1. Introduction and overview of cyclooxygenase pathways
Prostanoids are cyclic, oxygenated products of ω3 and ω6 20- and 22-carbon essential fatty acids (FAs) that are formed enzymatically through “cyclooxygenases”. Prostaglandin endoperoxide synthases -1 and -2 (PGHS-1 and -2)a, which are also known as cyclooxygenases -1 and -2 (COX-1 and -2), catalyze the committed step in the biosynthesis of prostanoids (Figure 1). These compounds include what are sometimes referred to as the “classical” prostaglandins (PGs) PGD, PGE, and PGF as well as prostacyclins denoted as PGI's and the thromboxanes abbreviated Tx's; monohydroxy acids can also be formed via PGHSs, but information on the possible physiologic importance of such compounds is incomplete.1-3 The most abundant prostanoids are the “2-series” compounds (e.g. PGE2) that are formed from arachidonic acid (AA; 5Z, 8Z, 11Z, 14Z- eicosatetraenoic acid; 20:4 ω6; Figure 1). The “2” denotes the number of carbon-carbon double bonds in the product.
Figure 1. Biosynthetic pathway for the formation of prostanoids.
Generally, a given cell type forms only one or two of these products in abundance. For example, circulating human platelets form primarily thromboxanes. Abbreviations include: cPLA2, cytosolic phospholipase A2; sPLA2, nonpancreatic, secretory phospholipase A2; PG, prostaglandin; PGHS, prostaglandin endoperoxide H synthase; COX, cyclooxygenase; POX, peroxidase; H-PGDS, hematopoietic PGD synthase; L-PGDS, lipocalin-type PGD synthase; cPGES, cytosolic PGE synthase; mPGES-1, microsomal PGE synthase-1; PGFS, PGF synthase; PGIS, PGI (prostacyclin) synthase; TXAS, TxA synthase. DP1, DP2, EP1, EP2, EP3, EP4, FP, IP and TP are designations for the G protein linked PG receptors. Reprinted with permission from Reference 9; copyright 2008 Elsevier.
PGHSs catalyze two distinct reactions that occur at physically distinct but functionally interacting sites. The cyclooxygenase (COX) reaction is a bis-oxygenation in which two O2 molecules are inserted into the carbon backbone of AA to yield PGG2 (Figure 1). The peroxidase (POX) reaction is a transformation in which the 15-hydroperoxyl group of PGG2 undergoes a net two electron reduction to PGH2 plus water. The POX reaction is important in the enzyme mechanism, but other peroxidases such as glutathione peroxidase may contribute importantly to the reduction of PGG2 to PGH2 in vivo.
PGH2 is thought not to accumulate in cells but rather to be converted quickly to what are considered the biologically relevant, downstream products. There are specific synthases involved in forming PGD2, PGE2, PGF2α, PGI2 and TxA2 from PGH2. Except for the case of PGF2α, which is formed by a two electron reduction of PGH2, these enzymes catalyze non-oxidative rearrangements. Finally, there is a PGH 19-hydoxylase that converts PGHs to their corresponding 19-hydroxy derivatives which themselves are substrates for PGE synthase(s). There are one or more specific G protein-linked receptors for each prostanoid, and in some cases prostanoids may also act through nuclear peroxisome proliferator activated receptors.
AA and other 20- and 22-carbon, highly unsaturated FAs are found esterified at the sn2 position of glycerophospholipids. Basal prostanoid formation generally occurs at a low rate relative to stimulated formation. A major factor limiting prostanoid formation is AA availability, which is controlled through the net rates of deacylation and reacylation of glycerophospholipids. Prostanoid formation is enhanced when phospholipase A2 (PLA2) activity is increased, and thus PLA2s play a substrate-limiting role in regulating prostanoid biosynthesis. Although reacylation may also be important, its possible role in regulating prostanoid biosynthesis is largely unexplored.
In this chapter we review the biochemistry and the biochemical pharmacology of the enzymes involved in converting AA to various prostanoid products. These enzymes include PGHS-1, PGHS-2, hematopoietic PGD synthase (H-PGDS), lipocalin-type PGD synthase (L-PGDS), microsomal PGE synthase-1 (mPGES-1), microsomal PGE synthase-2 (mPGES-2), cytosolic PGE synthase (cPGES), PGF synthase (PGFS), PGI synthase (PGIS) and TXA synthase (TXAS). The PLA2s involved in mobilizing AA and the receptors through which prostanoids function are surveyed in other chapters of this volume.
2. Prostaglandin endoperoxide H synthases (PGHSs)
We last provided a comprehensive review of PGHSs in 2000.4 Since then several cogent reviews have appeared that describe specifics of enzyme mechanism, regulation of expression, COX pharmacology and roles of PGHSs in disease processes.5-13 Here we focus on advances in studies of enzyme mechanism and biochemical pharmacology that have occurred during the last ten years with particular emphasis on the functioning of PGHSs as conformational heterodimers.
2.1. Primary structure and numbering PGHSs
PGHS-1 and PGHS-2 are the products of different genes. Ovine (ov) PGHS-1 was the first PGHS whose cDNA was cloned and sequenced.14-16 The sequence encoded a protein with an open reading frame of 600 amino acids (Figures 2 and 3). Following removal of the signal peptide, the mature protein has 576 amino acids;17,18 this is also true of other mammalian versions of this isoform. Based on the N-terminal sequence of purified PGHS-219 and assuming there is no further processing, the mature forms of PGHS-2 have 587 amino acids. Figure 2 compares the deduced amino acid sequences of several mammalian PGHS. More detailed amino acid sequence comparisons of PGHSs from multiple sources and a discussion of evolutionary relationships among PGHSs and related plant oxygenases can be found in a review by Simmons et al.6. There is 60%–65% sequence identity between PGHS-1 and 2 from the same species and 85%–90% identity among individual isoforms from different mammalian species.4
Figure 2. Comparison of amino acid sequences of ovine, murine and human PGHSs.
Numbering of PGHSs utilizes the sequence of ovPGHS-1 as a reference for alignment. For ovPGHS-1 the signal peptide is shown in yellow, the EGF-like domain is shown in green and the membrane binding domain with its four helices is shown in cyan. Numbering of ovPGHS-1 from 1-600 begins with the Met at the translation start site. Ala-25 is the N-terminal residue of the mature protein. Beginning with Phe-107 all of the sequences are appropriately aligned until approximately residue 584. Immediately upstream of Phe-107, there is an additional residue in PGHS-2. In addition the signal peptides differ in lengths. Finally, PGHS-2 contains eight more residues at the C-terminus than PGHS-1. The net result is that mature forms of PGHS-1 contain 576 residues while mature forms of PGHS-2 contains 587 residues. Immediately upstream of the C-terminal ER-retention signal STEL, which is highlighted in brown, PGHS-2 contains an 19 amino acid sequence (underlined) that contains Asn-594, a consensus glycosylation site unique to PGHS-2; the 27-amino acid instability motif (27-AA IM) found near the C-terminus of PGHS-2 and shown in dark yellow includes this 19 amino acid insert. There are three N-glycosylation sites common to both isoforms including Asn-68, Asn-144 and Asn-410 shown in red. Some other important residues are also shown in red. Arg-120 is involved in interacting with the carboxyl group of fatty acid substrates, Tyr-385 is the tyrosyl residue involved in abstracting a hydrogen from the substrate fatty acid, His-388 is the proximal heme ligand at the POX active site, Tyr-504 is a tyrosine radical reservoir, and Ser-530 is the site of acetylation by aspirin. Cys-37 links the EGF-like domain to the catalytic domain via Cys-159. Ovine PGHS-1, NCBI Reference Sequence: NP_001009476.1; murine PGHS-1, NCBI Reference Sequence: NP_032995.1; human PGHS-1, NCBI Reference Sequence: NP_000953.2; human PGHS-2, NCBI Reference Sequence: NP_032995.1; murine PGHS-2, NCBI Reference Sequence: NP_035328.2.
Figure 3. Domain structures of human PGHSs.
Comparison of the domain structures and numbering of huPGHS-1 and huPGHS-2. Numbering is based on the ovPGHS-1 template noted in the text and the legend to Figure 2. The mature forms of huPGHS-1 and huPGHS-2 contain 576 and 587 residues, respectively. Both isoforms have a signal peptide (S), an epidermal growth factor (EGF)-like domain, a membrane-binding domain (MBD), and a catalytic domain. PGHS-2 has a shorter signal peptide (17 amino acids) than PGHS-1 (23 amino acids). The C-terminal STEL sequences of PGHS-1 and PGHS-2, respectively, are weak ER targeting signals. The asparagine N-glycosylation sites are shown; note the additional N-glycosylation site at Asn-594 in PGHS-2. Arg-120, Tyr-385 and His-388 are important in catalysis. Tyr-504 can serve as a reservoir for the tyrosine radical. Ser-530 is the aspirin acetylation site. The amino acid sequence of the 27 amino acid instability motif (IM) comprising residues 586-612 is shown.
The convention for numbering amino acid residues in proteins is to number the methionine at the translational start site as residue number 1. This system is useful in comparing human (hu) PGHS variants associated with disease risk such as those arising from single nucleotide polymorphisms.20-23
There is a major limitation to using the conventional numbering system in describing the results of biochemical and structural studies that frequently involve comparing homologous residues among PGHSs. The problem arises because PGHS-1 forms from different species have signal peptides of different lengths and because PGHS-2 has a proline residue, not found in PGHS-1, located 75 residues in from the N-terminus of mature mammalian PGHSs. Therefore, a second, alternative numbering system has developed that uses the numbering of processed ovine (ov) PGHS-1 as the template (Figure 2). In this alternative system, the N-terminal alanine of mature ovPGHS-1 is designated Ala-25 for all PGHSs. This numbering system functions adequately for mature forms of the enzymes from the most widely studied species although one is left with a spare residue at what, in this alternate numbering system, is position 105 of the PGHS-2 sequence. Beyond residue 107, the alternate system provides one with the ability to compare the same residues among different species and isoforms. For example, to apply the ovPGHS-1 numbering system to huPGHS-1, all that is needed is to add one integer to the conventional numbering (i.e. the true Ser-529 in huPGHS-1 is numbered as Ser-530 because huPGHS-1 has one less amino acid in its signal peptide than does ovPGHS-1). To number huPGHS-2 using the ovPGHS-1 system, one adds 14 residues (i.e. the true Arg-106 in huPGHS-2 is denoted as Arg-120 in the ovPGHS-1 system). Unless otherwise noted, we have used the ovPGHS-1 system (Figure 2) in this chapter.
An mRNA splice variant of PGHS-1 that retains intron-1 was identified in canine brain and dubbed “COX-3”.6 A canine protein reported to correspond to this variant sequence was described as responding in an unusual manner to acetaminophen; however, the results were not compelling, and related splice variants in most species including humans are out of frame.6
2.2. Co-translational and post-translational modifications
One can infer from the N-terminal amino acid sequence17,18 and the sequence of cDNAs of ovPGHS-114-16 and the fact that PGHSs are N-glycosylated24,25 that signal sequences are cleaved co-translationally during the synthesis of PGHSs and their entry into the lumen of the endoplasmic reticulum.
PGHS-1 and PGHS-2 have three common N-glycosylation sites at Asn-68, Asn-144 and Asn-41025-30 that appear to be glycosylated co-translationally30,31 (Figures 2 and 3). NMR and mass spectral analyses of the carbohydrate chains of PGHSs indicate that the structures of the N-glycosyl moieties are of the form (Man)n(GlcNAc)2(n = 6-9)32-34. N-glycosylation of Asn-410 appears to be important in the folding of ovPGHS-1 to an active structure.26 As detailed below, PGHSs function as conformational heterodimers with a single, catalytically active monomer defined by its ability to bind heme with high affinity.35 It is not clear whether differences in co-translational N-glycosylation of the monomers comprising a dimer contribute to the existence of the conformational heterogeneity.
A fourth N-glycosylation site present only in PGHS-2 is located at Asn-594 (Figures 2 and 3). This residue is seen in its unglycosylated form in one muPGHS-2 crystal structure36 (PDB 1CVU). N-glycosylation of this site appears to occur post-translationally as opposed to co-translationally and is associated with enzyme degradation and/or enzyme trafficking.30,31,37
2.3 Domain structures of PGHSs
Figure 3 compares the domain structures of huPGHS-1 and huPGHS-2. The numbering of the amino acids corresponds to the numbering in Figure 2, which uses the ovPGHS-1 numbering as the template. Figure 4 is a ribbon diagram of the x-ray crystal structure of ovPGHS-1 (PDG 1CQE) complexed with an inhibitor flurbiprofen (FBP). The major domains are color coded to match the sequence and domain diagrams in Figures 2 and 3.
Figure 4. Structures of ovine PGHS-1.
Ribbon diagrams of (A) ovPGHS dimer with flurbiprofen bound in the COX active sites (PDB 1CQE).28 (B) Monomer of the structure in (A). Epidermal growth factor (EGF) domains in green; membrane binding domains (MBD) in cyan; catalytic domains in dark and light blue; heme groups at the POX active sites are shown in red; carbohydrate groups (N-acetyl-glucosamines) are in black/gray; flurbiprofen in yellow.
2.3.1 Signal peptides and the N-termini
The signal peptides of PGHS-1 are 22-26 amino acids long while those of PGHS-2 are all 17 amino acids long (Figures 2 and 3). There is relatively little sequence conservation between signal peptides from the two isoforms. The rate of translation has been reported to be greater for PGHS-1 than PGHS-2 protein in an in vitro system, and this has been attributed to differences in the signal peptides;6 however, this issue has not been addressed in sufficient detail to make any generalizations. A signal nucleotide polymorphism in the signal peptide of huPGHS-1 that changes the amino acid sequence (P17L) shows a modest association with a lack of an effect of traditional nonsteroidal anti-inflammatory drugs (NSAIDs) use on the development of colorectal polyps related to colon cancer.22
Mature PGHS-1 has eight amino acids at its N-terminus that are absent in mature PGHS-2 (Figures 2 and 3). A mutant in which the first seven amino acids of the mature ovPGHS-1 are deleted is reported to have the same KM but a much reduced COX VMAX compared to native ovPGHS-1.38 This eight amino acid segment is not seen in ovPGHS-1 crystal structures28,39,40 and so is apparently unstructured. It is possible that this segment plays some regulatory role perhaps in cross talk between the monomers comprising a PGHS dimer.
2.3.2. Epidermal growth factor (EGF)-like domains
PGHSs contain an EGF-like domain of about 50 amino acids (Figures 2-4). In ovPGHS-1 the EGF-like domain begins at residue 33 and extends to at least residue 72 with additional homology beyond that point.28 There is about 59% amino acid sequence identity between the EGF-like domains of PGHS-1 and PGHS-2 within a species. The EGF-like domains contain seven cysteine residues. Six of these are involved in intrachain disulfide bonds that generate a small β-sandwich very similar to that of EGF.28 The other cysteine in the EGF-like domain, Cys-37, forms a disulfide bond with Cys-159 of the catalytic domain thereby linking the EGF-like and catalytic domains. A Cys69Ser mutant of ovPGHS-1 lacks activity suggesting, not surprisingly, that an intact EGF-like domain is essential for the structural integrity of the protein.4 One surface of the EGF-like domain forms a part of the interface between the monomers comprising the PGHS homodimer. The carbohydrate moiety found at Asn-68 faces away from the catalytic domain and towards the solvent. Disruption of N-glycosylation of ovPGHS-1 at Asn-68/67 diminishes but does not eliminate COX or POX activities.26 EGF-like domains are often involved in protein-protein interactions. Garavito and colleagues provide more details about the EGF-like domain and its characteristics and an erudite discussion of the potential role of this domain in the interactions between the two monomers in their landmark paper describing the first crystal structure of a PGHS-1.28
2.3.3. Membrane binding domains (MBDs)
PGHSs are integral membrane proteins in the empirical sense that detergent is required for their solubilization.41 The topic of how PGHSs interact with membranes was a confusing one until the ovPGHS-1 crystal structure was determined (Figure 4).28 It was clear from the structure that there are no transmembrane domains of the helical or β-barrel type. Rather, there are four consecutive short amphipathic α-helices on one surface of each monomer. The hydrophobic residues point away from the body of the protein and form the outlines of a rectangle with the center of the rectangle providing an opening from the membrane into the COX active site28 (Figure 4). This portion of the protein, which involves residues 72-116, was designated as the membrane binding domain (MBD). While, it was clear that this segment of the protein might anchor ovPGHS-1 in one surface of the bilayer, it was also conceivable that membrane binding also involved some associated, transmembrane protein. Subsequent studies using mutagenesis and reagents that label segments of protein or specific amino acids that are located in the membrane have provided evidence that the four α-helices of PGHSs insert directly into the membrane42-44 and that the protein can associate directly with liposomes.44 Although there are certainly proteins that interact with PGHSs during their synthesis, trafficking and degradation, there is as yet no compelling evidence for interacting proteins that anchor PGHSs to biological membranes. Thus, PGHSs are monotopic membrane proteins that insert into a single face of a lipid bilayer by virtue of their ability to bind lipids in membrane bilayers. Similar lipid-interacting motifs are found in squalene-hopene cyclase45 and fatty acid amide hydrolase.46 Like PGHSs both of these enzymes are dimers, their MBDs both point in the same direction, and each monomer provides an entrance to the active site.
It is worth noting that the major sequence difference between PGHS isoforms occurs in the MBD where there is 38% identity.42,43 This is of unknown biologic significance, but there are some differences in the sensitivities to isoforms from the same organelle to detergent solubilization.43 It is not known if differences in the sequences contribute to subtle differences in the subcellular distribution of the two isoforms, a topic that is discussed in further detail below.
2.3.4. Catalytic domains
The catalytic domains of PGHSs are large globular domains that encompass about 460 residues. This domain begins just before Arg-120 near the end of Helix D that contributes to the MBD. It had been known from sequence comparisons and the fact that PGHSs possess POX activity that these enzymes were somehow related to other peroxidases. The original x-ray structure demonstrated that the closest structural relative of ovPGHS-1 is myeloperoxidase.28 One can view PGHSs as peroxidases having a hydrophobic channel that extends about 25 Å from the MBD to Gly-533. This channel is the COX active site. COX inhibitors—traditional NSAIDs and COX-2 inhibitors called coxibs—and both substrate and non-substrate FAs bind within the COX channel. However, as will be discussed later the two COX channels differ in solution and each FA and each inhibitor has a preference for binding one of the two channels. As discussed below, differences between the two monomers comprising a dimer are not apparent in most x-ray crystal structures, most of which have been determined using relatively high concentrations of both COX ligands and Fe3+-heme (or Co3+-heme).
2.3.5. C-termini of PGHSs
The last six residues in the sequences of both isoforms can be considered to be the same or homologous (Figures 2 and 3). PGHSs have a sequence at the very C-terminus—PTEL in ovPGHS-1 and STEL in most other forms—that is involved in ER retention of the proteins. This is a “weak” ER retention signal47 and probably provides for the ability of PGHSs to traffic to the Golgi apparatus.
The last amino acid visible in the structure of ovPGHS-1 is Arg-586, which is 14 amino acids in from the C-terminus. If one considers Arg-586 to be the end of the catalytic domain of ovPGHS-1, then there remain eight residues near the end of ovPGHS-1 whose function is unknown (i.e. residues 587 to 594). There is no sequence homology with any contiguous eight residues in PGHS-2.
The C-terminal region of PGHS-2 is 19 amino acids longer than that of PGHS-1. These 19 residues and at least another nine or ten residues immediately upstream encompassing a 27 amino acid instability motif (Figures 2 and 3) are involved in aspects of protein degradation and/or trafficking. As noted above Asn-594 is post-translationally N-glycosylated in association with PGHS-2 degradation. Residues both upstream and downstream of Asn-594 appear to regulate the glycosylation.7,30,31,37 One structure of PGHS-2 is complete through Ser-59636 (PDB 1CVU) .
2.3.6. PGHS quaternary structures—PGHS homodimers
PGHSs are sequence homodimers comprised of tightly associated monomers with identical primary structures (Figure 4).24,28,29,48 Each monomer comprising a PGHS homodimer has a physically distinct COX and POX active site. The interface between the monomers is extensive,28,29 and dissociation of the dimers into monomers only occurs upon denaturation.48
Most of the x-ray crystal structures of PGHSs filed in the Worldwide Protein Data Bank (http://www.wwpdb.org/) have been determined using crystals formed in the presence of a minimum of two Fe3+-protoporphyrin IX (i.e. heme) or two Co3+-protoporphyrin IX groups per dimer and a 5-10 fold molar excess of either a COX substrate or inhibitor.28,29,39 Crystals prepared in this way have an asymmetric unit containing a biological dimer composed of monomers that have apparently identical structures related by a non-crystallographic two-fold symmetry axis. The highest resolution structure is at 1.7 Å.49 It is now clear that the two monomers comprising PGHS homodimers have different conformations and function cooperatively during catalysis in solution2,35,40,50-58. Recently, structures of ovPGHS-140,56 and muPGHS-235,57,58 have been determined in which the monomers are occupied by different ligands, ligands that are oriented differently in the two COX sites or by solvent molecules. In these cases, some subtle differences are apparent between the monomers.
