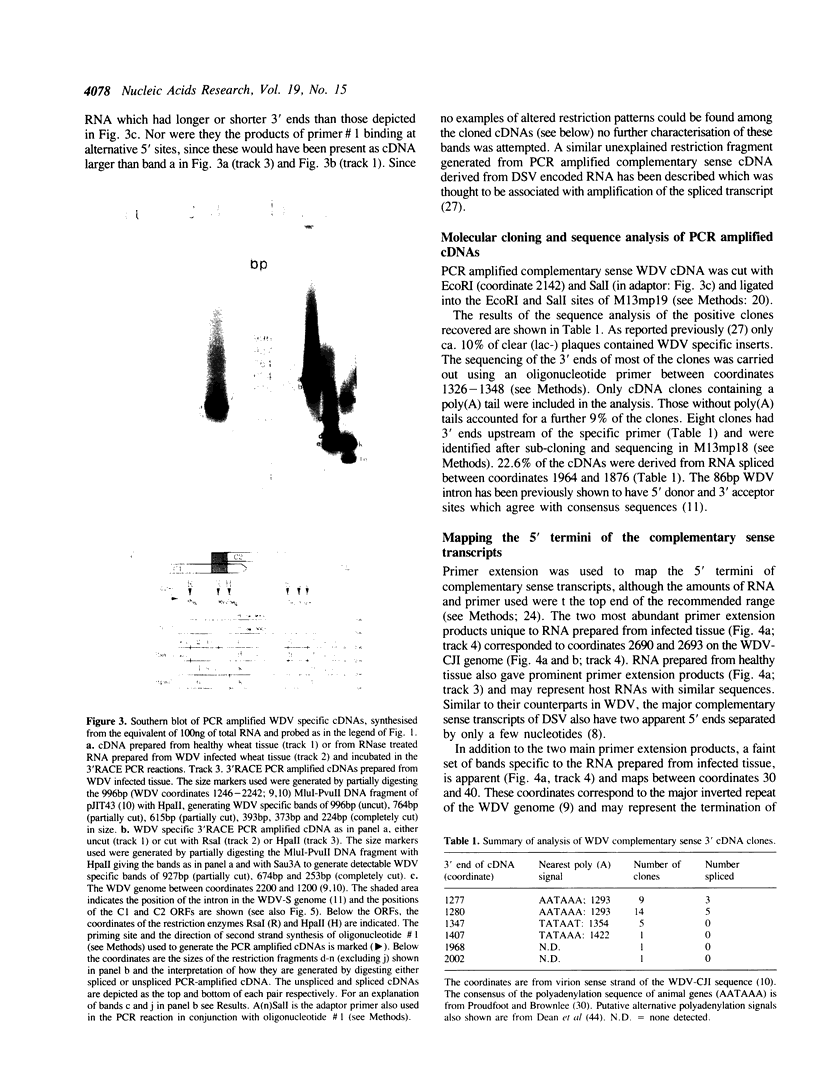
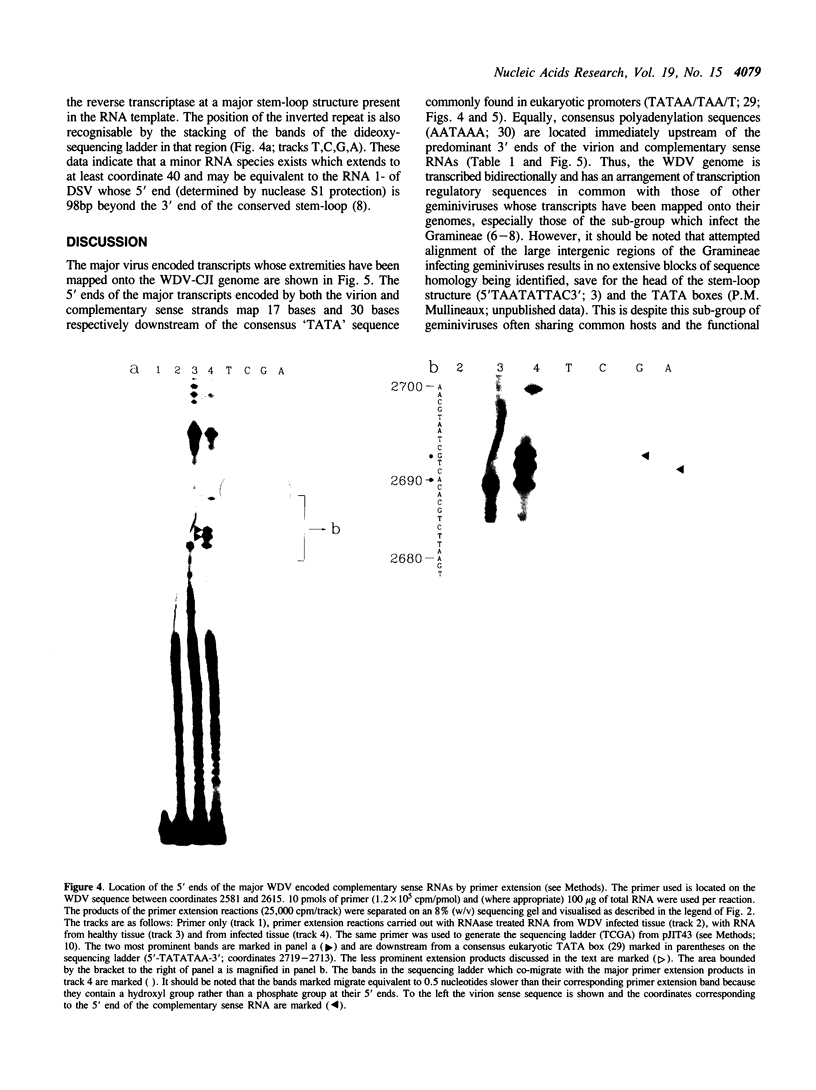
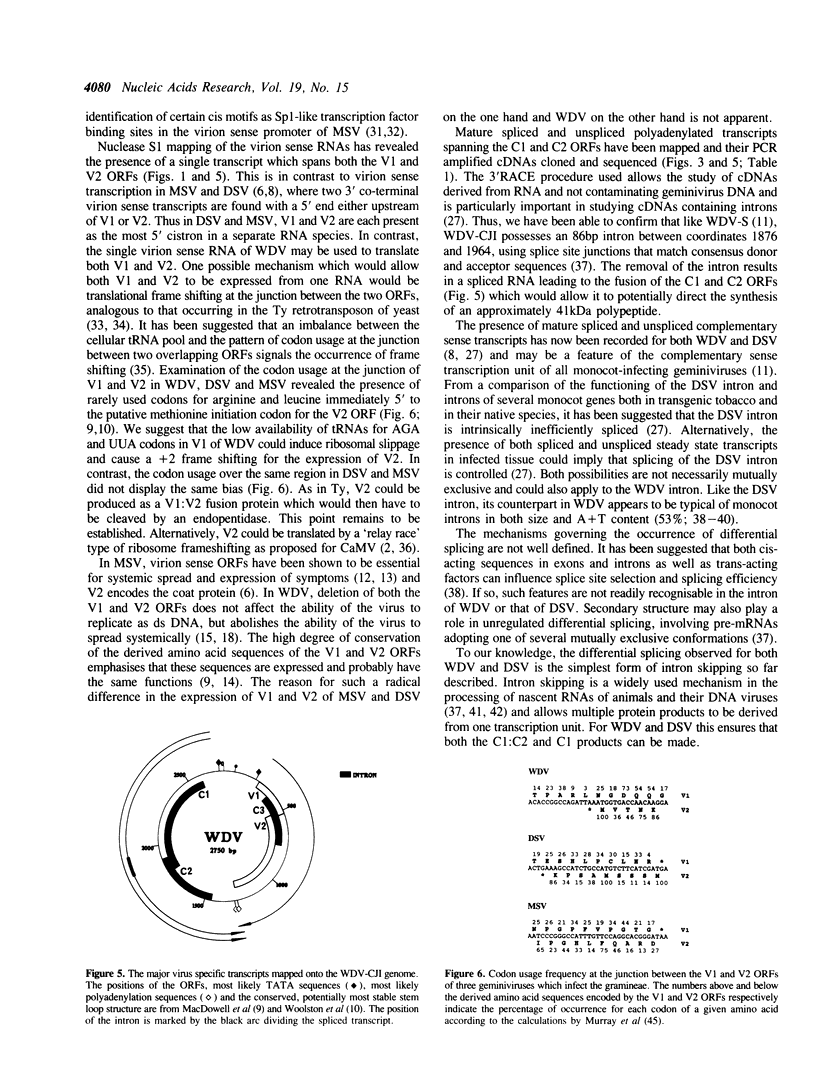
Abstract
We have characterised the major transcripts of the Czech isolate of wheat dwarf virus (WDV-CJI) which show that WDV uses two different mechanisms for expressing overlapping open reading frames (ORFs). Mapping of the virion sense RNAs identified a single polyadenylated transcript of 1.1kb spanning the overlapping ORFs V1 and V2 which encode cell-cell spread functions and the coat protein respectively. This finding distinguishes WDV from other monocot-infecting geminiviruses studied so far which were shown to encode two 3' co-terminal transcripts capable of expressing either the V1 or V2 ORF. A survey of codon usage at the junction between the V1 and V2 ORF has led us to propose that translational frame shifting analogous to that in the yeast Ty element may occur. Analysis of polymerase chain reaction (PCR) amplified complementary sense cDNA clones has revealed the presence of mature spliced and unspliced RNAs which could encode products of an intron mediated C1:C2 ORF fusion or the C1 ORF product alone. Mapping of the 5' and 3' extremities of the major WDV encoded transcripts has allowed us to identify putative transcription regulatory sequences and the presence of multiple overlapping transcripts may suggest temporal regulation of transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Accotto G. P., Donson J., Mullineaux P. M. Mapping of Digitaria streak virus transcripts reveals different RNA species from the same transcription unit. EMBO J. 1989 Apr;8(4):1033–1039. doi: 10.1002/j.1460-2075.1989.tb03470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