The earliest evidence that monomers of PGHS homodimers differ in solution was reported in 1984. Kulmacz and coworkers showed that maximal COX activity of ovPGHS-1 occurred with one heme per dimer;50 recent studies with PGHS-2 have indicated that this isoform is also maximally functionally with one heme per dimer.35 Additionally, FBP and other traditional NSAIDs were found to inhibit PGHS-1 at a stoichiometry of one NSAID per dimer.51 There was no ready explanation for these earlier results, and they were not immediately pursued. Twenty years later, evidence was obtained that huPGHS-2 exhibits half of sites COX activity with AA as the substrate53 and that the monomers comprising ovPGHS-1 and muPGHS-2 homodimers can have slightly different crystal structures.35,40,56,57 Indeed, the enzymes function as conformational heterodimers with an allosteric (Eallo) and a catalytic (Ecat) monomer. This introduces a level of complexity that we will consider later in discussing the COX and POX reactions and their regulation.
2.4. Cyclooxygenase reaction
The basic kinetic properties of PGHS-1 and PGHS-2 are summarized in Table 1. Both enzymes catalyze the same reactions and have similar structures. However, there are subtle kinetic differences between the isozymes as well as some major differences in substrate and inhibitor specificities some of which are discussed below.
Table 1.
Apparent kinetic parameters for prostanoid biosynthetic enzymes.
| Enzyme | Substrate(s)/Cofactor(s) | kcat (s-1) | KM (μM) | kcat/KM (μM-1 s-1) | Comments/References |
|---|---|---|---|---|---|
| ovPGHS-1 (COX) | AA | 95 | 2-15 | 11 | Km values for AA from Tsai and Kulmacz;11 kcat value is an estimate based on O2 consumption determined with 100 μM AA;54 kcat/KM calculated based on a KM of 8.5 μM |
| ovPGHS-1 (COX) | O2 | 5-12 | Km values for O2 from Tsai and Kulmacz11 | ||
| ovPGHS-1 (POX) | 15-HPETE | 120 ± 21 | 42 ± 14 | 2.7 | Values based on guaiacol (4.5 mM) used as reducing cosubstrate147 |
| ovPGHS-1 (POX) | H2O2 | 17 ± 1.6 | 1700 ± 260 | 0.01 | Values based on guaiacol (4.5 mM) used as reducing cosubstrate147 |
| huPGHS-1 (COX) | AA | 10 | 4-10 | Km values for AA from Tsai and Kulmacz;11 kcat value is an estimate based on O2 consumption determined with 100 μM AA (unpublished data); kcat/KM calculated based on a KM of 8.5 μM | |
| huPGHS-2 | AA | 100 | 1-15 | 12 | Km values for AA from Tsai and Kulmacz;11 kcat value is an estimate based on O2 consumption determined with 100 μM AA;54 kcat/KM calculated based on a KM of 8.5 μM |
| Human H-PGDS (None) | PGH2 GSH |
175 | 460 600 |
0.38 | kcat and kcat/KM values are estimated from the VMAX of 451 μmol/min/mg of protein;166 Erratum in: Nat. Struct. Biol. 10:409 (2003). |
| Human H-PGDS (2 mM MgCl2) | PGH2 GSH |
419 | 930 140 |
0.45 | kcat and kcat/KM values are estimated from the VMAX of 1,080 μmol/min/mg of protein;166 Erratum in: Nat. Struct. Biol. 10:409 (2003). |
| Human H-PGDS (2 mM CaCl2) | PGH2 GSH |
205 | 330 590 |
0.62 | kcat and kcat/KM values are estimated from the VMAX of 529 μmol/min/mg of protein;166 Erratum in: Nat Struct Biol. 2003;10:409. |
| Rat H-PGDS | PGH2 GSH |
21 | 200 300 |
0.10 | From162 |
| Rat L-PGDS | PGH2 GSH |
1.8 | 14 100 |
0.13 | VMAX = 4 μmol/min/mg of protein161,162 |
| Human L-PGDS | PGH2 | 0.04 | 2.8 | 0.014 | From403 |
| Mouse L-PGDS | PGH2 | 1.7 | 0.8 | 2.08 | KM = 0.8 μM, VMAX = 5.7 μmol/min/mg of protein (Y. Urade, unpublished data) |
| Human L-PGDS β-trace | PGH2 | 0.45 | 4 | 0.11 | From177 |
| Human mPGES-1 | PGH2 | 50 ± 6 | 160 ± 40 | 0.31 ± 0.040 | Human mPGES-1 purified from E-Coli300 |
| Human mPGES-1 | PGH2 | 7 | 10 | 0.700 | Human mPGES-1 purified from Sf9 cells301 |
| Mouse mPGES-1 | PGH2 | 10 | 130 ± 24 | 0.077 | Mouse mPGES-1 in E-Coli membrane fraction404 |
| Human mPGES-1 | PGG2 | 75 ±4 | 160 ± 3 | 0.470 ± 0.030 | Human mPGES-1 purified from E-Coli300 |
| Human mPGES-1 | GSH | 700 ± 200 | Human mPGES-1 purified from E-Coli300 | ||
| Human mPGES-1 | GSH | 750 ± 250 | Human mPGES-1 purified from Sf9 cells301 | ||
| Murine mPGES-1 | GSH | 37 ± 1 | Mouse mPGES-1 in E-Coli membrane fraction404 | ||
| Human cPGES | PGH2 | ~0.8* | 14 | ~0.057 | Human recombinant cytosolic PGES purified from E-Coli. *Calculated from VMAX multiplied by a factor of 10 since recombinant enzyme displays an order of magnitude lower activity. |
| Monkey mPGES-2 | PGH2 | 1.8 | 28 | 0.067 | Monkey recombinant mPGES-2 purified from E-Coli284 |
| PGFS (AKR1B family) | PGH2/NADPH | 0.014-0.032 | 1.9 | 0.0020-0.0082 | VMax = 26-53; MW 35.8 kDa; Prostaglandin F2alpha synthase activities of aldo-keto reductase 1B1, 1B3 and 1B7.346 |
| Human AKR1B1 | PGH2/NADPH | 0.016 | 1.9 | 0.0082 | From346 |
| Murine AKR1B3 | PGH2/NADPH | 0.032 | 9.3 | 0.0034 | From346 |
| Bovine AKR1B5 | PGH2/NADPH | 0.014 | 7.1 | 0.0020 | An aldose reductase with 20 α-hydroxysteroid dehydrogenase activity is most likely the enzyme responsible for the production of PGF2α in the bovine endometrium.349 |
| Murine AKR1B7 | PGH2/NADPH | 0.026 | 3.8 | 0.0069 | From346 |
| TbPGFS (AKR5A2) Trypanosoma brucei | PGH2/NADPH | 1.03 | 1.3 | 0.79 | VMAX = 2 μmol/min/mg, MW 36.8 kDa From357 |
| LmPGFS (AKR5A1) Leishmania major | PGH2/NADPH | 0.14 | 15 | 0.0096 | VMAX = 270 nmol/min/mg, MW 32 kDa From334 |
| TcOYE Trypanosoma cruzi | PGH2/NADPH | 0.54 | 5.0 | 0.11 | VMAX=766 nmol/min/mg protein, MW 42.3 kDa; From360 |
| Human PGIS (CYP8A1) | PGH2 | 13 | 13 | 1.0 | VMAX 15 μmol/min/mg381 and ~MW = 50 kDa |
| Human TXAS | PGH2 | 9.4 | 20 | 0.48 | VMAX 12 μmol/min/mg395 and ~MW = 50 kDa |
aRefer to references for specific details because even for a specific enzymes the values were determined under a variety of assay conditions.
2.4.1. Formation of the 11-hydroperoxyl arachidonyl radical from AA and O2
The generalities of the chemical changes occurring during the COX reaction catalyzed by both PGHS-1 and PGHS-2 derive from the classic studies of Hamberg and Samuelsson on ovine PGHS-1 preparations reported in 196759,60 and have most recently been described in detail in excellent reviews by Schneider et al.8 and by Tsai and Kulmacz.11 The first chemical step in the COX reaction is removal of the 13-proS hydrogen atom from AA to form an arachidonyl radical (Figures 5 and 6). The hydrogen abstraction reaction was originally demonstrated using mixtures of unlabeled 8Z, 11Z, 14Z-eicosatrienoic acid, [3-14C] 8Z, 11Z, 14Z-eicosatrienoic acid and 8Z, 11Z, 14Z-eicosatrienoic acid stereospecifically labeled at C-13 with tritium59 and was subsequently confirmed using specifically deuterated AA.11,61 Hydrogen abstraction is performed by a tyrosyl radical centered on the phenolic oxygen of Tyr-385.11,62-64 As discussed below, the formation of this radical involves an initial oxidation of the heme group.
Figure 5. Steps in the cyclooxygenase reaction.
(A) Removal of the 13-proS hydrogen atom from AA by a Tyr385 radical to form an arachidonyl radical. (B) Addition of O2 to C-11 to form an 11-hydroperoxyl radical. (C) Reaction of the 11-hydroperoxyl radical to form an endoperoxide with a carbon-centered radical at C-8. (D) Cyclization (C-8 to C-12 bond formation) to form a five membered ring and a carbon-centered radical on C-15. (E) Addition of a second O2 at C-15 and re-addition of the hydrogen atom abstracted in (A).
Figure 6. Removal of the 13-proS hydrogen from AA by the Tyr-385 radical.
Ribbon diagram of region of ovPGHS-1 (in green) encompassing Arg-120 and Tyr-385 (both in magenta) and showing the relationship of Tyr-385 to C-13 (and the 13-proS hydrogen of arachidonic acid (AA) in yellow. The carboxyl end of AA is shown interacting with Arg-120.
Crystallographic studies have been performed to determine the structure of both substrate and non-substrate FAs in the COX active sites of both ovPGHS-165-68 and murine PGHS-2.35,57,58 AA is bound in a catalytically competent L-shaped conformation in both COX active sites of a Co3+-protopohyrin IX-reconstituted ovPGHS-1 homodimer. The 13-proS hydrogen modeled from the structure of the carbon backbone of AA is appropriately positioned for abstraction about 2.8 Å from the phenolic oxygen of Tyr-385 (Figure 6), and a stepwise conversion of AA to PGG2 can be rationalized.65 The situation is more complicated in the case of PGHS-2. AA is found in a catalytically competent conformation similar to that in ovPGHS-1 in only one of the two monomers of muPGHS-236,57 but is in an inverted, nonproductive conformation in the other monomer. The nonproductive conformation in one monomer presumably relates to the allosteric regulation of PGHS-2 by various FAs, which is discussed below.
The segment of AA involving C-11 to C-15 is non-planar in AA/PGHS co-crystal structures.57,65 In contrast, evidence from EPR studies of the arachidonyl radicals formed anaerobically by ovPGHS-1 and huPGHS-269 and from AA having deuterium atoms positioned on C-11 through C-16 by ovPGHS-1 suggest that a planar 1,4-cis,cis-pentadienyl structure involving C-11 to C-1511,61 is present in the newly generated AA radical. Indeed, the lowest energy configuration of a pentadienyl radical is planar.8,70 It should be noted that it is not known if a planar radical would be formed in the presence of O2, which could, in principle, change the nature of the arachidonyl radical. While it is more difficult to remove a hydrogen atom from a non-planar pentadiene,8 a slightly out of plane structure could yield a radical preferentially localized on C-11, the site of O2 insertion. In the absence of information to the contrary, it seems reasonable to assume that formation of a planar structure in the AA radical involves a significant change in structure from the non-planar structure of AA that is observed in crystal structures and presumed to resemble the structure of AA at the time of hydrogen abstraction.
With regard to the structure of AA in the COX active site, it should be noted that native ovPGHS-1 forms not only PGG2 but also the monohydroperoxy acids 11R-HPETE and 15R- and 15S-HPETE. Similarly, mutants of PGHS-1 and PGHS-2 form PGG2 and various monooxygenated products in different proportions.8,67,71,72 With native ovPGHS-1 PGG2, 11R-HPETE and 15R- plus 15S-HPETE are formed in different proportions depending on the concentration of AA substrate. For example, at high AA concentrations up to about 10% of the product formed by ovPGHS-1 is comprised of monohydroxy FAs. This indicates that the VMAX values for the formation of the different products differ. Interestingly, the KM values of ovPGHS-1 for AA for the formation of the individual products also differ somewhat with the lowest KM being that for PGG2.71
These and other results suggest that different oxygenated products arise from different, catalytically competent conformations of AA within the COX active site.71 Crystallographic studies comparing native ovPGHS-1 with a V349A/W387F ovPGHS-1 mutant are consistent with this concept.73 Native ovPGHS-1 forms mainly PGG2 from AA whereas V349A/W387F ovPGHS-1 forms predominantly 11R-HPETE. AA is bound differently in the COX sites of native and V349A/W387F ovPGHS-1. There are significant differences in the positions of several of the carbon atoms of bound AA. Notably, in the V349A/W387F ovPGHS-1/AA complex, the relative locations of C-9 and C-11 of AA with respect to one another would make it difficult to form the endoperoxide group from the 11-hydroperoxyl radical suggesting that the COX reaction catalyzed by V349A/W387F ovPGHS-1 aborts to yield 11R-HPETE instead of PGG2.73
There is both a regio- and stereo-selectivity in the reaction of molecular O2 with the newly formed arachidonyl radical. O2 insertion occurs preferentially at C-11 and on the side of the carbon chain opposite that of hydrogen abstraction (antarfacial) to form an 11R-hydroperoxyl radical. The basis for the selectivity of O2 insertion is not known but as outlined by Schneider et al.8 could be due to one or more of several factors. One is steric shielding of the arachidonyl radical by the enzyme so that O2 has access only to the antarafacial surface at C-11. A second is a positioning of O2 perhaps through a combination of O2 channeling and binding such that O2 is appropriately situated for insertion at C-11. A third is a kinetically determined trapping of the 11-hydroperoxyl radical (hydroperoxyl radical formation being readily reversible), and a fourth is an enzyme-dependent stabilization of the radical at C-11 through, for example, formation of a non-planar pentadienyl radical.
2.4.2. Cyclooxygenase reaction—formation of PGG2 from the 11-hydroperoxyl arachidonyl radical
Once formed the 11-hydroperoxy radical reacts at C-9 to generate an endoperoxide group and a radical centered on C-8; the latter step is a 5-exo-cyclization and is readily reversible.8,11,74 This step is relatively favored vs. reduction of the 11-hydroperoxy radical to 11-HPETE. A second cyclization then occurs in which a bond is formed between C-8 and C-12 and another radical generated, which is delocalized over C-13 to C-15, and again this is a reversible process. Finally, a second O2 reacts at C-15 to form a 15-hydroperoxyl radical, which abstracts a hydrogen atom from Tyr385 to form PGG2.
2.4.3. Rate limiting step in COX catalysis
The dogma until recently has been that abstraction of the 13-proS hydrogen from the COX substrate is the rate limiting step in COX catalysis because it occurs with an isotope enrichment in the substrate. However, the tritium and deuterium kinetic isotopic effects are modest (e.g. the tritium/hydrogen KIE is about three fold) in comparison to those seen with related oxygenases.75,76 Additionally and more recently, it has been found that O2 insertion also occurs with an 16O-18O KIE of about 1.01374; that is, 16O-16O is incorporated more efficiently than 16O-18O into PGG2. These latter results have suggested that the rate limiting step occurs after hydrogen abstraction, most likely at the step involving formation of the cyclopentane ring in the reaction of the C-8 radical with C-12.8 Schneider et al.8 point out that the reaction of O2 with a carbon radical to give a hydroperoxyl radical is readily reversible while H-abstraction from a carbon is not. At this point, the question of which step is actually rate limiting in the COX reaction—hydrogen abstraction or a subsequent step is unresolved because of the potential reversibility of the various steps and the energy barriers calculated to exist in forming the intermediates.8,11 One reasonable explanation is that hydrogen abstraction and C-8 to C-12 bond formation occur at comparable rates.
2.4.4. Peroxide-dependent activation of COX activity and tyrosine radicals in PGHSs
It was found in the early 1970s that peroxides are required to activate and sustain the COX activity of PGHS-1.77 Purified PGHS-1 was subsequently established to have significant POX activity toward H2O2 and both primary and secondary alkyl peroxides78-80 and similar results have been found for PGHS-2.81 The inorganic oxidant peroxynitrite formed by inflammatory cells can also activate both PGHS-1 and PGHS-2 COX activities.82 Many compounds will serve as reducing cosubstrates for the POX activities of PGHSs13, but it is not known which compound(s) serves as the major reducing cosubstrate in vivo.
The relationships between peroxide activation of COX activity and the POX activity of PGHSs have evolved from studies by Ruf, Kulmacz, Tsai and Marnett and their coworkers. Studies performed during the mid 1980s showed that heme became oxidized and that both heme and non-heme free radical signals appeared in association with COX catalysis by ovPGHS-1.83-85 The key conceptual advance was provided with Ruf and coworkers who published spectral evidence for a tyrosyl radical being formed during the oxygenation of AA, and then importantly, suggested that a tyrosyl radical was involved in abstracting the 13-proS hydrogen from AA to initiate COX catalysis.86,87 The catalytically competent tyrosine radical was subsequently determined to be located at Tyr385.28,62-64,88
It is now clear that formation of the Tyr385 radical is initiated by oxidation by a peroxide of the heme group at the POX active site to a horseradish peroxidase Compound I-like, oxyferryl radical cation shown as Intermediate I in Figure 7. The oxidant can be hydrogen peroxide, an alkyl hydroperoxide or peroxynitrite.11,81,82 PGHSs show a marked preference for alkyl hydroperoxides vs. H2O2. For example with ovPGHS-1, Intermediate I formation occurs 103-104 faster with PGG2 or 15-HPETE than with H2O2.84,89,90 The POX site appears relatively exposed to solvent and so the preference for alkyl hydroperoxides is an enigma. The greatest change in rate constant (102-103 fold) occurs in going from H2O2 to ethyl hydroperoxide.89 MD simulations of PGG2 docking on ovPGHS-1 suggest that the carbons immediately adjacent to the one bearing the hydroperoxyl group of PGG2 have van der Waals interactions with the protoprophyrin IX ring and that these interactions may underlie the specificity of PGHSs for organic hydroperoxides.90
Figure 7. Relationship between the POX and COX activities of PGHSs.
A hydroperoxide oxidizes the heme group to an oxyferryl heme radical cation (Intermediate I) with concomitant formation of the corresponding alcohol. Intermediate I can undergo either intramolecular isomerization to form Intermediate II containing an oxyferryl heme and a tyrosine radical centered on Tyr385 or a one electron reduction to a Compound II-like species; Intermediate II and Compound II have the same UV-VIS heme spectrum. The Tyr385 radical can abstract the 13-proS hydrogen from AA to initiate COX catalysis. The POX and COX activities of PGHS-2 can undergo self-inactivation via different processes both of which appear to emanate from Intermediate II. Adapted from Reference 9 with permission; copyright 2008 Elsevier.
Intermediate I can be reduced to a Compound II or can undergo a facile intramolecular conversion to an Intermediate II having an oxyferryl heme and a Tyr385 radical (Figure 7). Thus, a POX reaction must precede the initial COX reaction. Afterwards there is only a loose coupling of the POX and COX reactions.70 Instead, COX catalysis involves a “branched chain” process in which PGG2 can accumulate and a Tyr385 radical, once formed, can be involved in multiple cycles of H abstraction from AA requiring only occasional regeneration of Intermediate I.
Multiple tyrosyl radical signals are observed in PGHSs. Careful, thorough and elegant analyses by Kulmacz, Tsai and coworkers over the past 10-15 yrs indicates that free radical signals can be associated with either Tyr385 or Tyr504 but not other tyrosines.11 Tyr504 is bridged via a water molecule to the imidazole group of His-388 that bonds to the iron on the proximal side of the heme group. There are some differences between PGHS-1 and PGHS-2, but, in general, Tyr385 is the source of a doublet radical signal and is kinetically competent for hydrogen abstraction. Tyr504 is the source of a long-lived singlet radical that is in equilibrium with the Tyr385 radical.11 A third signal is a narrow singlet associated with enzyme inactivation.11,91
It should be noted that PGHS-1 requires higher concentrations of hydroperoxide than does PGHS-2 to initiate and sustain its COX activity.81,92 Formation of Intermediate I occurs at approximately the same rates with PGHS-1 vs. PGHS-2, but formation of Intermediate II from Intermediate I is considerably faster with PGHS-2 than with PGHS-1, and PGHS-2 Intermediate II is more stable than PGHS-1 Intermediate II.81,93,94 Thus, to sustain the reaction cycle, PGHS-1 needs to generate more PGG2 more quickly than does PGHS-2.
At low AA concentrations (≤1 μM) 95,96, the rate of PGG2 formation by PGHS-1 proves to be too slow to sustain COX catalysis, and thus, a positive substrate cooperativity is seen with PGHS-1 and AA at AA concentrations of ≤1 μM.95,96 This kinetic cooperativity can be overcome by adding an exogenous peroxide. A similar kinetic hysteresis has been observed in titrating 3T3 cells expressing PGHS-1 with low concentrations of exogenous AA.97 One explanation for this behavior is that the 3T3 cells have relatively low peroxide concentrations that prevent PGHS-1 from functioning efficiently at low AA concentrations.81,94
2.4.5. Suicide inactivation of PGHSs
Prior to any understanding of the relationship between the POX and COX activities of PGHSs, PGHS-1 was observed to undergo suicide inactivation (i.e. self-inactivation) when incubated with various fatty acid substrates.77 It is now understood that there are both POX and COX inactivation processes and that each has different characteristics.11 Treatment of PGHSs with a peroxide leads to the rapid loss of POX activity, and moreover, POX inactivation occurs with a Y385F mutant lacking COX activity.98,99 COX activity is also lost rapidly when either PGHS-1 or PGHS-2 is incubated with a AA or any other FA substrate.99,100 POX inactivation is first order kinetically, is independent of the nature of the peroxide, and occurs at a slower rate than and independent of COX inactivation.98,101 COX inactivation is dependent on the nature of the peroxide, the POX reducing substrate and the fatty acid substrate and POX activity remains after COX activity is completely lost.99,100 Tsai and Kulmacz have proposed that both POX and COX inactivation proceed from Intermediate II and have speculated that POX inactivation involves the oxyferryl heme group while COX inactivation involves a tyrosyl radical.11 However, little is known about the chemical changes that occur in the enzymes related to POX or COX self-inactivation events other than that heme is oxidized in association with POX inactivation.11 It is also unclear whether there is any physiologic relevance to self-inactivation, which occurs during the course of hundreds to thousands of substrate turnovers. PGHS-2 self-inactivated in cultured cells by treatments with substrates appears to be rapidly degraded whereas self-inactivated PGHS-1 does not appear to be degraded.31
2.5. Allosteric regulation of PGHSs
2.5.1. Half of sites COX activity and allosteric regulation of COX activity by fatty acids
As noted above, there is considerable evidence that PGHS-2 exhibits half of sites COX activity (i.e. only one COX active site functions at a given time) and that there is also ligand-dependent cross-talk between the monomers of huPGHS-2. There is indirect evidence for cross-talk between monomers comprising PGHS-1 although the outcomes for the two isoforms with a given ligand are often different.40,50,51,56 For example, palmitic acid (PA) stimulates PGHS-2 and inhibits PGHS-1,54 and most COX inhibitors affect PGHS-1 vs. PGHS-2 differently.102
In addition to half of sites COX activity, there also appears to be a related half of sites POX activity. This is deduced from the fact that there is only one high affinity heme binding site per dimer. Maximal COX activity occurs with one heme bound per dimer,35,50 and the COX activity of a monomer will not function unless the POX site of that specific monomer is functional.90
One PGHS monomer designated Eallo functions allosterically to regulate its partner catalytic monomer Ecat (Figure 8). Evidence for cross-talk between monomers during catalysis first emerged from studies demonstrating that huPGHS-2 exhibits a kinetic preference for AA over eicosapentaenoic acid (EPA) when the two are tested together.103 This preference, which conflicted with predictions from Michealis-Menten kinetics determined with AA or EPA individually, suggested that EPA and AA both are allosteric regulators of PGHSs. It was subsequently found that even FAs that are not substrates, including common saturated FAs and monounsaturated FAs, are also allosteric regulators of PGHSs.54 Studies to date indicate that common 12-22 carbon FAs that are not substrates bind to Eallo with similar affinities.35,54 A given FA can elicit a stimulatory or inhibitory effect on AA oxygenation depending on the FA and the PGHS isoform.35,54
Figure 8. Eallo vs. Ecat in the conformationally heterodimeric structure of PGHS-2.
Ecat is the catalytically functional monomer, which is regulated by the allosteric monomer Eallo. Heme binds Ecat but not Eallo with high affinity; in essence, heme binding to Ecat significantly reduces the affinity of heme for Eallo. It is not known whether heme binds randomly to equivalent monomer in forming the heterodimer or whether heme binds a transiently pre-existing Ecat/Eallo dimer. AA binds to both Eallo and Ecat although it has a higher affinity for Eallo (see Figure 9). PA also has a higher affinity for Eallo. Some inhibitors (e.g. naproxen and ibuprofen) bind Eallo with higher affinity than Ecat whereas, as shown, many inhibitors bind Ecat with higher affinity and compete directly with AA to inhibit COX activity. Adapted from Reference 35 with permission; copyright 2011 American Society of Biochemistry and Molecular Biology.
Most common, non-substrate FAs stimulate the rate of oxygenation of AA by huPGHS-2. FAs bind to the COX active site of Eallo and promote the AA oxygenase activity of Ecat.35,54 Heme binds with high affinity only to one monomer of a dimer,35,50 and COX activity can only occur in the monomer to which heme is bound.90 Thus, the monomer to which heme is bound and that preferentially binds AA must be Ecat (Figures 8 and 9).35,51,53-56
Figure 9. Interplay between Eallo and Ecat during COX catalysis.
(A) Binding of AA to Eallo and Ecat and its oxygenation by PGHS-2. (B) Binding of AA and its oxygenation by PGHS-2 in the presence of an excess of palmitic acid (PA). Adapted from Reference 35 with permission; copyright 2011 American Society of Biochemistry and Molecular Biology.
There are two reasonably well-understood examples of FA interactions with Eallo/Ecat of huPGHS-2. The first involves the binding of AA alone to huPGHS-2 (Figure 9A). AA is bound in different conformations in the two COX sites of an AA/muPGHS-2 co-crystal.57 In one monomer, AA is “upside down” in the COX active site in a nonproductive conformation in which the carboxylate group of the AA interacts with Tyr385. This is Eallo as depicted in Figure 9A. In the other monomer, AA is “right side up” with its 13-proS hydrogen interacting with Tyr385 in a catalytically productive conformation57,65,67-- Ecat in Figure 9A; again, a Tyr385 radical abstracts the 13-proS hydrogen from AA in the rate determining step of the COX reaction. In experiments performed at high huPGHS-2/AA ratios, the rate of AA oxygenation drops to zero before all the AA is consumed but not as a result of self-inactivation.35 The model in Figure 9A predicts that this would happen if AA is bound nonproductively to the Eallo site but not bound to the Ecat site. This occurs because the KD for AA binding to the Eallo site is about 0.25 μM while the KD for AA binding to the Ecat site is significantly higher (8.4 μM).35 The orientations of AA shown in Figure 9A are based on crystal structures, and we note that the inverted conformation of AA in Eallo has not been shown to occur in solution.
The second example of allosteric regulation of huPGHS-2 by FAs involves interactions between AA and PA, a non-substrate FA (Figure 9B). PA can stimulate the rate of AA oxygenation by huPGHS-2 more than two fold.35 As determined by crystallography PA binds to Eallo in a right side up conformation with its carboxylate group interacting with Arg120. The KD of PA for Eallo is ~7.5 μM.35 When PA is in excess of AA, PA can displace AA from the Eallo site of native huPGHS-2, and AA can then be completely consumed by the enzyme.35 This occurs because PA does not effectively inhibit binding of AA to the Ecat site (i.e. the KD of PA for Ecat is ≥ 50 μM). The situation shown in Figure 9B is representative of the one that presumably occurs in vivo where AA would be a minor FA, and PGHS-2 would be in a milieu having high concentrations of various non-substrate FAs including PA.
We now suspect that allosteric regulation of PGHS-2 by non-substrate FAs involves increasing the VMAX for AA without affecting the KM. This presumption is based on the finding that naproxen, an inhibitor that functions allosterically by binding Eallo,35 reduces the VMAX without changing the KM for AA (unpublished observation). Unfortunately, determining whether PA affects the KM or VMAX for AA is difficult because decreases in AA substrate concentrations change the relative amounts of Eallo to which PA is bound; it is Eallo with non-substrate FA bound that is the true “E” in the Michealis-Menten equation (ie. Eallo-PA/Ecat (Figure 9B)). While perhaps overly simplistic, we will assume for the purposes of discussion that only changes in VMAX values toward AA occur upon binding of non-substrate FAs or inhibitors to Eallo.
2.5.2. Regulation of PGHSs by COX inhibitors
COX inhibitors include traditional NSAIDs that inhibit the COX activities of both PGHS-1 and PGHS-2 and COX-2 specific inhibitors, often referred to as coxibs, that have greater selectivity toward PGHS-2.12,102 Until recently, COX inhibitors were thought to fall into two mechanistic categories depending on whether the effects were time-dependent or time-independent.
The defining features of time-dependent inhibition are (a) that when a PGHS is preincubated with an inhibitor, there is a time-dependent loss of COX activity and (b) that upon reducing the concentration of inhibitor, COX activity is recovered at a slow, sometimes almost undetectable rate (Figure 10).104,105 The rapid, reversible binding of a time-dependent inhibitor can be monitored by measuring “instantaneous” inhibition immediately after enzyme is added to an assay chamber containing substrates and inhibitor. The resulting kinetic data can be used to compute a KI value for what is an apparent, simple competitive inhibition. Time-dependent inhibition can be measured by preincubating the enzyme with an inhibitor and then adding an aliquot of the preincubation mixture to an assay chamber containing substrates and measuring COX activity; under these assay conditions the free inhibitor is diluted to an ineffective concentration. Figure 10 depicts how time-dependent inhibition is measured.
Figure 10. Diagram of time-dependent inhibition of PGHSs.
(A) Time-dependent loss of COX activity observed when either PGHS-1 or PGHS-2 is pre-incubated with a time-dependent inhibitor. (B) Recovery of COX activity that occurs following time-dependent loss of activity.
It is now useful to expand the earlier categorization of COX inhibitors as being time-independent or time-dependent to include the observations that some inhibitors function through Eallo, some through Ecat and some through both Eallo and Ecat; furthermore, there can be differences in how the inhibitors act depending of the COX substrate.55 Briefly, there are (a) time-independent inhibitors that can function by binding Eallo (e.g. ibuprofen and mefanamate with 2-arachidonylglycerol and PGHS-2) or both Eallo and Ecat (e.g. ibuprofen and mefanamate with AA and PGHS-2);55 (b) time-dependent inhibitors that function by binding Eallo (e.g. naproxen with AA and PGHS-2)35 and (c) time-dependent inhibitors that function by binding Ecat (e.g. celecoxib with AA and PGHS-2) and in some instances covalently modifying Ecat (e.g. aspirin with AA and PGHS-2).35
Ibuprofen and mefanamic acid are considered to be time-independent inhibitors of PGHS-2 35,55. Each inhibitor binds to Eallo with relatively high affinity and to Ecat with lower affinity. The binding of ibuprofen or mefanamate to Eallo of PGHS-2 causes ~75% inhibition of activity toward the alternate substrate 2-arachidonylglycerol (2-AG; Figure 11A) but at most a modest ~15% inhibition of activity with AA (Figure 11B). It is only when one of the inhibitors is also bound to Ecat that complete inhibition of AA oxygenation by ibuprofen or mefanamate occurs (Figure 11B). Thus, from a kinetic perspective ibuprofen and mefenamate are mixed inhibitors. Noncompetitive inhibition involving binding of an inhibitor to Eallo is the major contributor to inhibition of 2-AG oxygenation. Competitive inhibition involving binding of an inhibitor to Ecat is the main contributor to inhibition of AA oxygenation. Observations consistent with these conclusions are (a) that ibuprofen and mefenamate act at relatively low concentrations as noncompetitive inhibitors of 2-AG oxygenation,55 (b) that noncompetitive inhibition of 2-AG oxygenation is incomplete (ca. 75%) and involves decreasing the VMAX but not the KM with 2-AG, (c) that ibuprofen behaves as a mixed or competitive inhibitor of AA oxygenation by PGHS-2 and (d) that PA interferes with inhibition of AA oxygenation by ibuprofen by competing with ibuprofen for binding to Eallo.35
Figure 11. Binding of various inhibitors to PGHS-2 and inhibition of AA and 2-AG oxygenation.
(A) Representation of ibuprofen (or mefenamic acid) inhibition of 2-AG oxygenation by PGHS-2; in this example of time-independent allosteric inhibition, Eallo-I/Ecat-2-AG functions at only 25% of the rate of Eallo/Ecat-2-AG.55 This is a form of (B) Representation of ibuprofen (or mefenamic acid) inhibition of AA oxygenation by PGHS-2; in this example of time-independent inhibition, there is a modest allosteric inhibition (~15%) with both Eallo-I /Ecat-AA and Eallo- AA /Ecat-AA being active but functioning at slightly different rates.35,55 (C) Time-dependent, allosteric inhibition of PGHS-2 mediated AA oxygenation such as that seen with naproxen. In this situation *Eallo-I /Ecat is time-dependently inhibited and has relatively low but still appreciable COX activity.35 (D) Time-dependent inhibition by of AA oxygenation by PGHS-2 involving the binding of the inhibitor to Ecat. In this case, which is representative of what is seen with celecoxib, rofecoxib, indomethacin and diclofenac,35 *Eallo- AA /Ecat-I is a time dependently inhibited and completely inactive form of the enzyme.
Time-dependent COX inhibitors are generally visualized as functioning in two phases although there may be a third phase with some inhibitors105 (Figures 11C and 11D). The first phase is a rapid, reversible binding to the COX active site of either Eallo or Ecat. The second phase involves a slow conformational change to an “*E” form of the enzyme (Figures 11C and 11D). The latter process occurs in seconds or minutes and is even more slowly reversible.4,35,105 Modeled in Figure 11C is the case for an inhibitor (e.g. naproxen) that binds preferentially to Eallo. Figure 11D illustrates the case for an inhibitor (e.g. celecoxib, diclofenac) that preferentially binds to Ecat. aspirin is an agent that at least in the case of PGHS-2 acetylates Ecat.2 Most of the information on interactions of inhibitors with the two subunits has been determined for PGHS-2.35
The discovery that non-substrate FAs such as palmitic acid bind preferentially to Eallo permits discrimination between binding of inhibitor to Eallo vs. Ecat. FAs uniformly interfere with inhibitors that bind to Eallo while they generally have no effect or even potentiate the actions of inhibitors that bind to Ecat.35 In many, if not all cases, a given inhibitor will interact differently with PGHS-1 vs. PGHS-2. A distinctive feature of coxibs is their ability to cause time-dependent inhibition of PGHS-2 but time-independent inhibition of PGHS-1.56,105,106 Rofecoxib and celecoxib both appear to interact preferentially with Ecat of PGHS-2. The extent to which different nonselective inhibitors interact with Eallo vs. Ecat of PGHS-1 and PGHS-2 is unresolved.
2.5.3. Molecular basis for cross-talk between PGHS monomers
As discussed above, a rate determining step in COX catalysis is abstraction of the 13-proS hydrogen from AA by a Tyr-385 radical.8,59 Thus, it is logical that allosteric regulation involves changing the juxtaposition of the 13-proS hydrogen and the phenolic oxygen of Tyr385 in Ecat so as to slow or increase the rate of hydrogen abstraction. However, more complex changes such as redistributing the radical between Tyr-385 vs. Tyr-504107,108 or stabilizing the radical(s)109 could also affect the rate. Alternatively, a structural change that alters the rate of O2 insertion, which has also been considered to be another rate limiting step,74 may affect catalysis. In short, different non-substrate FAs 35 and different inhibitors may influence catalysis in different ways.11,109
Structural changes do occur in PGHS-1 and PGHS-2 upon binding of a FA or inhibitor to the enzymes.18,35,40,54,110 For example, binding of inhibitors or heme decreases the sensitivity of ovPGHS-1 to cleavage by trypsin apparently by decreasing the availability of the peptide bond involving Arg-277.18,110 A subtle change in the position of Tyr-385 (≤0.4 Å) in Ecat would have a major impact on catalysis. Unfortunately, small changes in the location of Tyr-385 cannot be delineated in any of the structures that have been published because, event the most highly resolved structures are at relatively moderate resolution (≤ 2.0Å).40,49,57,65,66,68,111
Cross-linking studies in which cysteine residues have been inserted at different positions at the interface between the two monomers of PGHS-2 identified a region of cross-talk involving residues 125-127 from one monomer and residues 542-543 in the adjoining monomer.54 As illustrated in Figure 12, structural studies show that the loop involving residues 125-127 can assume one of two conformations depending on the occupancy of the COX active site.40,56 The loop involving residues 125-127 emanates from a helix involving Arg-120, a residue that interacts with FA substrates and many inhibitors of PGHSs. One can imagine that a ligand-induced change in the position of Arg-120 in Eallo could change the structure of the loop involving residues 125-127 in Eallo and that this structural change could be transmitted to the partner Ecat through interactions with residues 542-543 in the partner monomer. It is not obvious how this change would alter the COX site of Ecat (e.g. the juxtaposition of Tyr-385 and the 13-proS-hydrogen of AA).
Figure 12. The two forms of the 125-127 loop of PGHS-2 at the dimer interface of ovPGHS-1.
Alternate conformations of residues 121–129 in one monomer at the interface with another monomer. The monomer shown with the residues in orange is a monomer of ovPGHS-1 to which celecoxib is bound. The sidechains of the other monomer occur in two conformations. One is from a monomer to which no celecoxib is bound (blue) and the other is from a monomer to which celecoxib is bound in the COX site (magenta). Reprinted with permission from Reference 56; copyright 2010 National Academy of Sciences.
2.5.4. Physiologic and pharmacologic consequences of PGHS allosterism
The biologic significance of PGHS allosterism remains to be established, but allosteric regulation of PGHSs is likely to have important in vivo consequences. As summarized above, COX activities of purified PGHS-1 and PGHS-2 are modulated in vitro by all common FAs, including both those that are PGHS substrates and those that are not.35,54 We hypothesize that in vivo PG production is influenced by the effective concentration and FA composition of the milieu in which PGHSs operate—the FA tone. Presumably every individual establishes a fairly constant FA tone as a consequence of dietary habits and genetic profile (e.g. 112,113). We imagine that having a FA tone outside some normal range would change PG formation to an extent sufficient to affect the health of an individual. For example, having a FA tone that enhances PG formation via PGHS-2 might well exacerbate inflammatory conditions believed to underlie many chronic diseases. Dietary studies designed to investigate this possibility have not been performed in either humans or laboratory animals.
FAs modulate responses of PGHSs to COX inhibitors. For example, in vitro palmitic, stearic and oleic acids potentiate to different degrees the inhibition of PGHS-2 by aspirin and of celecoxib on PGHS-2.35 And FAs can interfere with the actions of naproxen and FBP. The existence of interactions among FAs and COX inhibitors with PGHSs suggests that there are unrecognized relationships between FA tone and the actions of COX inhibitors. These drugs are the most widely used pharmaceutical agents in the U.S., but they have adverse side effects responsible for an estimated 20,000 deaths in the U.S. annually.12 We envisage that some side effects, which are generally of unknown origin, are related to diet/drug interactions and could be ameliorated by altering diet or drug. For example, responses of patients to traditional NSAIDs or coxibs likely vary among groups of individuals on typical Western vs. Mediterranean vs. ω3 fish oil-enriched diets, and the differences are probably drug specific.
2.6. Regulation of PGHS-1 vs. PGHS-2 in cells by peroxide tone and FA tone
As noted earlier PGHS-1 and PGHS-2 are products of different genes. Further, there are now many examples indicating that each isoform acts independently to subserve different, non-overlapping biologies.114-116 Funk and coworkers characterized transgenic mice in which the PGHS-1 coding region was substituted for the PGHS-2 coding region behind the PGHS-2 promoter.117 Their studies provide direct evidence that differences in both the patterns of expression and the enzymatic properties of the two isoforms contribute to their unique biologies.
Although not addressed in detail here, there are major differences between PGHS-1 and PGHS-2 expression. PGHS-1 is typically expressed constitutively9,118,119 and in an estimated 10% of mammalian cell types.120 PGHS-2 is usually expressed transiently, perhaps by all cells, during replication and/or differentiation.9,118-121 There are also significant differences between PGHS-1 and PGHS-2 protein degradation.31,37
From an enzymatic perspective, the most interesting situation occurs when PGHS-1 and PGHS-2 are co-expressed in cells, and the activity of PGHS-1 becomes effectively latent. An example of this is seen with so-called “late phase” murine NIH 3T3 fibroblasts. Growth-arrested, senescent murine NIH 3T3 fibroblasts express PGHS-1 but not PGHS-2.9,122,123 When 3T3 cells are stimulated with a mitogen or phorbol myristate acetate (PMA), PGHS-1 levels remain constant, but PGHS-2 gene expression is induced with peak PGHS-2 protein expression occurring 2-3 hr later. PGHS-2 protein levels then decrease rapidly to near basal levels.122-125 Correspondingly, PGE2 production takes place in two phases.9,126-128 One is an early phase that occurs immediately upon addition of a mitogen or PMA to senescent 3T3 cells, plateaus within minutes and is due to the sequential actions of AA (and EPA)-specific cPLA2,126,127 PGHS-1122-125,128 and cytosolic PGES129 or mPGES-1.130,131 The amount of PGE2 formed in the early phase is in the range of that observed when about 1-5 μM exogenous AA is added to senescent 3T3 cells.97,128 The second or late phase involves a slower but continuous PGE2 production that coincides with the appearance of PGHS-2 beginning ~0.5 hr after adding a mitogen or PMA. sPLA2(s), which shows no preference among fatty acids at the sn2 position of phospholipids,132 appears to be the major, operative PLA2 in late phase PGE2 synthesis.133,134 In the late phase, PGHS-2 is responsible for more than 95% of the PGE2 formed.135 This occurs despite PGHS-1 being present at more than twice the level of PGHS-2136, both enzymes apparently being co-localized25,137,138 and the two isozymes having very similar kinetic properties (i.e. KM, VMAX).