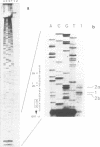
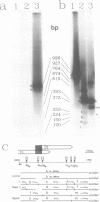
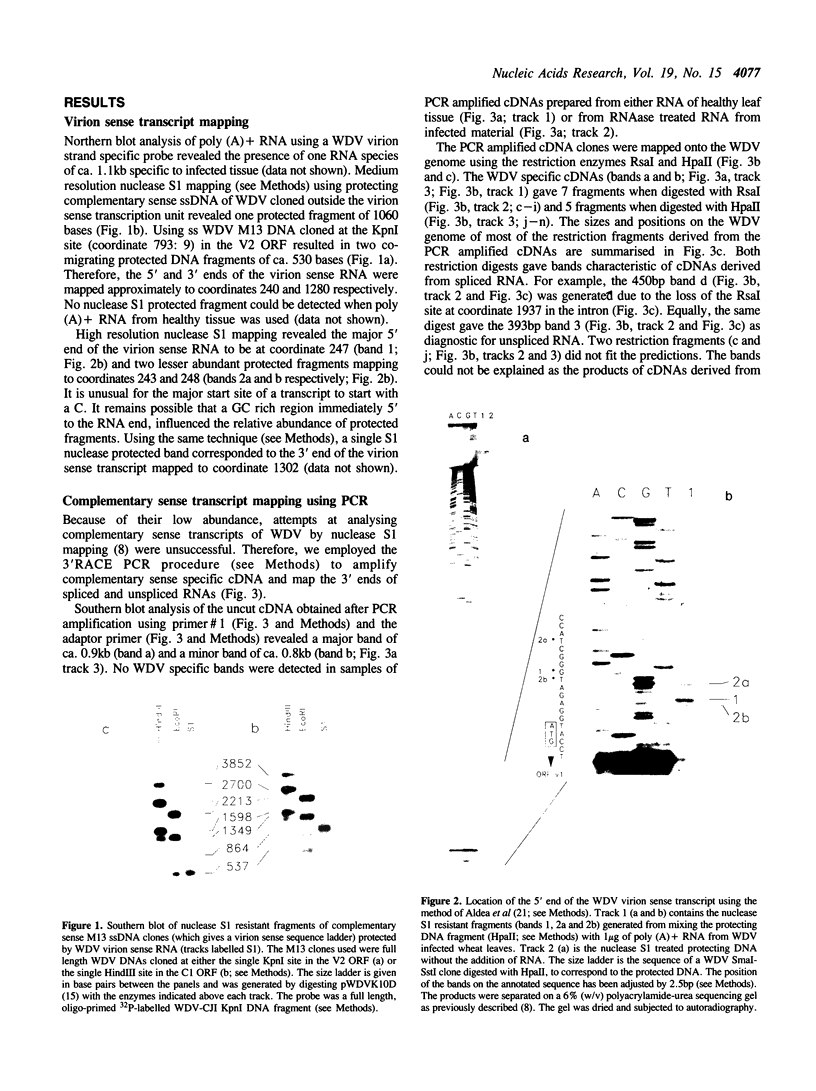
- Aldea M., Claverie-Martín F., Díaz-Torres M. R., Kushner S. R. Transcript mapping using [35S]DNA probes, trichloroacetate solvent and dideoxy sequencing ladders: a rapid method for identification of transcriptional start points. Gene. 1988 May 15;65(1):101–110. doi: 10.1016/0378-1119(88)90421-0. [DOI] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brooks-Gunn J., Warren M. P. Mother-daughter differences in menarcheal age in adolescent girls attending national dance company schools and non-dancers. Ann Hum Biol. 1988 Jan-Feb;15(1):35–43. doi: 10.1080/03014468800009441. [DOI] [PubMed] [Google Scholar]
- Davies J. W., Stanley J., Donson J., Mullineaux P. M., Boulton M. I. Structure and replication of geminivirus genomes. J Cell Sci Suppl. 1987;7:95–107. doi: 10.1242/jcs.1987.supplement_7.7. [DOI] [PubMed] [Google Scholar]
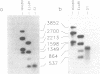
- Dean C., Tamaki S., Dunsmuir P., Favreau M., Katayama C., Dooner H., Bedbrook J. mRNA transcripts of several plant genes are polyadenylated at multiple sites in vivo. Nucleic Acids Res. 1986 Mar 11;14(5):2229–2240. doi: 10.1093/nar/14.5.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L. K., Hohn T. Initiation of translation of the cauliflower mosaic virus genome from a polycistronic mRNA: evidence from deletion mutagenesis. EMBO J. 1984 Dec 1;3(12):2731–2736. doi: 10.1002/j.1460-2075.1984.