There are at least two subtle differences between the kinetic properties of PGHS-1 and PGHS-2 that could explain why PGHS-2 functions in late phase 3T3 cells whereas PGHS-1 is effectively latent. First, as discussed earlier, PGHS-1, but not PGHS-2 exhibits positive substrate cooperativity (i.e. unexpectedly low oxygenation rates with a combination of low AA and hydroperoxide activator concentrations).96 Second, PGHS-2 is stimulated by ambient non-substrate FAs whereas PGHS-1 is slightly inhibited.35,54 Additionally, the oxygenation of AA by PGHS-1 is inhibited to a significantly greater extent than PGHS-2 by EPA and docosahexaenoic acid (DHA) which are present at the sn2 position of common phospholipids.54,103
The inability of PGHS-1 to function in late phase 3T3 cells may result in part from intracellular conditions that promote positive cooperativity.94 There is indirect evidence that hydroperoxide promotes PGHS-1 activity in platelets,139 and this suggests that, at least in some cells, changing the concentrations of activating hydroperoxide can modulate PGHS-1 activity. Thus, in vivo PGHS-1 may be regulated by ambient peroxide concentrations or peroxide tone.94 Additionally, as noted in the discussion of PGHS allosterism, the concentration and composition of the ambient FA pool (i.e. the FA tone) at the time of AA mobilization from phospholipid precursors may have the effect of promoting PGHS-2 and attenuating PGHS-1 activities.
2.7. Alternative substrates of PGHSs
North American and Western European diets have relatively high levels of omega (ω) 6 fatty acids (e.g. linoleic acid).140,141 As a result, the most common prostanoid precursor in these populations is the 20 carbon ω6 fatty acid, arachidonic acid (AA). Adrenic acid, which is the elongated version of ω6 AA is also an effective substrate.54
EPA, an ω3 “fish oil” FA can function as a precursor of “3-series” prostanoids in vivo.142-145 However, it is not clear whether 3-series PGs can be formed via PGHS-1 and/or PGHS-2; that is, the impact of increases in fish oil FAs including both EPA and DHA, on PGHS-1 vs. PGHS-2 pathways in vivo is unresolved. EPA is a very poor substrate for PGHS-1 and is only oxygenated in the presence of high levels of hydroperoxide;103,146,147 additionally, EPA is a reasonably good inhibitor of AA oxygenation by PGHS-1 and at equimolar concentrations EPA inhibits AA oxygenation by ~50%.103 In contrast to PGHS-1, PGHS-2 oxygenates EPA to an approximately equimolar mixture of 15-hydroperoxyeicosapentaenoic acid and PGH3 at about 30% of the rate of conversion of AA to PGH2.2 EPA is a weak inhibitor of AA oxygenation by PGHS-2.103 DHA is a very poor substrate for PGHS-2 and is not oxygenated at appreciable rate by PGHS-1.2,54 As described by Rouzer and Marnett in an accompanying chapter there is considerable evidence that AA esters, notably 2-arachidonylglycerol, are PGHS-2 substrates involved in endocannabinoid metabolism.10
2.8. Subcellular localization and trafficking of PGHSs
The subcellular locations of PGHS-1 and PGHS-2 seem to be qualitatively similar but quantitatively different. Both PGHS-1 and PGHS-2 are embedded in the luminal surface of the ER and the inner membrane of the nuclear envelope25,137,138 and associated lipid bodies.148 Recent results suggest that a significant fraction of PGHS-2 is present in the Golgi apparatus (unpublished results).
3. Prostaglandin D synthases (PGDSs)
PGD2 is a mediator of allergic and inflammatory responses149 and is produced by mast cells150,151 and Th2 cells152 in a variety of tissues and is an endogenous sleep-promoting substance in the brain.153 PGD2 is formed from PGH2. PGD synthases (PGDSs; PG endoperoxide D-isomerase, EC 5.3.99.2) catalyze the isomerization of the 9,11-endoperoxide group of PGH2 to PGD2, which has a 9-hydroxy and an 11-keto group (Figure 1). There are two distinct types of PGDSs. One is hematopoietic PGDS (H-PGDS), which is found in mast cells,154 Th2 cells,152 and microglia,155 and the other is lipocalin-type PGDS (L-PGDS), which is localized in the brain,156 male genital organs,157,158 and cardiovascular tissues including the human heart.159
3.1. Catalytic and molecular properties
3.1.1. Purification and properties of H-PGDS
H-PGDS was originally purified from rat spleen by Christ-Hazelhof and Nugteren in 1979 as a 26 kDa, cytosolic, monomeric glutathione-requiring enzyme.160 The Mr of H-PGDS is the same as that of L-PGDS,161 but the biochemical characteristics of H-PGDS are distinct from L-PGDS in terms of kinetic parameters, amino acid composition, and immunological properties, and H-PGDS is associated with glutathione-S-transferase (GST) activity.162 Based on molecular and x-ray crystallographic studies of recombinant rat H-PGDS expressed in E. coli, H-PGDS was identified as the vertebrate homolog of the σ class of GSTs.163 Sequences of full length cDNAs for the human and mouse H-PGDS have subsequently been obtained.164 The cDNAs encode a protein composed of 199 amino acid residues with calculated Mr values of 23,297, 23,343 and 23,226 for the rat, human and mouse enzymes, respectively. The N-terminal methionine is cleaved from the mature protein. The cDNA for the chick homolog was isolated by Thompson et al.165 The x-ray crystal structure of the human recombinant H-PGDS was reported in 2003.166 Human H-PGDS is now known to be a homodimeric protein with an Mr of 45,000 to 49,000, that binds one molecule of reduced glutathione (GSH) per monomer and one Mg2+ ion per dimer, as described below in Section 3.2.1. in detail. Mg2+ and Ca2+ ions increase the activity of H-PGDS about 1.5-fold over basal levels in a concentration-dependent manner, with half-maximal effective concentrations of 50 μM for Mg2+ and 400 μM for Ca2+. Mg2+ increases the affinity of the enzyme for GSH, decreasing the KM value four-fold from 0.6 mM in the absence of MgCl2 to 0.14 mM in the presence of 1 mM MgCl2; Ca2+ does not change the affinity of human H-PGDS for GSH (KM = 0.6 mM). Because H-PGDS is localized in the cytosol, where the concentration of Mg2+ is estimated to be >1 mM, H-PGDS likely exists as the Mg2+-bound form in vivo.
3.1.2. Purification and properties of L-PGDS
L-PGDS was isolated from rat brain in 1985 as a GSH-independent PGDS with an Mr of 26,000.161 The enzyme had previously been misidentified as a protein with an Mr of 80,000 to 85,000.167 L-PGDS does not absolutely require GSH for its catalytic reaction; it is active with various other sulfhydryl compounds, such as dithiothreitol, β-mercaptoethanol, cysteine, and cysteamine.168 In 1989, the cDNA for L-PGDS was isolated from a rat brain cDNA library169 and subsequently from many other mammalian species, including human,170,171 and also from non-mammals, including the chicken, frog, and fish.172,173 The cDNA for L-PGDS encodes a protein composed of 189 and 190 amino acid residues in the mouse and human enzymes, respectively. L-PGDS is post-translationally modified by cleavage of an N-terminal signal peptide of 24 and 22 amino acid residues from the mouse and human enzymes, respectively. Two N-glycosylation sites, at positions of Asn-51 and Asn-78, in the mouse and human enzymes are conserved in all mammalian enzymes thus far identified but are not found in the amphibian homologs. Mammalian L-PGDS is highly glycosylated, having two N-glycosylated sugar chains, each with a molecular mass of 3,000 daltons.169 Homology searching in a database of amino acid sequences revealed that L-PGDS is a member of the lipocalin gene family,171 which consists of small, secretory, non-enzymic proteins that serve as transporters of various lipophilic ligands.174 L-PGDS is the first lipocalin recognized to have an enzymatic activity.
In 1993, Hoffman et al.175 purified a major protein of human cerebral spinal fluid (CSF), β-trace,176 and found that the partial N-terminal amino acid sequence of human β-trace is identical to that of human L-PGDS after truncation of the N-terminal signal sequence predicted from the cDNA sequence.170 Then in 1994, Watanabe et al.177 purified L-PGDS from human CSF and demonstrated that human L-PGDS is immunologically the same protein as β-trace. Human L-PGDS/β-trace is also present in the seminal plasma, serum, and urine, and the L-PGDS/β-trace concentration in various body fluids has been proposed to be useful as a clinical marker for many diseases, as described below in Section 3.4.4.
3.1.3. H-PGDS and L-PGDS as novel examples of functional convergence
The catalytic and molecular properties of H-PGDS and L-PGDS are summarized in Table 1 and Table 2. Urade, Hayaishi and coworkers have extensively studied the structural and functional properties of H-PGDS and L-PGDS and reported their tissue distribution, cellular localization, cloning of their cDNA, the chromosomal gene of the human and mouse enzymes, and the regulatory mechanisms controlling their gene expression. Knockout (KO) mice178 and transgenic mice overexpressing human H-PGDS179 and human L-PGDS180,181 have been used to identify functional abnormalities. Although both H-PGDS and L-PGDS catalyze the same reaction, these enzymes have evolved from different origins--H-PGDS from GST163 and L-PGDS from lipocalins.170,171 Therefore, H-PGDS and L-PGDS represent examples of functional convergence.182,183 H-PGDS/L-PGDS double KO mice have undetectable levels of the major urinary metabolite of PGD2, tetranor-PGDM,184 indicating that these two enzymes are responsible for the production of all of the PGD2 in mice.
Table 2.
The catalytic and molecular properties of human H-PGDS and L-PGDS.
| Hematopoietic PGD synthase (H-PGDS) | Lipocalin-type PGD synthase (L-PGDS) | |
|---|---|---|
| Molecular weight | ≈23,000 | ≈26,000 |
| Subunit | dimer | monomer |
| Co-factor | GSH | Sulfhydryl compounds (natural cofactors are unknown) |
| Activator | Mg2+, Ca2+ | --- |
| Chromosomal localization | 4q21-22 | 9q33-34 |
| Post translational modification | --- | N-glycosylation (β-trace) |
| Tissue distribution | Leptomeninges, Choroid plexus, Oligodendrocytes, Testis (Leydig cells, Sertoli cells), Heart | Microglia, mast cells, oviduct, Langerhans cells, dendritic cells, Kuppfer cells, Th2 cells |
| Inhibitor | HQL-79, TFC-007 | AT-56 |
Members of the α, μ, and π-classes of GSTs and squid GST(s) of the σ-class GSTs have the ability to convert PGH2 to a mixture of PGD2, PGE2, and PGF2α in the presence of GSH.185,186 However, their PGDS activities are lower than their PGE and PGF synthase activities. Therefore, the H-PGDS gene apparently evolved from a common ancestor of the invertebrate α-class GSTs, and acquired PGDS activity during evolution. Metal activation of H-PGDS is unique among members of the GST family in that it is not observed in other GST isozymes in the α, μ, π or σ classes or in the σ class GST from Shistosoma mansoni. A 28-kDa GST of S. mansoni (Sm28GST) is associated with PGDS activity (1 mol/min/mg protein with 40 μM PGH2 and 1 mM GSH) having about 3% of rat H-PGDS activity (30 mol/min/mg protein); Sm28GST does not catalyze the conversion of PGH2 to PGE2 or PGF2α.187 GST isozymes with PGDS activity are also found in the filarial nematode Onchocerca volvulus (Ov-GST1)188 and the porcine nodule worm Oesophagostomum dentatum.189 Those GSTs play important roles in the pathogenesis and physiology of protozoan and metazoan parasites, as reviewed by Kubata et al.190
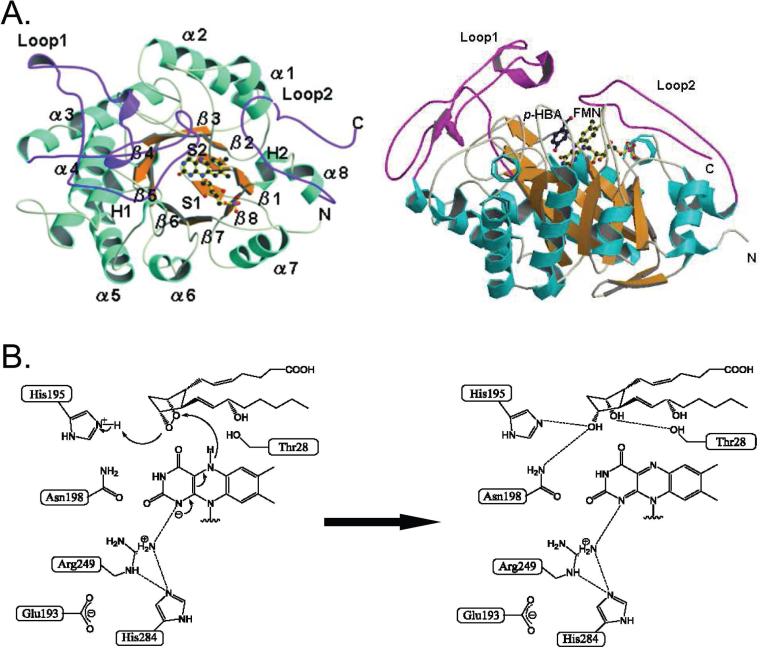
3.2. Tertiary structures of PGDSs
3.2.1. X-ray crystallographic structure of H-PGDS
X-ray crystallographic structures of H-PGDS have been determined for the following complexes: the rat H-PGDS-GSH complex at 2.3 Å resolution (PDB code: 1PD2);163 the human H-PGDS-GSH complex in the presence of Mg2+ or Ca2+ at resolutions of 1.7 Å and 1.8 Å, respectively (PDB codes:1IYH and 1IYI, respectively);166 and a quaternary complex of human H-PGDS with Mg2+, GSH, and an inhibitor (HQL-79) at a resolution of 1.45 Å (PDB code: 2CVD).179 The crystal structure of the human H-PGDS-HQL79 complex is shown in Figure 13.
Figure 13. X-ray crystallographic structure of human H-PGDS bound with HQL.
(A) The dimeric structure.166 (B) The Mg2+-binding site.166 (C) The Ca2+-binding site.166 (D) The monomeric structure.179 (E) Close-up view of the superimposed structures around the active site of H-PGDS in the presence (sky blue) and in the absence (gray) of HQL-79 shown as a space-filling model.179 GSH molecules in the presence (green carbon atoms) and absence (gray carbon atoms) of HQL-79 are also shown. (F) Schematic drawing of the binding mode of HQL-79.179 Hydrogen bonds and salt bridges are shown by yellow-dotted lines, weak hydrogen bonds by blue-dotted lines, and the coordinate bond for Mg2+ by the yellow arrow.
The structure is that of a homodimer with each monomer complexed with one GSH molecule (Figure 13A). The monomers are related by a non-crystallographic two-fold axis of symmetry. The dimer interaction involves a “lock-and-key” complementarity with a hydrophobic surface and a limited number of electrostatic interactions as is commonly observed in various GSTs.
The overall folding motif of H-PGDS is the same as that of other GSTs. The H-PGDS monomer is composed of two major domains with a prominent interdomain cleft; that is, the N-terminal domain (amino acid residues 1–71) and the C-terminal domain (82–199) are connected by residues 72–81, which include two turn structures (residues 72–75 and 76–79). The N-terminal domain contains a four-stranded β sheet and three α helices, arranged in a βαβαββα thioredoxin motif,191 in which the β2 and β1 strands are parallel and the β1 and β3, and the β3 and β4, are antiparallel. The α1 and α3 helices comprise the dimer interface with the α4, α6, and α8 helices of the counterpart in the dimer. The loop structure (residues 46–52) bends at the position of Pro52 to the outside of the enzyme, which is the GSH binding site. The angle between the directions of the loop and the β3 backbone is approximately 90°, so that the side chains of the loop residues are exposed to the solvent with the Ile-51 residue in a cis-conformation, resulting in the formation of a GSH binding dimple. The C-terminal domain is composed of five α helices, in which the α4, α5, and α6 helices make an α-helix bundle. The long α5 helix bends at position Gln-123. In the connecting loop of the α4 and α5 helices, the backbone between Ser-100 and Trp-104 is kinked to compensate for the α4 helix, which is shorter than that found in other GSTs.
GSH is bound to the side of the N-terminal thioredoxin-like domain by forming two and eight hydrogen bonds to the atoms of the protein backbone and side chain, respectively, similar to other GSTs. The cysteinyl backbone of GSH interacts with that of cis-Ile-51 of the N-terminal domain via hydrogen bonds in an anti-parallel β-sheet manner. The amino nitrogen of the γ-glutamate residue of GSH forms hydrogen bonds with both of the carboxyl oxygens of the Asp-97 side chain of the other H-PGDS molecule in the dimer. The Sγ atom of GSH and the Oη of Tyr-8 show a hydrogen bonding distance of 3.1 Å. All these residues are highly conserved among the members of the GST family.
The prominent cleft including the GSH-binding site between the two domains is the catalytic pocket. The interdomain cleft expands to a wide and deep pocket (pocket 1) behind the GSH-binding site with the longest loop eaves of the α4 and α5 helices. At the entrance of pocket 1, the indole ring of Trp-104 forms a ceiling on the C-terminal domain, because the indole ring is directed parallel to the α4 helix extended by the kinked backbone including Trp-104. Pocket 1 has a path to a branched cavity consisting of another pocket (pocket 2) and a narrow tunnel. Pocket 1 also opens to a third pocket (pocket 3) on the outer surface due to the short C-terminal end of H-PGDS. There is a straight path from the outside of the protein to pocket 2 via pockets 3 and 1.
The crystal structure of human H-PGDS in the presence of GSH and Mg2+ or Ca2+ has revealed that Mg2+ or Ca2+ resides at the center of the dimer interface (Figure 13B and 13C).166 The Mg2+- or Ca2+-binding site consists of pairs of Asp-93, Asp-96 and Asp-97 from each monomer. These six Asp residues form an acidic cluster at the dimer interface, and the residues from each pair are related via a non-crystallographic two-fold symmetry axis at the center of the dimer. The Mg2+- or Ca2+-binding site lies at the hinge portion between the N- and C-terminal domains of the subunit (Figure 13B and 13C). The Mg2+ ion is octahedrally coordinated by six water molecules (W1–W6), whereas the Ca2+ ion is coordinated by five water molecules (W1–W3, W5, and W6), because the ionic radius of Mg2+ (0.65 Å) is shorter than that of Ca2+ (0.99 Å). The hydrogen-bonding network between Arg-14 and Asp-96 changes from only one hydrogen bond to the N1 of Arg-14 in the Mg2+-bound form to two hydrogen bonds between both O2 (Asp-96) atoms and the guanidinium nitrogen atoms of Arg-14 (N1 and N2) in the Ca2+-bound form. Although the conformations of GSH, Ser-100, and Trp-104 are identical in both metal-bound forms, the angle between the guanidinium plane of Arg-14 and the carboxyl plane of Asp-96 is 11.9° and 11.1°, respectively. The side chain of Arg-14 rotates upon Mg2+ binding, producing the metal ion effect on the reactivity described below. The Asp93Asn, Asp96Asn, and Asp97Asn mutants of human H-PGDS are not activated by the Mg2+ ion. Neither the Asp93Asn nor the Asp97Asn mutant shows increased affinity for GSH in the presence of Mg2+. The affinity of the Asp96Asn mutant for GSH is already increased irrespective of Mg2+, differing from the wild-type enzyme and the two other mutants. The Asp97Asn mutation causes a decrease in the activity to ~1% of that of the wild-type enzyme when the transformant E. coli cells are cultured at 37° C. However, when the transformants are cultured at 16° C, the purified enzyme shows almost the same activity as the wild-type enzyme, indicating that the Asp-97 residue is important for the stabilization of the enzyme in E. coli grown at the higher temperature.
The HQL-79 molecule becomes inserted in the catalytic cleft between Trp-104 and GSH (Figure 13D and 13E).179 No steric hindrance is detected between HQL-79 and the GSH molecule, consistent with the kinetic analyses, described below, showing that HQL-79 is a competitive inhibitor against the substrate, PGH2, and a non-competitive inhibitor of GSH. Among the three pockets (pockets 1, 2, and 3) in the catalytic cleft of H-PGDS, phenyl-1 and phenyl-2 of the diphenyl of HQL-79 penetrate into pocket 1 and pocket 2, respectively. The HQL-79 molecule becomes stabilized by weak hydrogen bonding with Met-99, Phe-102, Trp-104, and Tyr-152 located within a distance of 3.5 Å (Figure 13F), and by Arg-14, Thr-159, and Leu-199, including GSH (by non-bonding interactions including salt bridges and hydrogen bonding) through water molecules. The tetrazole ring of HQL-79 is located at the entrance of pocket-3 and does not directly interact with the positively charged amino acid cluster of Lys-112 and Lys-198 in this pocket. No direct interaction is detected around the tetrazole ring, suggesting that the tetrazole group of HQL-79 interacts with Lys-112 and Lys-198 via diffusible water molecules in pocket-3.
In the catalytic cleft, a phenyl ring of the diphenyl of HQL-79 exhibits van der Waals interaction with the indole ring of Trp-104, including weak hydrogen bonding with the ring nitrogen. In comparison with the native structure of the enzyme, the HQL-79 molecule penetrates into the ceiling of the catalytic cleft and pushes out the indole ring of Trp-104, resulting in the rotation of the indole ring by 48 degrees with a 4.3 Å shift. The movement of Trp-104 induces twisting of loop 7, which is linked to the long, kinked α5-helix. However, in the cases of H-PGDS complexes with more potent inhibitors, neither penetration of the inhibitor molecule into the ceiling of the catalytic cleft nor pushing out of the indole ring of Trp-104 is observed.192,193
3.2.2. Catalytic mechanisms of H-PGDS
Site-directed mutagenesis of rat H-PGDS194 has indicated that Tyr-8, Arg-14, and Trp-104 residues are important for the catalytic activity of the enzyme. Tyr8Phe, Arg14Lys/Glu, and Trp104Ile mutants completely lack both PGDS and GST activities. All these mutant enzymes bind to GSH-Sepharose, indicating that the loss of PGDS and GST activities cannot be attributed to a complete lack of GSH binding. The reactivity of the thiol group of GSH bound to the mutants was examined using the thiol-modifying agent, 5,5'-dithiobis-(2-nitrobenzoic acid) (DTNB). When wild-type rat H-PGDS was incubated with DTNB in the absence of GSH, no reaction was observed at pH 5.0, indicating that the two free cysteine residues in the wild-type protein cannot be titrated with DTNB under relatively acidic conditions. In acidic solution with 0.5 mM GSH, DTNB was converted to 5-thio-2-benzoic acid at an initial rate of 15 mmol/min. The addition of the wild-type enzyme or the Trp104Ile mutant increased the reaction rate exactly two-fold, indicating that GSH had been activated to the thiolate anion even in the Trp104Ile mutant that lacks both PGDS and GST activity. However, no enhancement of thiol reactivity was observed with Tyr8Phe and Arg14Lys/Glu mutants, suggesting that Tyr-8 and Arg-14 residues are essential for activation of GSH to the thiolate anion within the catalytic cleft of H-PGDS. Ultraviolet resonance Raman spectroscopy of human H-PGDS revealed that the Y8a Raman band of Tyr-8 at pH 8.0 is shifted from 1615 cm-1 to 1600 cm-1 by the addition of Mg2+ and to 1611 cm-1 upon subsequent addition of GSH. This result indicates that Tyr-8 is deprotonated in the presence of Mg2+ and reprotonated by the abstraction of H+ from the thiol group of GSH and that the reprotonated Tyr-8 forms a hydrogen bond with the thiolate anion of GSH.195
There are several reaction models that have been proposed for the conversion from PGH2 to PGD2 (Figure 14).196 The catalytic mechanism of H-PGDS is shown in Figure 15. Mutagenesis studies revealed that Arg-14 and Tyr-8 are involved in the stabilization of the thiolate anion of GSH bound to H-PGDS. The activated thiolate anion attacks the O-11 of the endoperoxide group of the substrate PGH2. A hydrogen bond between the amide nitrogen of glycine from GSH and the O-9 of the endoperoxide group of PGH2 may stabilize the bound PGH2 molecule. Arg-14 forms a hydrogen bond with the thiolate group of GSH to stabilize the negative charge, which accelerates the nucleophilic attack on the endoperoxide group of PGH2.
Figure 14. Reaction models for the conversion of PGH2 to PGD2.196.
Several reaction models have been proposed for the conversion of PGH2 to PGD2. C-9 and C-11 substituents are omitted for clarity. B represents a base including a hydroxyl ion (OH-). Path A is hypothetical and Paths B and C seem plausible.
Figure 15. Catalytic mechanism of H-PGDS.163.
A schematic drawing of the catalytic pocket of H-PGDS with PGH2. Pockets 1, 2, and 3 are shaded. The thiolate anion of bound GSH attacks the oxygen at C11 of PGH2 (panel 2). The putative reaction intermediate of PGH2 with GSH is attacked by a certain base-like bulk GSH in solvent (panel 3) to produce PGD2 in a sterically restricted manner (panel 4). A GS- in step 3 of the reaction is accessible from the outside of the catalytic cleft of H-PGDS. Adapted from from Reference 163 with permission; copyright 1997 Cell Press.
3.2.3. NMR solution structure and x-ray crystallographic structure of L-PGDS
The structure of L-PGDS was first determined by NMR analysis of the recombinant mouse Δ1-24 Cys89,186Ala mutant lacking the intramolecular disulfide bond (PDB code: 2E4J).197 This mutant enzyme exhibits the typical lipocalin fold, consisting of an eight-stranded, anti-parallel β-barrel and a long α-helix associated with the outer surface of the barrel (Figure 16A). In the NMR structures, the first ten residues at the N-terminus, the E-F loop, and G-H loop are undefined because of the signal broadening of these flexible loop regions. The interior of the barrel forms a hydrophobic cavity opening to the upper end of the barrel. The central cavity of L-PGDS is larger in size than that of other lipocalins and contains two pockets, the relatively hydrophilic upper pocket 1 and the hydrophobic lower pocket 2 (Figure 16B). The NMR structure of the Δ1-24 Cys65Ala mutant that has an intramolecular disulfide bond (PDB code 2RQ0) was recently determined to be essentially identical to that of the Cys89,186Ala mutant,198 suggesting that the overall structure of wild-type L-PGDS is very similar to that of those mutants.
Figure 16. Tertiary structure of L-PGDS.
(A) NMR structure of mouse Δ1-24Cys89,186Ala L-PGDS.197 (B) The central cavity of L-PGDS.197 Inner accessible surface of the cavity with the polypeptide backbone is shown in gray. (C) X-ray crystallographic structure of mouse Δ1-24Cys65Ala L-PGDS.200 The conformational differences between the open and closed forms are colored in sky blue and pink, respectively, in the E-F loop. (D) Open and closed lids of the mouse L-PGDS cavity; E-F loop is shown in sky blue (open form) and pink (closed form).200 (E) X-ray crystallographic structure of human Δ1-28Cys65Ala L-PGDS bound with oleic acid (yellow) and palmitoleic acid (green).201
Molecular docking studies based on the results of NMR titration experiments with a stable PGH2-analog (U-44619) and retinoic acid suggest that PGH2 almost fully occupies the upper pocket 1, in which Cys65 is located and that all-trans-retinoic acid enters the lower pocket 2, in which amino acid residues important for retinoid-binding in other lipocalins are well conserved.197 Small-angle x-ray scattering analysis of the mouse Δ1-24 Cys65Ala mutant revealed that L-PGDS has a globular shape with the radius of gyration of 19.4 Å and that the radius is reduced to 18.8, 17.8, and 17.3 Å after binding to all-trans-retinoic acid, biliverdin, and bilirubin, respectively, indicating that the L-PGDS molecule becomes compact after binding of these non-substrate ligands.199
X-ray crystallographic structures of mouse Δ1-24 Cys65Ala L-PGDS were determined at 2.0 to 2.1 Å resolution and showed that the enzyme exists in two different conformations, one with an open calyx of the β-barrel and the other with a closed calyx due to a conformational change in the E-F loop (PDB codes: 2CZT and 2CZU, respectively).200 Their overall profiles (Figures 16C) are similar to the NMR solution structures and characterized by having a large cavity consisting of the two pockets described above. The upper compartment of the central large cavity contains the catalytically essential Cys-65 and its network of hydrogen bonds made with polar residues of Ser-45, Thr-67, and Ser-81; whereas the lower compartment is composed of hydrophobic amino acid residues highly conserved among other lipocalins. The pyrrolidine ring of Pro-110 in the E-F loop changes from its up-puckering configuration in the open conformer to a down-puckering one in the closed conformer; this change is accompanied by the synergetic rearrangement of the hydrogen bonds in the E-F loop (Figures 16C and 16D). X-ray crystallographic structures of human Δ1-28 Cys65Ala L-PGDS were recently determined in the complex with oleic acid and palmitoleic acid at 1.45 to 1.70 Å resolution (PDB codes: 3O19, 3O2Y, and 3O22).201 In this complex, the oleic acid binds in proximity to the catalytically critical Cys-65 and palmitoleic acid resides in a relatively neutral region of the upper part of the central cavity (Figure 16E). The overall structure is essentially the same as the structure of mouse L-PGDS.
3.2.4. Reaction mechanism of L-PGDS
L-PGDS is inactivated by sulfhydryl modifying agents, suggesting that a free cysteine residue is involved in the catalytic reaction of L-PGDS.168 Three cysteines, Cys-65, Cys-89, and Cys-186, in L-PGDS are conserved in all mammalian species.168 Cys-89 and Cys-186 are highly conserved among most, but not all, lipocalins and form a disulfide bridge, whereas Cys-65 is unique to L-PGDS and absent from other lipocalins. Chemical modification and site-directed mutagenesis revealed that the Cys-65 residue is the key residue in the catalytic reaction of L-PGDS.168 L-PGDS requires free sulfhydryl compounds, such as β-mercaptoethanol, dithiothreitol or GSH, for its reaction, suggesting that both endogenous and exogenous sulfhydryl groups are essential for the catalysis.168
Titration with a specific thiol-reacting agent, 2,2’-dithiodipyridine, under conditions of low reactivity (i.e. pH 4) revealed that Δ1-24-L-PGDS undergoes the chromogenic reaction 16-fold faster than GSH. This indicates that the SH group of Cys-65 is activated to a thiolate anion.200 Among various mutants of Δ1-24 L-PGDS, the sulfhydryl activation is decreased to 9% in the Ser45Ala mutant and to 7% in the Thr67Ala mutant. The Ser45Ala/Thr67Ala/Ser81Ala triple mutant shows the same rate of chromogenic reaction as that of GSH, whereas the Δ1-24Cys65Ala L-PGDS shows no chromogenic reaction. These results indicate that the hydrogen bond network of the catalytic Cys-65 with the hydroxyl cluster of Ser-45, Thr-67, and Ser-81 synergistically contributes to a decrease in the pKa of the thiol of Cys-65 and stabilizes the thiolate anion as the reactive group in the PGDS reaction at physiological pH.
Based on the similarity of the reaction mechanism between L-PGDS and H-PGDS, a reaction mechanism of L-PGDS was proposed (Figure 17); PGH2 is bound to the open form of L-PGDS and induces the closed conformation of the catalytic pocket with the proper geometry to allow the endoperoxide oxygen atom of C-11 of PGH2 to face the sulfur atom of Cys-65 for the PGDS reaction (Step 1). The endoperoxide oxygen atom at C-11 undergoes a nucleophilic attack by the thiolate anion of Cys-65 at the base of the closed cavity (Step 2) yielding the putative S-O adduct as an unstable reaction intermediate (Step 3). The labile S-O bond breaks autonomously with the proton rearrangement at the hydroxyl of Ser-45 and the C-11 to form the carbonyl group in a concerted manner (Steps 3 and 4). The product PGD2 is released after the opening of the Trp-54/His-111 gate together with a conformational change in the E-F loop associated with the change in up- and down-puckerings of the pyrrolidine ring conformations of Pro-110.202 The thiol proton of Cys-65 then dissociates to form the thiolate anion again as the reactive specimen.
Figure 17. Catalytic mechanism of L-PGDS.200.
Details of the various steps are presented in the text.
3.2.5. Conserved catalytic structures among H-PGDS, L-PGDS, and GSTs
An activated thiol is the common reactant among H-PGDS, L-PGDS, and GSTs (Figure 18). Thiol activation by a hydrogen bonding network also occurs in the case of H-PGDS and the θ class of GSTs. In H-PGDS, the phenolic hydroxyl group of Tyr-8 activates the cysteinyl thiol of enzyme-bound GSH and decreases the pKa from 8.5 to 7.8,194 in a manner similar to many other members of the GST family. However, in the θ class of GSTs,202 the activated thiol group of the bound GSH is also stabilized by hydrogen bonding with hydroxyl groups of serine residues. In L-PGDS, the Cys-65 residue is activated by hydrogen bonding interactions with the surrounding hydroxyl side chains of Ser-45, Thr-67, and Ser-81 in the catalytic pocket.200 Based on thiol titration analyses, the relative contribution of those amino acids in activating Cys-65 is in the order of Ser-45 ≈ Thr-67 > Ser-81. In a model of the hydrogen cluster of Δ1-24 Cys65Ala L-PGDS, Cys-65 forms a hydrogen bond with Ser-45 and Thr-67 and secondarily with Ser-81 to facilitate synergistically the thiolate formation of Cys-65.
Figure 18.
Thiol activation by hydrogen bond networks of L-PGDS, H-PGDS the σσσ class of GST, and the θθθ class of GST.200
H-PGDS and L-PGDS have distinct protein folds as members of the σ class of GSTs and the lipocalin family, respectively. However, because both enzymes catalyze the same reaction, they are expected to possess a similar tertiary structure of their catalytic pocket. When ones compare the catalytic pockets of H-PGDS and L-PGDS, the amino acid residues involved in the catalysis and the substrate binding of both enzymes can be geometrically superimposed at similar positions. The active thiol of GSH in H-PGDS interacts with the phenolic oxygen of Tyr-8; whereas the active thiol of Cys-65 in L-PGDS is surrounded by the hydroxyl cluster involving Ser-45, Thr-67 and Ser-81, which overlaps the position of the Tyr-8 phenolic oxygen of H-PGDS. The 15(S)-hydroxyl-bearing ω-chain of PGH2 is aligned with the hydrophilic polar residues of Arg-14, Tyr-152, and Cys-156 in H-PGDS and by those of Ser-133, Thr-147, and Tyr-149 in L-PGDS. The α-chain of PGH2 with its negatively charged carboxylate is considered to be anchored by a pair of positively charged groups, Lys-112 and Lys-198, in the exterior of the H-PGDS protein and by those of Arg-85 and Lys-92 in L-PGDS. In the superimposed model, Trp-104 of H-PGDS, which plays a role in opening the catalytic cleft to solvent, overlaps Pro-110 of L-PGDS. In the docking model of PGH2 within the catalytic pocket, the cyclopentane group of PGH2 with its endoperoxide is enwrapped by hydrophobic residues such as Phe-83 and Pro-110 of L-PGDS; the side-chain conformation of the latter residue being altered in the flexible E-F loop between the open-closed conformers, as described earlier (Figures 16C and 16D).
3.3. Inhibitor development
Knowledge of the tertiary structures of H-PGDS and L-PGDS has accelerated the development of inhibitors selective for each PGDS. Figure 19 shows the structures of several H-PGDS and L-PGDS inhibitors.
Figure 19. Structures of H-PGDS and L-PGDS inhibitors.
(A) Non-selective inhibitors (B) H-PGDS-selective inhibitors.179,192 (C) L-PGDS-selective inhibitor AT-56.207
3.3.1. Non-selective inhibitors
Stable PGH2 analogs such as U-44069 and U-46619 (Figure 19A) are competitive inhibitors of both H-PGDS and L-PGDS.200 Inorganic quadrivalent selenium (Se4+) compounds are noncompetitive and reversible inhibitors of L-PGDS.203 SeCl4 inhibits both H-PGDS and L-PGDS by modifying the catalytically important thiolate anion, and inhibition by SeCl4 is reversed by the addition of sulfhydryl compounds such as dithiothreitol.
3.3.2. H-PGDS inhibitors
HQL-79, chemically defined as 4-benzhydryloxy-1-[3-(1H-tetrazol-5-yl)-propyl]-piperidine (Figure 19B) is an orally active inhibitor of human H-PGDS. HQL-79 was the first H-PGDS inhibitor to be characterized.179 It was originally developed as a histamine H1 receptor antagonist, but a part of the anti-allergic and anti-asthmatic effects of HQL-79 was proposed to be mediated by the inhibition of PGD2 production.204 HQL-79 inhibits the activity of purified, recombinant human H-PGDS with an IC50 of 6 μM, but has almost no effects on purified PGHS-1, PGHS-2, L-PGDS or mPGES-1 when tested at concentrations of up to 300 μM.179 Kinetic analyses have revealed that HQL-79 inhibits the H-PGDS activity in a manner competitive with PGH2 (KI = 5 μM), and in a non-competitive manner with GSH (KI = 3 μM in the presence of 1 mM MgCl2).179 These results indicate that HQL-79 binds to the PGH2-binding site but not to the GSH-binding site, subsequently confirmed by the X-ray crystallographic analysis (Figure 16). The development of novel H-PGDS inhibitors, such as TFC-007 (Figure 19B), with increased selectivity and inhibitory potency is currently being extensively pursued192,193,205,206 based on crystal structures of the H-PGDS/HQL-79 complex.
3.3.3. L-PGDS inhibitors
AT-56 (4-dibenzo[a,d]cyclohepten-5-ylidene-1-[4-(2H-tetrazol-5-yl)-butyl]-piperidine) is an orally active selective inhibitor of L-PGDS (Figure 19).207 AT-56 was found in testing derivatives of HQL-79, an inhibitor of H-PGDS, based on the assumption that some of these derivatives might have an inhibitory activity toward L-PGDS, because both enzymes catalyze the same reaction. AT-56 is competitive with PGH2 (KM = 14 μM) with a KI value of 75 μM in enzyme assays conducted with purified human L-PGDS/β-trace or the mouse Δ1-24 Cys89,186Ala L-PGDS. AT-56 does not affect the activities of H-PGDS, PGHS-1, PGHS-2 or mPGES-1 at concentrations up to 250 μM.207 AT-56 inhibits the production of PGD2 by human TE-671 cells expressing L-PGDS after their stimulation with Ca2+ ionophore (5 μM A23187); the IC50 of ~3 μM. AT-56 does not affect PGD2 production by human megakaryocytes expressing H-PGDS.207 Orally administered AT-56 (30 mg/kg) decreases PGD2 production to 40% of control levels in brains of H-PGDS KO mice, in which PGD2 is produced by L-PGDS, and suppresses the accumulation of eosinophils and monocytes in the broncho-alveolar lavage fluid from an antigen-induced lung inflammation model using human L-PGDS-transgenic mice.207
3.4. Physiological and pathological involvement of PGDSs
3.4.1. Receptors and metabolism of PGD2
PGD2 binds to two distinct G protein linked receptors.208 One is the Gs-coupled DP receptor (DP1) identified as a homolog of receptors for various other prostanoids.209 The other is the Gi-coupled DP receptor (DP2 or CRTH2), a chemoattractant receptor for PGD2.210 DP1 are constitutively expressed in arachnoid trabecular cells of the mouse brain211 and are involved in the regulation of sleep;212 those found in human basophils and eosinophils and in mouse pulmonary and airway epithelial cells are induced by allergens and inflammation.149 DP2 are expressed in human Th2 cells, eosinophils, and basophils, and mediate the chemotaxis of these cells toward PGD2.213,214 PGD2 can also be recognized by Gq-coupled PGF receptors (FP) under certain conditions associated with a burst production of PGD2.208 The structures of several agonists and antagonists of DP1, DP2, as well as PGF receptors (FP) are shown in Figure 20.
Figure 20.
Structures of agonists and antagonists of DP1, DP2, and FP receptors.
PGD2 can be further metabolized by an 11-keto PGD2 reductase (a PGF synthase), belonging to the aldo-keto reductase (AKR) family, to 9α,11β-PGF2, a stereoisomer of PGF2α, which exerts various pharmacological actions different from those induced by PGF2α (reviewed by Smith et al.215). PGD2 is chemically unstable in aqueous solution and non-enzymatically dehydrated to produce PGs of the J series including PGJ2, Δ12-PGJ2, and 15-deoxy-Δ12,14-PGJ2 that have a cyclopentenone structure. 15-Deoxy-Δ12,14-PGJ2 acts as a ligand for a nuclear receptor, peroxisome proliferator-activated receptor γ (PPARγ), and promotes adipocyte and macrophage differentiation.216,217 However, the J series of PGs have never been detected in fresh tissue samples or body fluids,218,219 indicating that these PGs are artificially produced in vitro during incubation or storage of samples containing PGD2.