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckworth M. L., Gait M. J., Goelet P., Hong G. F., Singh M., Titmas R. C. Rapid synthesis of oligodeoxyribonucleotides VI. Efficient, mechanised synthesis of heptadecadeoxyribonucleotides by an improved solid phase phosphotriester route. Nucleic Acids Res. 1981 Apr 10;9(7):1691–1706. doi: 10.1093/nar/9.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fenoll C., Black D. M., Howell S. H. The intergenic region of maize streak virus contains promoter elements involved in rightward transcription of the viral genome. EMBO J. 1988 Jun;7(6):1589–1596. doi: 10.1002/j.1460-2075.1988.tb02984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989 Aug 11;58(3):473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Goodall G. J., Filipowicz W. The minimum functional length of pre-mRNA introns in monocots and dicots. Plant Mol Biol. 1990 May;14(5):727–733. doi: 10.1007/BF00016505. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hanley B. A., Schuler M. A. Plant intron sequences: evidence for distinct groups of introns. Nucleic Acids Res. 1988 Jul 25;16(14B):7159–7176. doi: 10.1093/nar/16.14.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi C. P. Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res. 1987 Dec 10;15(23):9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Regulation of protein synthesis in virus-infected animal cells. Adv Virus Res. 1986;31:229–292. doi: 10.1016/S0065-3527(08)60265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski F. A., Rio D. C., Rubin G. M. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986 Jan 17;44(1):7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S. G., Pinder A. J., Damsteegt V. D., Rogers S. G. Maize streak virus genes essential for systemic spread and symptom development. EMBO J. 1989 Apr;8(4):1023–1032. doi: 10.1002/j.1460-2075.1989.tb03469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDowell S. W., Macdonald H., Hamilton W. D., Coutts R. H., Buck K. W. The nucleotide sequence of cloned wheat dwarf virus DNA. EMBO J. 1985 Sep;4(9):2173–2180. doi: 10.1002/j.1460-2075.1985.tb03912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morris-Krsinich B. A., Mullineaux P. M., Donson J., Boulton M. I., Markham P. G., Short M. N., Davies J. W. Bidirectional transcription of maize streak virus DNA and identification of the coat protein gene. Nucleic Acids Res. 1985 Oct 25;13(20):7237–7256. doi: 10.1093/nar/13.20.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux P. M., Guerineau F., Accotto G. P. Processing of complementary sense RNAs of Digitaria streak virus in its host and in transgenic tobacco. Nucleic Acids Res. 1990 Dec 25;18(24):7259–7265. doi: 10.1093/nar/18.24.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E. E., Lotzer J., Eberle M. Codon usage in plant genes. Nucleic Acids Res. 1989 Jan 25;17(2):477–498. doi: 10.1093/nar/17.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Schalk H. J., Matzeit V., Schiller B., Schell J., Gronenborn B. Wheat dwarf virus, a geminivirus of graminaceous plants needs splicing for replication. EMBO J. 1989 Feb;8(2):359–364. doi: 10.1002/j.1460-2075.1989.tb03385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G., Gardiner W. E., Bisaro D. M. Identification of tomato golden mosaic virus-specific RNAs in infected plants. Virology. 1989 May;170(1):243–250. doi: 10.1016/0042-6822(89)90372-3. [DOI] [PubMed] [Google Scholar]