L-PGDS and H-PGDS double KO mice have been generated and found to grow normally,184,212 indicating that neither enzyme is of the housekeeping type but is involved in luxury and/or emergency processes. However, the KO mice show several functional abnormalities. Pharmacological studies with inhibitors and RNAi's for H-PGDS or L-PGDS have demonstrated that both enzymes are involved in various physiological and pathological events, as described in the next sections.
3.4.2. H-PGDS in inflammation and muscular dystrophy
H-PGDS contributes to the production of PGD2 in the spleen, thymus, intestine, and various peripheral tissues of rats.220 Northern blot analyses show that the tissue distribution profile of the mRNA for H-PGDS varies greatly among various species including rats,163 humans, mice,164 and chickens.165 H-PGDS is highly expressed in the oviduct of all three of the mammalian species, suggesting that H-PGDS plays an important role in female reproduction. However, a specific function of H-PGDS in the oviduct has yet to be established. H-PGDS is localized to Langerhans cells and dendritic cells in the skin,221,222 Kuppfer cells in the liver, the dendritic cells in the thymus and intestine,223 mast cells in a variety of tissues,154 human megakaryocytes,224 activated Th2 cells152 and eosinophils,225 and microglia of the brain.155,178,226,227
PGD2 produced by H-PGDS is involved in a variety of allergic and non-allergic disorders. For example, H-PGDS is expressed in infiltrates of mast cells, eosinophils, macrophages, and lymphocytes in the nasal mucosa of patients with polyposis225 or allergic rhinitis,228,229 in necrotic muscle fibers of patients with Duchenne's muscular dystrophy or polymyositis,230 and in microglial cells around the region of demyelination in twitcher mice,178 an animal model of human Krabbe's disease.
Orally administered HQL-79 inhibits antigen-induced production of PGD2 and ameliorates airway inflammation in wild-type and human H-PGDS-transgenic mice.179 PGD2 produced by H-PGDS in human mast cells and Th2 cells accelerates allergic and inflammatory responses by stimulating DP2 on Th2 cells in an autocrine manner, and DP1 and DP2 on the mast cells in a paracrine manner. Accordingly, human H-PGDS is a promising target for designing drugs to alleviate allergies and inflammation. HQL-79 administration also suppresses the astrogliosis, neuroinflammation, and demyelination seen in the genetic demyelinating twitcher mouse,178 the expansion of muscular necrosis in mdx mice,231 an animal model of Duchenne's muscular dystrophy, and secondary tissue damage after spinal cord contusion injury.232 These results suggest that blockade of H-PGDS/PGD2/DP signaling would be an effective therapy for neuroinflammation and muscular dystrophy. In short, H-PGDS inhibitors are new-concept drugs against a variety of allergic and non-allergic diseases.
3.4.3. L-PGDS in sleep regulation, neuroprotection, male genital development, cardiovascular and renal function, adipocyte differentiation, and bone formation
L-PGDS was originally identified in the brain and later found in male genitalia and in cardiovascular and other tissues. The physiological and pathological functions of this enzyme has previously been reviewed.233
The involvement of L-PGDS in sleep regulation has been extensively studied and this topic reviewed elsewhere.234 Human L-PGDS overexpressing transgenic mice sleep excessively after noxious stimulation such as tail clipping.181 L-PGDS- or DP1-null mice fail to exhibit a rebound from excessive sleep after sleep deprivation.235 The intraperitoneal administration of SeCl4, an inhibitor of PGDS, decreases the PGD2 content in the brain and induces almost complete insomnia for one hr after the administration.212 The SeCl4-induced insomnia is observed in H-PGDS KO mice but not in L-PGDS KO mice or DP1-KO mice.212 All these results indicate that the L-PGDS/DP1 system plays a crucial role in the regulation of physiological sleep; furthermore, the balance between PGD2 and PGE2 seems to be important in regulating sleep where PGE2 produced via mPGES1 is responsible for wakefulness.236 In the CNS, the L-PGDS/DP1 receptor system is also considered to be involved in pain sensation180 and food intake.237 Upregulation of L-PGDS gene expression was recently reported in brains of patients with various neurodegenerative diseases and in brains of animals having related neurologic abnormalities. Examples include twitcher mice,230,238 and patients with multiple sclerosis,239,240 Tay-Sachs or Sandohof diseases241 and mouse models of lysosomal storage diseases including Tay-Sachs’ and Sandhof's diseases, GM1 gangliosidosis, and Niemann-Pick disease.242 L-PGDS is also induced in the brain after hypoxic ischemic injury as a critical beneficial factor.243-245 L-PGDS is present in the cochlea246 and the retina,247 from which L-PGDS/β-trace is secreted into the perilymph and the interphotoreceptor matrix, although the functional significance remains elucidated.
The importance of L-PGDS in the male sexual differentiation has recently been studied in terms of the regulation of the sex-determining factor SOX9. PGD2 produced by L-PGDS induces nuclear import of SOX9248 and amplifies SOX9 activity in Sertoli cells during male sexual differentiation,249 while SOX9 enhances L-PGDS gene transcription.250,251
In the heart, L-PGDS is induced in hypoxia252 and mediates the protective effect of glucocorticoids on ischemia/reperfusion injury.253 L-PGDS gene expression is upregulated in vascular endothelial cells upon stimulation by shear stress254 and in asymptomatic atherosclerotic plaques obtained by carotid endarterectomy.255 A genetic polymorphism of L-PGDS has been identified in Japanese hypertensive patients with carotid atherosclerosis.256 The cells of Henle's loop and the glomeruli of the kidney also actively produce L-PGDS.257 PGD2 produced by L-PGDS is involved in adipocyte differentiation.258-261 As a consequence, L-PGDS KO mice exhibit nephropathy, atherosclerosis and obesity.262,263
In other tissues, L-PGDS is localized in melanocytes of the skin,263 acting as a regulator of the retinoic acid signaling.264 In the bone, L-PGDS is expressed in osteoblasts265 and the serum level increases after bone fracture.266 L-PGDS is induced in macrophages after treatment with bacterial endotoxin or Pseudomonus infection, and the clearance of Pseudomonus from the lung is improved in transgenic mice overexpressing human L-PGDS and impaired in L-PGDS KO mice, suggesting that L-PGDS plays a protective role in the host immune response.267 In Helicobacter pylori-induced gastritis, L-PGDS is induced in the gastric mucosa and plays protective roles against the inflammatory changes.268 L-PGDS is increased in the lamina propria infiltrating cells and muscular mucosa in patients with ulcerative colitis. In a dextran sodium sulfate-induced colitis model, L-PGDS KO mice show lower disease activity than the wild-type mice, suggesting the exacerbating role of L-PGDS in this model.269
3.4.4. L-PGDS/β-trace as a clinical marker and an extracellular transporter
As noted earlier in Section 3.1.2., L-PGDS is the same protein as β-trace,175,177 a major protein of human CSF176 that was later identified in the seminal plasma, serum, and urine. The L-PGDS/β-trace concentration in body fluids is proposed to be useful as a clinical marker for various diseases.
Several groups reported a relationship between L-PGDS and human physiological and pathological sleep. Jordan et al.270 reported that the serum L-PGDS/β-trace concentration shows a circadian change with a nocturnal increase, which is suppressed during total sleep deprivation but not affected by rapid eye movement sleep deprivation. More recently they also demonstrated that narcolepsy increases the serum L-PGDS/β-trace level, an increase that correlates with excessive daytime sleepiness but not with cataplexy.271 Bassetti et al.272 reported that CSF levels of L-PGDS are significantly lower in patients (n = 34) with excessive daytime sleepiness, when compared with levels in healthy controls (n = 22). Barcelo et al.273 reported that serum levels of L-PGDS are higher in obstructive sleep apnea patients with excessive daytime sleepiness than those without excessive daytime sleepiness. Both reports proposed that L-PGDS measurements could provide a neurochemical assay for excessive daytime sleepiness.
Serum L-PGDS levels have been proposed as a biomarker for the severity of stable coronary artery disease274 and subclinical atherosclerosis in untreated, asymptomatic subjects.275 Urinary secretions of L-PGDS are enhanced after injury to glomerular capillary walls. A cross-sectional and prospective multicenter study in Japan revealed that urinary secretions of L-PGDS are a better predictor of the future status of renal injury in type-2 diabetes associated with <30 mg/g creatinine albuminuria than other markers including urinary excretions of type-IV collagen, beta-2-microglobulin and N-acetyl-β-D-glucosaminidase and serum creatine levels.276
The substrate of L-PGDS, PGH2, is produced by PGHS-1 or PGHS-2, both of which are membrane-binding enzymes localized in the ER and/or outer nuclear membrane. PGH2 is relatively unstable in aqueous solution with a t1/2 of < 5 min; it is nonenzymatically degraded to several products including PGE2. Therefore, L-PGDS/β-trace is unlikely to act as an enzyme in those body fluids. L-PGDS binds retinoids (KD = 70 - 80 nM),264 bilirubin and biliverdin (KD = 33 - 37 nM),277 as well as GM1 and GM2 gangliosides (KD = 65 - 210 nM)242 (that accumulate in neurons in certain lysosomal storage diseases). Thus, like other lipocalins L-PGDS/β-trace may act as an extracellular transporter for various hydrophobic compounds.
L-PGDS/β-trace purified from human CSF tightly binds to amyloid β (Aβ) peptide (1-40), Aβ(1-42), and their fibrils with high affinity (KD = 18-50 nM) and inhibits both the spontaneous and the seed-dependent aggregation of Aβ(1-40) or Aβ(1-42) with a KI of 0.75 μM,278 which is in the physiological range (1-5 μM) of the L-PGDS/β-trace concentration in human CSF. Moreover, the inhibitory activity toward seed-dependent Aβ aggregation in human CSF is decreased by 60%, when L-PGDS/β-trace is removed from human CSF by immunoaffinity chromatography. This suggests that L-PGDS/β-trace is a major amyloid β-chaperone in human CSF.278 When Aβ(1-42) is injected into the lateral ventricle of the brain, Aβ deposition is 3.5-fold higher in L-PGDS-KO mice and reduced to 23% in L-PGDS-over-expressing transgenic mice, as compared with the wild-type deposition. Thus, L-PGDS/β-trace appears to be an endogenous Aβ-chaperone in the brain, and disturbance of this function of the protein may be involved in the onset and progression of Alzheimer's disease. In fact, L-PGDS activity is decreased in the CSF of patients with HIV dementia.279
4. Prostaglandin E synthases (PGESs)
The existence of enzymes that catalyze the formation of PGE2 from PGHS-derived PGH2 was recognized many years ago, and attempts to purify the membrane-associated and glutathione-dependent enzyme(s) from seminal vesicles were reported in the 1970s and 1980s.280,281 In 1999, 2000 and 2002, three distinct PGESs were cloned and characterized, namely the inducible microsomal prostaglandin E synthase-1 (mPGES-1),282 cytosolic prostaglandin E synthase (cPGES)283 and microsomal prostaglandin E synthase-2 (mPGES-2).284 Before that, certain soluble GSTs had been identified that catalyzed the formation of PGE2 from PGH2; however, no biological function has been attributed this activity of these enzymes.185,285 mPGES-1 has been demonstrated to play an important role in several human diseases by promoting inflammation, pain and fever; this topic has been the subject of several recent reviews.286-288 This enzyme also seems to participate in the hemostasis promoting atherosclerosis.289 An increasing number of articles also deal with a role for mPGES-1 in various types of cancers.290,291 In contrast, cPGES and mPGES-2 are for the most part constitutively expressed and therefore more likely to play a role in homeostasis;287 a possible role for cPGES in nociception has been described292 while the importance of mPGES-2 is unclear. Genetic deletion studies in mice have been unable to support a role of either cPGES or mPGES-2 as PGESs in vivo.293,294 In this section, we will review the biochemical characterizations of these enzymes and at the end provide a brief update on the development of mPGES-1 inhibitors.
4.1. Characterization of PGESs
mPGES-1 belongs to a protein superfamily termed MAPEG (Membrane Associated Proteins in Eicosanoid and Glutathione metabolism), a family of proteins, all whose members except 5-lipoxygenase activating protein (FLAP), possess glutathione conjugating capacity.295,296 After the cloning of FLAP and leukotriene (LT) C4 synthase, both of which are involved in leukotriene biosynthesis and are discussed elsewhere in this issue, it was realized that these proteins were related with an amino acid sequence identity of about 30%, similar size (17 kDa) and had similar hydropathy profiles. This motivated a search for other members of this protein family and subsequent work revealed two novel microsomal GSTs (MGST2 and MGST3).297,298 At this time it was also recognized that MGST1 must also be part of this protein family, and a third novel protein was identified as an orphan homologue of MGST1 with 38% amino acid sequence identity and named MGST1-like 1 (MGST1-L1). At the point where the identity of these six human proteins were known, the MAPEG superfamily was defined based on enzymatic activities, structural properties and sequence similarities (Figure 21).295,296 Encouraged by the identification of MGST2 and MGST3 investigations were performed to determine if any of the MAPEG members exhibited PGH2 metabolizing activity because previous reports had shown that prostaglandin synthases might be membrane proteins and be sensitive to glutathione. Subsequently, MGST1-L1 was found to possess prostaglandin (PG) E2 synthase activity and was renamed microsomal prostaglandin E synthase 1 (mPGES-1).282 mPGES-1 is a glutathione-dependent, inducible enzyme that was shown to couple to both PGHS-1 and PGHS-2 but preferentially to PGHS-2, using a HEK293 cell system in which mPGES-1 was overexpressed together with either of the PGHS isoenzymes.129
Figure 21. Multiple sequence alignment of the six human MAPEG members.
Every 10th residue is underlined and the total numbers of residues are given for each protein. Conserved residues of the MAPEG superfamily are highlighted with red letters. Note that some of these residues are not conserved in FLAP, which is consistent with its lack of GSH binding and enzymatic activity. For mPGES-1 the positions of the four transmembrane helices are indicated and crucial residues located at the active site or important for structural integrity, respectively, are highlighted with blue or gray background. These residues are further discussed in the text and shown in detail in Figure 25. Adapted from Reference 309.
At the same time and using a different approach, Tanioka et al.283 were looking for the identity of a prostaglandin E synthase that acted in concert with PGHS-1 in the so-called “immediate” phase of PGE2 biosynthesis. They purified a 50 kDa (26 kDa on SDS PAGE) glutathione-dependent enzyme from brain cytosol isolated from LPS treated rats. The enzyme was identified as p23, a component of the steroid hormone receptor/hsp90 complex and the protein was referred to as cytosolic (c) PGES. By northern and western blotting, cPGES was found to be induced in the brain of rats treated with LPS; otherwise, the enzyme was detected in many tissues and cells with an overall constant level of expression independent of LPS challenge. Using the HEK293 cell system, cPGES was demonstrated to function in the immediate phase of PGE2 release in conjunction with cPLA2 and PGHS-1 following calcium ionophore stimulation.283
The third prostaglandin E synthase to be identified was first isolated from the microsomal fraction of bovine heart and subsequently the monkey orthologue was cloned, expressed in E. coli and enzymatically characterized as membrane-associated prostaglandin E synthase-2 (mPGES-2).284 This highly active 33 kDa protein requires reducing agents but is not restricted to glutathione. mPGES-2 has an N-terminal 87 amino acid sequence that anchors it to the membrane, but after cleavage the bulk of the active protein becomes more soluble. Again, using the HEK293 system, mPGES-2 was found to couple with either PGHS-1 or PGHS-2 nonselectively.299
4.2. PGES substrates and enzyme activities
The conversion of PGH2 into PGE2 is catalyzed by prostaglandin E synthases (Figure 1) in the presence of GSH or, in the case of mPGES-2, which is of questionable relevance as a PGES in vivo, also by other reducing equivalents. The apparent kinetic parameters for all terminal prostaglandin synthases, including PGESs, using PGH2 as the substrate are summarized in Table 1. Purified recombinant mPGES-1 also catalyzes the formation 15-hydroperoxy-PGE2 from the PGHS intermediate PGG2300 (Figure 22), in line with early observations using semi-purified enzyme.280 The physiological significance of the formation of 15-hydroperoxy-PGE2 has not been thoroughly investigated.
Figure 22. Formation of PGE2 from PGG2.
The COX activity of PGHSs forms PGG2. It has been demonstrated that mPGES-1 catalyses the intra-molecular oxido-reduction at similar rates regardless of the nature of the substituent at carbon 15, forming either PGE2 or 15-hydroperoxy-PGE2 which, if formed, is probably rapidly reduced into PGE2 either non-enzymatically or via various peroxidases.
Table 1 shows the apparent kinetic parameters (kcat, KM and kcat/KM) for PGH2, PGG2 and GSH as determined with purified, recombinant mPGES-1 by different laboratories. The disparity in the results most likely reflects difficulties in assaying mPGES-1. It should be noted that the expression systems, purification procedures, activity assays, including temperatures and detergents used, were substantially different among laboratories. In particular, differences in detergents may significantly influence kinetic parameters. It is interesting to note that the KM determination for PGH2 was similar in the two studies using E. coli as source of enzyme while substantially lower using insect cells. Nonetheless, the catalytic efficiency as judged from the kcat/KM values of mPGES-1 is high, although possibly somewhat lower with the mouse enzyme.
Due to the close homology between mPGES-1 and MGST1, it was also of interest to study some typical glutathione-S-transferase (GST) substrates. mPGES-1 catalyzes the conjugation of CDNB with GSH at a relatively slow but significant rate (0.81 ± 0.04 μmol min-1 mg-1); furthermore, like MGST1, mPGES-1 catalyzed the reduction of lipid peroxides such as cumene hydroperoxide and 5-hydroperoxy eicosatetraenoic acid with specific activities of 0.17 ± 0.02 and 0.043 ± 0.001 μmol min-1 mg-1, respectively.300 The physiological role(s), if any, of these activities have not been further investigated. These results were not reproduced using mPGES-1 over expressed and purified from insect cells.301 Also, no GST related activities were reported for mPGES-2302 or cPGES.283
As discussed above in the context of PGHS substrate specificities, ω3 fatty acids are considered beneficial for human health. A high intake of fish oil leads to accumulation of EPA over AA. EPA is converted to PGH3 by PGHS-2, and mPGES-1 is able to produce PGE3 from PGH3 although at a third of the rate of conversion of PGH2 to PGE2.103 Hypothetically, an increased formation of PGE3 relative PGE2 could diminish inflammation and evoke a positive effect on, for instance, the cardiovascular system. Yet another group of substrates for the PGHS-2/mPGES-1 pathway are the endocannabinoids 2-arachidonylglycerol (2-AG) and N-arachidonylethanolamide (AEA or anandamide). These compounds are oxidized via PGHS-2 into their corresponding fatty acid prostaglandin endoperoxide H2 homologs, and, in turn, isomerized into the respective PGE2 moieties by mPGES-1.303 Endogenous 2-PGE2-glycerolester has been identified in rat paws by mass spectrometry and shown to evoke nociceptive pain304 although questions remain about its biosynthesis and physiological importance.
4.3. PGES structure-function relationships
After the initial characterizations of FLAP and LTC4 synthase, the quaternary structures of these proteins were discussed relating to their hydropathy profiles and inhibitor binding profiles. Hydrodynamic studies and electron crystallography were employed to investigate the macrostructure of detergent solubilized mPGES-1.300 Analytic centrifugation demonstrated a sedimentation coefficient of 4.1 S, partial specific volume of 0.891 cm3/g and a Stokes radius of 5.09 nm. The Svedberg equation was used to calculate the molecular mass of the mPGES-1-Triton X-100 complex, which was found to be 215 kDa. The detergent content of the mPGES-1-Triton X-100 complex was 2.8 g Triton X-100/g protein and after subtracting the values for the detergent content the calculations matched best with a trimeric quaternary structure. The trimeric structure also matched with early data from electron crystallography studies. The first MAPEG protein to be structurally defined using electron crystallography was MGST1 in complex with glutathione.305 The 3.2 Å structure indicated that the protein was a homotrimer. A year later the structures of FLAP306 and LTC4 synthase307,308 were reported using x-ray crystallography, and in 2008 the structure of mPGES-1 was reported by electron crystallography.309 The known 3D structures of MAPEG proteins are summarized in Figure 23. The structure of cPGES/P23 has been known since 2000,310 and the structure of mPGES-2 was reported in 2005.311 The detailed structure of mPGES-2 led to a comprehensive understanding of the substrate binding pocket and a plausible reaction mechanism was outlined. Recombinant monkey mPGES-2 was crystallized with an N-terminal extended His tag at position 88 (i.e. without the N-terminal 87 amino acid signal sequence) in complex with indomethacin.311 mPGES-2 was observed to be a homodimer. Despite a quite low degree of sequence identity, the structure revealed striking similarities with hematopoietic prostaglandin D synthase (H-PGDS) and several classes of GSTs. A V-shaped cavity in each monomer represents the active surface by harboring indomethacin and a cysteine residue at position 110, which had previously been shown to be part of a thioredoxin active site motif and also experimentally determined to be essential for mPGES-2 catalytic activity.312 A glutathione peroxidase like mechanism reaction mechanism was suggested involving an attack of a thiol on the O-9 of the prostaglandin endoperoxide bridge (Figure 24). The authors proposed that the endoperoxide moiety of PGH2 is oriented near Cys-110 which acts as a proton donor to the peroxide oxygen at C-11 and then forms a sulfenate ester with the oxygen at C-9. A polarized water molecule subsequently abstracts a proton from C-9, the ester bond between Cys-110 and the C-9 oxygen is broken, and the keto group of PGE2 is formed.