- Townsend R., Stanley J., Curson S. J., Short M. N. Major polyadenylated transcripts of cassava latent virus and location of the gene encoding coat protein. EMBO J. 1985 Jan;4(1):33–37. doi: 10.1002/j.1460-2075.1985.tb02313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpfer R., Gronenborn B., Schell J., Steinbiss H. H. Uptake and transient expression of chimeric genes in seed-derived embryos. Plant Cell. 1989 Jan;1(1):133–139. doi: 10.1105/tpc.1.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugaki M., Ueda T., Timmermans M. C., Vieira J., Elliston K. O., Messing J. Replication of a geminivirus derived shuttle vector in maize endosperm cells. Nucleic Acids Res. 1991 Jan 25;19(2):371–377. doi: 10.1093/nar/19.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolston C. J., Reynolds H. V., Stacey N. J., Mullineaux P. M. Replication of wheat dwarf virus DNA in protoplasts and analysis of coat protein mutants in protoplasts and plants. Nucleic Acids Res. 1989 Aug 11;17(15):6029–6041. doi: 10.1093/nar/17.15.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Boeke J. D. Host genes that influence transposition in yeast: the abundance of a rare tRNA regulates Ty1 transposition frequency. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8360–8364. doi: 10.1073/pnas.87.21.8360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zhang H., Scholl R., Browse J., Somerville C. Double stranded DNA sequencing as a choice for DNA sequencing. Nucleic Acids Res. 1988 Feb 11;16(3):1220–1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]