Figure 23. MAPEG proteins with known 3-D structures.
The overall structures of microsomal PGES-1 (mPGES-1), microsomal glutathione-S-transferase-1 (MGST1), leukotriene C4 synthase (LTC4S) and 5-lipoxygenase activating protein (FLAP) are compared. Each monomer of the trimeric proteins is represented with a unique color with its N- and C-terminals facing downwards and cytosolic loops facing upwards. Membrane boundaries are indicated with horizontal lines. The four transmembrane helices are shown with GSH occupying the space between subunits 1 and 4 of mPGES-1, MGST1 and LTC4S. In FLAP, the corresponding space is occupied by the inhibitor MK-591. GSH and MK-591 are depicted in spherical models with carbon atoms colored yellow. Note that in MGST1 GSH adopts an elongated conformation while it is more condensed in mPGES-1 and LTC4S. Structures and in parenthesis PDB ID numbers are MPGES1 (3DWW), MGST1 (2H8A), LTC4S (2UUI), FLAP (2Q7M).
Figure 24. The proposed reaction mechanism for prostaglandin E synthase catalysis.
The mechanism could involve either a thiol attack by via a glutathione peroxidase like mechanism, or alternatively, the reaction could occur via acid-base catalysis.
The structures of MAPEG proteins reported to date are very similar, and some of these structures have been previously reviewed.313,314 They are homotrimers and each monomer contains four transmembrane (TM) alpha helices (Figure 23). The three GSH molecules per homotrimer are all localized between two adjacent monomers corresponding to the polar space between TM1/2 and TM3/4 of the respective monomer as shown in MGST1,305 LTC4 synthase307,308 and mPGES-1.309 In FLAP, the leukotriene biosynthesis inhibitor MK-591 was situated in a corresponding space limited by TM1 and TM4.306 Interestingly, in one of the structures of LTC4 synthase, a detergent molecule was aligned along the cleft between TM1 and TM4, in an area suggested to correspond to the volume occupied by its second substrate LTA4.307 Furthermore, Arg-104, which is in close proximity to the LTA4 epoxide group, was suggested to lower the pKa of the GSH sulfhydryl group307,308 allowing it to attack the allylic C-6 resulting in the formation of the GSH conjugate. The important roles of Arg-31 and Arg-104 were recently experimentally verified.315,316 In marked contrast, the cleft between TM1 and TM4 of adjacent monomers was not recognizable in the structure of mPGES-1.309 Thus, the most likely entry point for PGH2 reaching the glutathione was blocked and no alternative paths were found. Based on the similarities with LTC4 synthase, a homology model was built that displayed an open configuration. mPGES-1 seems to shift between open and closed conformations. The relevance of this observation remains to be elucidated, but one can speculate about a role in substrate specificity or in the handling of the GSH molecules, which are not consumed during catalysis301 as compared with the cases for GSTs, like LTC4 synthase or MGST1. In the active site of mPGES-1 Arg126 seems to correspond to Arg104 of LTC4 synthase. This arginine points towards the GSH309, has been shown critical for enzyme activity317 and is therefore likely to stabilize the glutathione thiolate anion supporting the glutathione peroxidase like mechanism depicted in Figure 24. Using backbone amide H/D exchange mass spectrometry, the inhibitor binding sites of different types of mPGES-1 inhibitors were investigated. The results provide further support for the outlined location of the active surface in mPGES-1.318
Figure 25 summarizes some of the known structure-function aspects of mPGES-1. Further mechanistic studies are required in order to verify the suggested reaction mechanism catalyzed by mPGES-1. Also, the membrane topology of mPGES-1 needs further investigation. It is anticipated, based on charge distribution algorithms that the N and C terminals face the luminal side of the endoplasmic reticulum and the soluble loops connecting TM1-2 and TM3-4 face the cytosolic side. Biochemical determination of the membrane topology as well as the intracellular localization of mPGES-1 in relation to the suggested localization of PGHS could lead to a better understanding how AA-derived substrates and products flow between cellular compartments and how PGHS may interact with mPGES-1.
Figure 25. Summary of the mPGES-1 structure.309.
(A) The human mPGES-1 homotrimer in an open configuration. Monomers are colored green, gray, and white, respectively. GSH is depicted in a stick representation with its carbon atoms colored in yellow. (B) The inhibitor binding site as observed via the V-shaped cleft between subunits 4 and 1 of neighboring monomers (colored in green and gray, respectively). Residues Thr-131, Leu-135, and Ala-138 (in stick representation with carbon atoms colored in dark blue) corresponding to Val-131, Phe-135, and Phe-138 in rat mPGES-1 together account for the major differences in inhibitor efficacies. Arg-126 (in stick representation with carbon atoms colored in light blue) is suggested to be the catalytic residue lowering the pKa of the glutathione sulfhydryl group. (C) The residues that function in subunit stabilization between TM2 and TM3, Arg-110 (in stick representation with carbon atoms colored in orange) makes polar contacts to residues Asn-74, Glu-77, and Thr-78 (all in stick representation with carbon atoms colored in magenta) as well as to GSH (in stick representation with carbon atoms colored in yellow). (D) The same part of the protein as in the upper right but rotated 180°.
4.4. mPGES-1 inhibitors
Since the discovery of mPGES-1 in 1999 and despite the fact that many companies and academic groups have worked to develop mPGES-1 inhibitors,319 no clinical trials have yet been reported. This signals that problems have hampered the development process. One such problem is the species differences as first described by Merck scientists.320 Potent inhibitors against the human enzyme may partly or completely lose their potencies against the rodent isozymes. The combined role of three individual amino acids, different among human and rodent mPGES-1, located in TM4 and pointing to the substrate entry point, seems to account for most of the observed species differences for the mPGES-1 inhibitor tested.321 Another problem may be related to efficacy. Theoretically, the shunting of PGH2 into, for instance PGI2, may in cases override the analgesic effect of selective mPGES-1 inhibition as argued.322 Therefore, it will be very important to develop inhibitors that potently inhibit mouse and rat mPGES-1 in order to test mPGES-1 inhibitors in various experimental models of nociception.
One may argue that inhibition of mPGES-1 will bring fewer side effects than traditional NSAIDs or PGHS-2 specific drugs (coxibs). For instance, in contrast to mice lacking PGHS-2, which develop kidney problems during intra uterine life as well as develop heart fibrosis as they grow older, mice devoid of mPGES-1 are seemingly healthy in the absence of any provocation. It also seems that mPGES1 inhibition may exhibit a positive cardiovascular effect due to more systemic prostacyclin formation.289 Thus, although it is anticipated that mPGES-1 inhibitors will be safer than traditional NSAIDs, published data from using knockout mice show that there may be certain safety problems. The most serious side effects of NSAIDs including coxibs are related to the cardiovascular system. Mice devoid of mPGES-1 have been shown to be protected from developing atherosclerosis; however, a rise in blood pressure was observed in some but not all cases following provocations, a matter further discussed along the terms of genetic backgrounds of the mice.323 Also after myocardial infarction, mPGES-1 seems to function in the healing process.324 So far, most publications support a role of mPGES-1 as a target for treating symptoms like pain and fever as well as diseases such as rheumatic diseases,288 cancer,291 inflammatory neurological disorders,325,326 ischemic stroke327 and some additional diseases and conditions like sickness behavior including anorexia and breathing dysfunctions in infants.328,329 Thus, mPGES-1 is a complex target, and there are even several more conditions reported in which targeting mPGES1 might be useful. Inhibition will undoubtedly lead to side effects, the seriousness of which are unknown. How will an mPGES-1 inhibitor compare with a PGHS inhibitor? The side effects and perhaps also the effects will depend on the actual organs and cells behind the disease. For instance, macrophages in the arthritic synovial membrane produce PGE2 but also TxA2, PGI2 and perhaps PGD2. If more TxA2 is produced this may damage the vascular system; if more PGD2 is produced it may enhance the anti-inflammatory effect obtained by reducing PGE2; and lastly, if more PGI2 is produced, nociception may persist. In summary, it remains to be determined whether selective pharmacological inhibition of mPGES-1 will result in the same effects as conferred by mPGES-1 gene deletion studies. Direct comparisons between mPGES-1 inhibitors and COX-2 inhibitors will provide important insights about the relative and putative side effects elicited by mPGES-1 inhibition.
5. Prostaglandin F synthases (PGFSs)
Prostaglandin (PG) F2α and PGE2 were the first prostanoids to be isolated from human semen, and their chemical structures were also the first to be determined among various prostanoids. PGF2α represses ovarian steroidgenesis and initiates parturition in mammals. It also induces potent smooth muscle contraction of the human uterus, and so this prostanoid and its derivatives are widely used clinically for induction of delivery. However, despite the long history of research on the physiological and pathological functions of PGF2α, the identity of the enzyme(s) that catalyzes PGF2α synthesis in vivo is unclear.
There are two pathways for PGF2α production (Figure 26)330 involving two electron reductions of the 9,11-endoperoxide group of PGH2 or the 9-keto group of PGE2; in addition, reduction of the 11-keto group of PGD2 yields 9α,11β-PGF2, a PGF2α stereoisomer.331 PGFS (EC 1.1.1.188) catalyzes the first reaction, the reduction of PGH2 to PGF2α; whereas PGE2 9-ketoreductase and PGD2 11-ketoreductase are responsible for the other two reactions. Most of these enzymes belong to the aldo-keto reductase (AKR) superfamily.
Figure 26. Reactions and enzymes involved in forming different PGF isomers from PGH2.346.
AKR, aldo-keto reductase.
AKRs are soluble monomeric proteins having a molecular mass of 37 kDa and NADPH-dependent oxido-reductase activity. AKR proteins are widely distributed in prokaryotes and eukaryotes, fall into 15 families,332 and metabolize a number of substrates including aldehydes, monosaccharides, steroids, polycyclic hydrocarbons, isoflavonoids, and PGs in the presence of NADPH.333 Members of AKR1C and 1B subclasses have been characterized as PGFSs in mammals; and those of the 5A subclass, in protozoan parasites such as Trypanosoma brucei, Leishmania major, and L. donovani; the first mentioned of which causes African sleeping sickness and the other two cause Leishmaniasis. PGFS purified from the New World parasite T. cruzi, which causes Chagas’ disease, is classified as a member of the “old yellow enzyme” family.334 PGF ethanolamide synthase, which belongs to the thioredoxin-like superfamily, was the most recently found enzyme to catalyze the reduction of PGH2 to PGF2α.335
5.1. Catalytic and molecular properties
5.1.1. Purification and properties of PGFS in the AKR1C subfamily
Watanabe et al.336 first isolated PGFS from bovine lung in 1985 as a cytosolic monomeric enzyme with a Mr of 30,500 and found it to catalyze the reduction of both the 9,11-endoperoxide of PGH2 (KM = 10 μM; VMAX = 57 nmol/min/mg protein) and the 11-keto group of PGD2 (KM = 120 μM; VMAX = 126 nmol/min/mg protein) to produce PGF2α as well as the reduction of 9,10-phenanthrenequinone (KM = 0.7 μM; VMAX = 378 nmol/min/mg protein), in the presence of NADPH. The product of PGFS from PGD2 was later determined to be 9α,11β-PGF2,331 not PGF2α.336 In 1988, they isolated the cDNA for bovine lung PGFS and showed that this enzyme is composed of 323 amino acids with a Mr of 36,666 and is highly homologous to human liver aldehyde reductase and ε-crystallin of the European common frog,337 both of which belong to the AKR family. Bovine lung PGFS is now classified as AKR1C7.338,339
Among various bovine tissues, the liver has the highest PGD2 11-ketoreductase activity, followed by lung and spleen. The PGD2 11-ketoreductase activity in the latter two tissues is catalyzed by AKR1C7, whereas the activity in the liver is due to another enzyme that is partially cross-reactive with anti-(bovine lung PGFS) antibody.340 Bovine liver PGFS has been purified to apparent homogeneity and shown to be associated with the activities of PGFS (KM for PGH2 = 25 μM; VMAX = 3 nmol/min/mg protein), PGD2 11-ketoreductase (KM for PGD2 = 10 μM; VMAX = 95 nmol/min/mg protein), and 9,10-phenanthrenequinone reductase (KM = 2 μM; VMAX = 339 nmol/min/mg protein).341 cDNA cloning of bovine liver PGFS showed that this enzyme is composed of 323 amino acids with an Mr of 36,742. It is highly homologous to bovine lung PGFS (87% amino acid sequence identity) and human liver dihydrodiol dehydrogenase isozymes (76 to 79% identity).342 Bovine liver PGFS is now classified as AKR1C11.
The human homolog (AKR1C3) of bovine lung PGFS (AKR1C7) is the same as 3α-hydroxysteroid dehydrogenase type II and is composed of 323 amino acids with an Mr of 36,842. AKR1C3 has the activities of PGFS (KM for PGH2 = 10 μM; VMAX = 100 nmol/min/mg protein), PGD2 11-ketoreductase (KM for PGD2 = 1.7 μM; VMAX = 258 nmol/min/mg protein), and 9,10-phenanthrenequinone reductase (KM = 0.8 μM; VMAX = 3,000 nmol/min/mg protein). This enzyme can also catalyze the reduction of prostaglandin D2 ethanolamide (prostamide D2) to 9α,11β-PGF2 ethanolamide (9α,11β-prostamide F2; KM for prostamide D2 = 50 μM; VMAX = 490 nmol/min/mg protein).343,344 A prostamide F2 analog and an anti-glaucoma agent, Bimatoprost, potently inhibit PGFS and PGD2 11-keto reductase, and prostamide F2 synthase activities of AKR1C3.343
PGE2 9-ketoreductase was first purified from the corpus luteum of pseudopregnant rabbits by Wintergalen345 and showed to be a member of the AKR family. This enzyme catalyzes the reduction of the 9-keto group of PGE2 to produce PGF2α (KM for PGE2 = 122 μM; VMAX = 3,250 nmol/min/mg protein) as well as the dehydrogenation of 20α-hydroxysteroid (KM for 20α-hydroxypregn-4-en-3-one = 28 μM; VMAX = 10,350 nmol/min/mg protein) and the reduction of 9,10-phenanthrenequinone (KM = 4.5 μM; VMAX = 210 nmol/min/mg protein). Rabbit corpus luteum PGE2 9-ketoreductase is now classified as AKR1C5.
The PGFS properties of the above-mentioned AKR1C subclass members were previously reviewed by Watanabe.330 These members display, in general, a PGFS activity with lower affinity for PGH2 and lower VMAX value than do members of the AKR1B subfamily346 and are associated with a PGD2 11-ketoreductase activity that is higher than the PGFS activity. In a COX/AKR1C enzyme co-expression study using HEK293 cells, AKR1C7 failed to couple with either COX isoform.347,348 Thus, members of the AKR1C subfamily though having PGFS activity in vitro seem not to contribute to in vivo of PGF2α production.
5.1.2. Characterization of AKR1B enzymes as the primary PGFSs
Aldose reductases (EC 1.1.1.21), such as human placental aldose reductase (AKR1B1) and mouse kidney aldose reductase (AKR1B3), are considered to be the prototypical members of the AKR superfamily. Beside these canonical aldose reductases, a second group named aldose reductase-like proteins (ARLP) have been characterized on the basis of sequence homology (at least 60 - 70% identity with aldose reductase). AKR1B7, initially identified as a mouse vas deferens, androgen-dependent protein, belongs to the ARLP group. These members of the AKR1B subfamily catalyze the conversion of PGH2 to PGF2α with apparent affinities for PGH2 (KM = 1.9-9.3 μM) and VMAX (26-53 nmol/min/mg protein) values higher than those of the AKR1C subclass.346 AKR1B1, AKR1B3, and AKR1B7 catalyze the reduction of both p-nitrobenzaldehyde (KM = 9-12 μM, VMAX = 52 to 480 nmol/min/mg protein) and the 9,11-endoperoxide group of PGH2 (to PGF2α) but do not reduce PGD2 to 9α,11β-PGF2 nor PGE2 to PGF2α; thus, AKR1B isoforms differ from members of the AKR1C subfamily.
20α-Hydroxysteroid dehydrogenase (AKR1B5), the bovine homolog of AKR1B1, also exhibits PGFS activity (KM for PGH2 = 7 μM, VMAX = 24 nmol/min/mg protein), a reductase activity comparable of that toward 17-hydroxyprogesterone (33.6 nmol/min/mg protein at a substrate concentration of 20 μM) or 17-hydroxypregnenolone (11.9 nmol/min/mg protein also at a substrate concentration of 20 μM) and higher than the PGFS activities of AKR1C enzymes.349 Bovine endometrium strongly expresses AKR1B5 at the time of luteolysis, when the PGF2α production is active, but does not contain members of AKR1C subclasses, suggesting that AKR1B5 is most likely the enzyme responsible for the production of PGF2α in the bovine endometrium.349
In the human endometrium, AKR1B1 is expressed at a high level during the menstrual cycle, with a peak at the secretory phase in both epithelial and stromal cells, whereas AKR1C3 is found only in the epithelial cells.350 A decrease in AKR1B1 elicited by specific siRNA knockdown is associated with a significant reduction in PGF2α production by human endometrial stromal and epithelial cell lines.350 Bresson et al.350 found that Ponalrestat (Statil), a specific inhibitor of AKR1B1, reduces PGF2α production in response to IL-1β in both cultured endometrial cells and endometrial explants obtained by biopsy or hysterectomy. Based on these findings, they concluded that AKR1B1 is the primary PGFS in the human endometrium and suggested that this enzyme may also be a potential target for the treatment of menstrual disorders. However, AKR1B1 was recently shown to be associated with complications of diabetes.351
Fujimori et al. recently demonstrated that the siRNA for AKR1B3 suppresses PGF2α production by mouse 3T3-L1 preadipocytes352 or during adipogenesis of mouse ST2 mesenchymal stem cells,353 indicating that AKR1B3 is the primary PGFS in mouse preadipocytes. Functional coupling between COX-2 and AKR1B7 to produce PGF2α has been demonstrated in HEK293 cells.354 Therefore, AKR1B3 and AKR1B7 qualify as the primary candidates as PGFSs in mice.
There are two AKR inhibitors, tolrestat and sorbinil, that efficiently inhibit the PGFS activity of purified AKR1B1 (KI = 3.6 and 21.7 μM, respectively) and AKR1B3 (KI =0.26 and 0.89 μM, respectively) in a non-competitive or mixed-type manner, whereas these inhibitors do not affect the appreciably affect the PGFS activity of AKR1B7 (KI = 9.2 and 18 mM, respectively).346 Ponalrestat, a specific inhibitor of AKR1B1, reduces PGF2α production in response to IL-1β in both cultured endometrial cells and endometrial explants.350
5.1.3. Purification and properties of T. brucei PGFS (AKR5A2) and L. major PGFS (AKR5A1)
Infection of mammals by African trypanosomes is characterized by increased levels of PGF2α and PGD2 in plasma355 and CSF,356 respectively. Kubata et al.357 demonstrated that the protozoan parasite T. brucei produces PGs enzymatically from AA and PGH2, and they purified a novel T. brucei PGFS (TbPGFS) and cloned its cDNA. TbPGFS is composed of 323 amino acids with an Mr of 36,842, and has the activities of PGFS (KM for PGH2 = 1.3 μM; VMAX = 2000 nmol/min/mg protein) and 9,10-phenanthrenequinone reductase (KM = 0.4 μM; VMAX = 48,000 nmol/min/mg protein) but does not exhibit PGD2 11-ketoreductase or PGE2 9-ketoreductase activities. TbPGFS exhibits remarkably high affinity for PGH2 and a high PGFS turnover number, as compared with mammalian PGFSs.336,341,343,346,349 TbPGFS is now classified as AKR5A2. The TbPGFS activity is weakly inhibited by the stable PGH2 analog U-46619 (40% inhibition at 2 mM) but is unaffected by 2 mM U-44069.358
Kabututu et al.334 found PG production in lysates of L. donovani and identified the developmentally-regulated P100/11E gene product from L. major359 to exhibit high homology (61%) with TbPGFS. They cloned the cDNA for this gene product as L. major PGFS (LmPGFS) and showed that it is composed of 284 amino acids with an Mr of 32,000 and has the activities of PGFS (KM for PGH2 = 15 μM; VMAX = 270 nmol/min/mg protein) and 9,10-phenanthrenequinone reductase (KM = 12.5 μM; VMAX = 4,780 nmol/min/mg protein) but is not associated with PGD2 11-ketoreductase activity. LmPGFS is now classified as AKR5A1. The LmPGFS activity is inhibited by the stable PGH2 analog U-46619 with an IC50 of about 2 mM but like TbPGFS is unaffected by 2 mM U-44069.334
5.1.4. Purification and properties of T. cruzi PGF2α synthase (TcOYE), a member of the “old yellow enzyme” superfamily
In searching for PGFS homologs, Kubata et al.360 identified a novel “old yellow enzyme” from T. cruzi, a parasite that causes Chagas’ disease in South America. They purified PGFS from crude lysates of T. cruzi to apparent homogeneity. It is a 42 kDa protein containing one mole of FMN cofactor per mole of enzyme. cDNA cloning reveled T. cruzi PGFS to be a member of the old yellow enzyme superfamily, and it is named the T. cruzei old yellow enzyme (TcOYE). The protein contains 379 amino acid residues with a calculated Mr of 42,260. TcOYE not only has PGFS activity (KM for PGH2 = 5 μM; VMAX = 766 nmol/min/mg protein) but also reduces a variety of trypanocidal drugs in the presence of a NAD(P)H-generating system. Under anaerobic conditions, TcOYE also reduces peroxides, such as H2O2 (KM = 2.3 μM; VMAX = 99 nmol/min/mg protein) and t-butyl hydroperoxide (KM, not yet determined; VMAX = 282 nmol/min/mg protein), and trypanocidal agents such as menadione, β-lapaphone, nifurtimox, and 4-nitroquinoline-N-oxide (KM = 0.17 to 19 μM; VMAX = 290 to 1,110 nmol/min/mg protein) in the presence of NADPH or NADH. Electron-spin resonance spectral analyses revealed that TcOYE catalyzes the one-electron reduction of menadione or β-lapaphone to produce their semiquinone radicals, which cause cell death in susceptible parasites, and also the two-electron reduction of nitroheterocyclic compounds such as 4-nitroquinoline-N-oxide and nifurtimox.360
5.1.5. Purification and properties of swine brain PGFS/prostamide F2α synthase, a new member of the thioredoxin superfamily
Moriuchi et al.335 purified prostaglandin F ethanolamide (prostamide F) synthase from swine brain. The enzyme catalyzes the reduction of prostamide H2 to prostamide F2α. cDNA cloning of mouse prostamide F synthase revealed that the enzyme is composed of 201 amino acids with an Mr of 21,669 and that the enzyme is a member of the thioredoxin superfamily containing the CXXC motif. This enzyme not only catalyzes reduction of prostamide H2 to prostamide F2α, (KM for prostamide H2 = 7.6 μM; VMAX = 250 nmol/min/mg protein) but also the reduction of PGH2 to PGF2α (KM for PGH2 = 6.9 μM; VMAX = 690 nmol/min/mg protein) as well as reductions of H2O2 (KM = 1.6 μM; VMAX = 281 nmol/min/mg protein), t-butyl hydrroperoxide (KM = 0.4 μM; VMAX = 248 nmol/min/mg protein), and cumene hydroperoxide (KM = 0.026 μM; VMAX = 281 nmol/min/mg protein) but does not reduce either PGD2 nor PGE2. The enzyme activity is activated about nine-fold by a thioredoxin-generating system (Figure 27), more efficiently than the activation observed with NADPH, NADH or GSH. The KM value for reduced thioredoxin is about 1 μM. The active site cysteine is Cys-44 in the 44CXXC47 thioredoxin motif. This enzyme is distributed mainly in the brain and spinal cord and localized in motor neurons and oligodendroglia.361 The tertiary structure and physiological significance of this enzyme remain unclear.
Figure 27.
Reaction scheme for prosamide/PGFS activity of the thioredoxin system.335
5.2. Tertiary structural characteristics
X-ray crystallographic structures of members of the AKR superfamily have already shown these enzymes to have a common fold known as the (α/β)8-barrel fold.358,362-364 The nucleotide cofactor binds in an extended conformation at the top of the α/β-barrel, with the nicotinamide ring projecting down in the center of the barrel and the pyrophosphate straddling the barrel lip.365
5.2.1. X-ray crystallographic structure of human AKR1C3
Komoto et al.366 crystallized human lung PGFS (AKR1C3) along with the cofactor NADP+ and the substrate PGD2, and with the cofactor NADPH and the inhibitor rutin, and determined these complex structures at 1.69 Å resolution (PDB codes: 1RY0 and 1RY8) (Figure 28). They found that the cofactor binds deep in a cavity at the C-terminal end of the (α/β)8 barrel and that the substrate and the inhibitor are both located at the same site above the bound cofactor.366 Within the catalytic pocket, the conserved Tyr-55 and His-117 residues form hydrogen bonds with the carbonyl O-11 of PGD2 and the 13-hydroxyl group of the rutin. The cyclopentane ring of PGD2 and the phenyl ring of rutin face the re-side of the nicotinamide ring of the cofactor, suggesting a direct hydride transfer from NADPH to PGD2 (Figure 28).
Figure 28. X-ray crystallographic structure of human AKR1C3.366.
(A) AKR1C3 bound with NADP+ and PGD2 and (B) with NADPH and rutin. The α-helices and β-strands in the α/β base are colored aquamarine and magenta, respectively. The loops are shown in yellow and labeled with numbers. The bound NADP+/NADPH and PGD2/rutin are colored cyan and light pink, respectively. (C) The possible hydrogen bond donors and acceptors. Distances between atoms are noted in black in (C).
5.2.2. X-ray crystallographic structure and catalytic mechanism of human AKR1B1
Kubiseski et al.367 established that the AKR enzyme follows a sequential ordered mechanism in which NADPH binds before the aldehyde substrate and NADP+ is released after the alcohol product is formed. When, in 1992, the first complete crystal structure of human AKR1B1 (PDB code: 2ACU) was solved,363 the conserved Tyr-48 was shown to function as a catalytic acid for NADPH-dependent reduction.363 However, site-directed mutagenesis of the catalytic tetrad of AKR1B1 revealed that His-110, not Tyr-48, is crucial for the PGFS activity in the presence of NADPH.368 Furthermore, AKR1B1 and AKR1B3, but not AKR1B7 and AKR1C3, also catalyze the isomerization of PGH2 to PGD2 in the absence of NADPH or NADP+.368
Nagata et al.368 demonstrated that the PGFS activity of AKR1B1 decreases with increasing pH, with an optimum of pH 5.5 (KM for PGH2 = 29 μM; VMAX = 169 nmol/min/mg protein), and a pKb of 6.19 in the presence of NADPH, whereas the PGDS activity increases with increasing pH, with a pH optimum of 8.5 (KM for PGH2 = 23 μM; VMAX = 151 nmol/min/mg protein) and a pKa of 7.94 in the absence of NADPH or NADP+. Site-directed mutagenesis of the catalytic tetrad of AKR1B1 revealed that the triad of Asp-43, Lys-77, and His -10, but not Tyr-48, acts as a proton donor in most AKR activities and is crucial for PGFS and PGDS activities. These mutagenesis and pH titration analyses indicate that His-110 acts as a base to generate PGD2 in the absence of NADPH or NADP+ and as an acid to generate PGF2α in the presence of NADPH (Figure 29).368
Figure 29. X-ray crystallographic structure of human primary PGFS (AKR1B1) with NADP+ bound and the formation of PGD2 and PGF2α.
(A) In this structure PGH2 has been docked in the catalytic site using a molecular simulation program. AKR1B1 (green), NADP+ (yellow), the substrate, PGH2 (orange), Tyr-48 (purple), Asp-43 (green), Lys-77 (cyan) and His-110 (magenta). The hypothetical catalytic mechanisms of PGDS (B) and PGFS (C) activities of AKR1B1 in the absence and the presence of NADPH, respectively. In the absence of NADPH, the concerted reaction of Asp-43, Lys-77 and His-110 increases the basicity of His-110 and extracts the proton C-11 of PGH2. Another proton is provided to the O-9 atom of PGH2 from an unidentified proton donor (EnzA-H) to produce PGD2. However, in the presence of NADPH, the hydride ion is transferred from NADPH to the O-9 atom of the peroxide oxygen of PGH2, and a proton is provided from His-110 to O-11 to produce PGF2a. In the presence of NADP+, Asp-43 forms a hydrogen bond with NADP+ and disrupts the catalytic triad, which is essential for the production of PGD2. However, the function of Tyr-48 is not clear at present. C-9 and C-11 chiralities of PGH2 have been corrected from Reference 369.
5.2.3. X-ray crystallographic structure and catalytic mechanism of TbPGFS (AKR5A2)
Kubata et al. crystallized TbPGFS (AKR5A2) with the cofactor NADP+ and citrate, and determined the structure of the complex at 2.1 Å resolution (PDB code: 1VBJ).358 TbPGFS also adopts a parallel (α/β)8-barrel fold but lacks the protruding loops seen in the mammalian AKRs. TbPGFS contains a catalytic tetrad of tyrosine, lysine, histidine, and aspartate, highly conserved among AKRs, in its hydrophobic-core active site. Site-specific mutagenesis of the catalytic tetrad revealed that a dyad of Lys-77 and His-110 is essential for the reduction of PGH2 and a triad of Tyr-52, Lys-77 and His-110, for the reduction of 9,10-phenanthrenequinone. Structural and kinetic analysis uncovered that His-110 acts as the general acid catalyst for PGH2 reduction and that Lys-77 facilitates His-110 protonation through a water molecule, while exerting an electrostatic repulsion against His-110 that maintains the special arrangement allowing the formation of a hydrogen bond between His-110 and O-11 of PGH2 (Figure 30). On the other hand, Tyr-52 acts as a general acid catalyst for 9,10-phenanthrenequinone.
Figure 30. X-ray crystallographic structure of TbPGFS (AKR5A2) (A) and the proposed catalytic mechanism of the enzymic reduction of PGH2 (B).358.
(A, left) The overall structure of TbPGFS with bound NADP+, showing the overall structure viewed down the COOH-terminal end of the central β-barrel with the NADP+ molecule drawn in ball-and-stick model. The α-helices (in blue) and β-sheets (in yellow) are separated among them by loops (in orange). One citrate molecule is also depicted as a ball-and-stick model within the putative catalytic cleft. (A, right) Side view of the overall structure of TbPGFS_NADP+_citrate ternary complex (in light blue), in which the extended loops (β1-α1, β4-α4, and β7-H1) in other AKRs are absent in TbPGFS. (B) Proposed catalytic mechanism for the enzymatic reduction of PGH2. When a substrate binds to the active site of TbPGFS, a NADPH hydride attacks the C-9 carbonyl of PGH2, whereas a proton from His-110 will be transferred to the negatively charged C-11 carbonyl. Lys77 appears to have a role of maintaining the position of His-110 and stabilizing Asp-47. C-9 and C-11 chiralities of PGH2 have been corrected from those in Reference 358.
5.2.4. X-ray crystallographic structure of TcOYE
Yamaguchi et al.369 crystallized TcOYE with its cofactor FMN in the absence and presence of menadione, and determined the structures of the complexes at 1.7 and 2.5 Å. The atomic coordinates were finally determined with the crystals of the TcOYE/FMN binary complex and TcOYE/FMN/p-hydroxybenzaldehyde at 1.7 and 2.05 Å, respectively, (PDB code: 3ATY and 3ATZ, respectively).370
TcOYE has an (α/β)8 barrel fold, like other members of the OYE family, in which the cylindrical core composed of eight parallel β-strands (β1-β8) is surrounded by eight α-helices (α1-α8). In addition, an N-terminal β-hairpin (S1 and S2) closes the bottom of the barrel and two extra helices (H1 and H2) lie in the loops connecting β4 to α4 and β8 to α8, respectively. FMN is tightly bound at the C-terminal ends of the eight β-strands in the barrel, on which both menadione and p-hydroxybenzaldehyde reside above the isoalloxazine ring of FMN via a π-π interaction. The structure of the complex of TcOYE with FMN and p-hydroxybenzaldehyde is well superimposed on that of 12-oxophytodienoate reductase from Lycopersicon esculentum (tomato) with FMN and 12-oxophytodienoate, whose chemical structure is similar to that of a prostaglandin. The reaction mechanism for the reduction of PGH2 by TcOYE is proposed to be initiated by a hydride transfer from the N5 of FMN to the C-9 oxygen of PGH2 followed by proton transfer from His-195 to C-11 of PGH2 (Figure 31).368
Figure 31. X-ray crystallographic structure of TcOYE and the proposed catalytic mechanism for the enzymic reduction of PGH2.
(A) In the TcOYE structure,370 the α-helices (cyan) and β-sheets (orange) are separated by loops (light yellow). Loop 1 and Loop 2 are shown in magenta. FMN is shown as a ball-and-stick model. (B) Proposed catalytic mechanism for the enzymatic reduction of PGH2. A hydride is expected to be transferred from N-5 of FMN to C-9 oxygen of PGH2, whereas a proton would be from His-195 to C-11 oxygen of PGH2.368
5.3. Physiological and pathological properties
PGF2α is involved in many physiologic functions including contraction of uterine, bronchial, vascular, and arterial smooth muscles,330 regulation of ocular pressure,371 renal filtration,372 stimulation of hair growth,373 and regulation of the ovarian cycle through the induction of luteolysis.374 These actions of PGF2a are mediated via the PGF2α receptor FP, which is a Gq-coupled rhodopsin-type receptor.375 FP KO mice are healthy and grow normally.376 No gross abnormalities are found in the KO mice in terms of general behavior, general appearance or tissue histology, nor are there major biochemical and hematological abnormalities. Male FP KO mice are normal in their reproductive activity. Female KO mice also exhibit normal estrous cycles, ovulation, fertilization, and implantation. These results show that FP is not involved in “housekeeping” functions. However, female FP receptor KO mice are unable to deliver normal fetuses at term and do not respond to exogenous oxytocin because of the lack of induction of the oxytocin receptor and do not show the normal decline in serum progesterone concentrations that precedes parturition.376 Ovariectomy at day 19 of pregnancy restores induction of the oxytocin receptor and permits successful delivery in FP KO mice.376 These results indicate that the FP system is essential for initiation of parturition in mammals. The role of PGs in parasite pathogenesis and physiology were recently reviewed by Kubata et al.190
6. Prostacyclin synthase (PGIS)
PGI2 is typically involved in mediating events through a Gs-coupled G protein linked receptor designated IP. Generation of cAMP via IP is associated with relaxation of both vascular and nonvascular smooth muscle and inhibition of platelet aggregation.377 PGI2 is also reported to function through PPARδ and PPARβ, respectively, to modulate transcription events in the uterus 378 and the lung.379 These and the many other various physiologic actions of PGI2 are described in detail in a separate review on prostanoid receptors.
Prostacyclin synthase (PGIS) catalyzes the isomerization of PGH2 to PGI2 (Figure 1). Ullrich and coworkers were the first to provide spectral evidence that the enzyme is a cytochrome P450;380 the enzyme is designated CYP8A1 in the P450 family.381 The heme is bound via a sulfide linkage (Cys-441 in huPGIS) to the iron. Unlike most P450s and consistent with the non-oxidative rearrangement catalyzed by PGIS, NADPH and O2 are not co-substrates.381,382 PGISs were originally purified and characterized from bovine383 and porcine384 aorta. Human PGIS was subsequently cloned, expressed and purified381,385. Recombinant huPGIS has a KM of 30 μM with PGH2 and a specific activity of 15 mmol/min/mg of protein at 24° C.381 Recent evidence suggests that PGIS undergoes a relatively specific, peroxynitrite-mediated nitration on Tyr-403 that is associated with enzyme inactivation386 and occurs under conditions of oxidative stress.382 Formation of a radical in a reaction involving the heme iron and C-11 endoperoxide oxygen of PGH2 has been proposed to be the initial step in the formation of PGI2 from PGH2.387 The kinetic properties of PGIS are summarized in Table 1.
7. Thromboxane A synthase (TXAS)
Thromboxane A2 functions through one of two thromboxane GPCR splice varients termed TPα and TPβ that have different cellular distributions and are involved in platelet aggregation and vasoconstriction.388 Like PGIS, thromboxane synthase (TXAS) is a cytochrome P450 (CYP5) although the enzymes share only 15% sequence identity.389 The kinetic properties of TXAS are summarized in Table 1.
TXAS catalyzes the isomerization of PGH2 to TxA2 with parallel production of malondialdehyde and 12-hydroxyeicosatrienoic acid. The mechanism of TXAS is related to that of PGIS, in both cases the enzymes catalyze a rearrangement of the endoperoxide oxygens of PGH2. TXAS binds PGH2 in a different orientation than PGIS, with the heme iron coupling to the C-9 endoperoxide oxygen as opposed to the C-11 oxygen in the case of PGIS.389,390 This frees the C-11 oxygen radical for intramolecular reaction across the C-11 to C-12 bond of the cyclopropane ring, ultimately leading to the characteristic oxirane ring of TxA2. TXAS forms 12-hydroxy-heptadecatrienoic acid and malondialdehyde in equimolar amounts with TxA2.389,390 TXAS inhibitors were originally considered to be promising anti-platelet agents,391 but clinical trials of various inhibitors yielded unsatisfactory results when compared with low dose aspirin. Nonetheless, TXAS inhibitors continue to evaluated for other specific conditions involving thromboxane synthesis including bronchial asthma392, inflammatory bowel disease,393 and pulmonary hypertension.394 Very little new biochemical work has been published on this enzyme during the past ten years.395
8. PGH 19-hydroxylase
18-, 19- and 20-hydroxylated versions of prostaglandins have been identified in various tissue preparations. 19-hydroxy-PGE1 and 19-hydroxy-PGE2 are found in high concentrations (ca. 100 μg/ml) in human semen and in some individuals are found at levels equivalent to PGE1 and PGE2.396,397 Studies by Oliw and coworkers have indicated that these compounds are formed primarily from PGH1 and PGH2 via a pathway involving 19-hydroxylation of the endoperoxides by P450 CYP4F8 to the corresponding 19-hydroxy-PGHs398 followed by mPGES-1 mediated rearrangements to the endproducts.397 When tested with various EP receptor preparations, 19-hydroxy-PGE2 interacts preferentially with EP2399 although it is about ten fold less potent than PGE2 with EP2;400,401 in contrast, 19-hydroxy-PGE1 is much more potent than PGE2 in causing interleukin-8 release from U937 cells.400 19-hydroxy-PGE1 and 19-hydroxy-PGE2 have different but incompletely characterized pharmacologic profiles399,402 and may even function through receptors other than the previously characterized prostanoid receptors.
9. Future Directions
There is now considerable structural, mechanistic and kinetic information about the many enzymes involved in prostanoid biosynthesis from AA. As noted in the review, a particular interest from a pharmacologic perspective will be the development and testing of enzyme inhibitors for possible clinical use. For example, mPGES-1 is an attractive target for the development of novel anti-inflammatory drugs. Clinical trials will need to test for putative adverse cardiovascular side effects, notably hypertensive effects. From a basic protein chemistry perspective, it is clear that many of the enzymes involved in prostanoid biosynthesis are dimers or higher oligomers, and it will be of interest to determine how the various monomers interact and how these higher order structures function in regulating enzyme activities in cells. In related work, the potential for interactions among the biosynthetic enzymes in transferring or channeling substrates and intermediates will necessarily receive much attention.
Acknowledgements
Aspects of the work described here were supported by U.S.P.H.S. NIH GM68848, the Program for Promotion of Fundamental Studies in Health Sciences of NIBIO (Japan), the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (Japan), Japan Aerospace Exploration Agency, Takeda Science Foundation, Takeda Pharmaceutical Co., Ltd., and Ono Pharmaceutical Co., Ltd., the Swedish Research Council, the Karolinska Institutet, the Swedish Rheumatism Association, the King Gustaf V 80-Year Fund and the Marianne and Marcus Wallenberg Foundation. Thanks to Sven-Christian Pawelzik for his kind assistance in drawing the MAPEG structure figures.
Abbreviations Used
- AA
arachidonic acid
- AKR
aldo-keto reductase
- AKRL
aldose reductase-like proteins
- COX
cyclooxygenase
- cPGES
cytosolic PGE synthase
- cPLA2
cytosolic phospholipase A2
- CSF
cerebral spinal fluid
- DHA
docosahexaenoic acid
- DP
receptor for PGD
- DTNB
5,5'-dithiobis-(2-nitrobenzoic acid)
- EP
receptor for PGE
- Eallo
allosteric subunit of the PGHS homodimer
- Ecat
catalytic subunit of the PGHS sequence homodimer
- EGF
epidermal growth factor
- FA
fatty acid
- FBP
flurbiprofen
- FLAP
5-lipoxygenase activity protein
- FP
receptor for PGF
- GSH
reduced glutathione
- GST
glutathione-S-transferase
- HPETE
hydroperoxyeicosatetraenoic acid
- H-PGDS
hematopoeitic PGD synthase
- hu
human
- KO
knockout
- L-PGDS
lipocalin PGD synthase
- LT
leukotriene
- LTC4
leukotriene C4
- MAPEG
Membrane Associated Proteins in Eicosanoid and Glutathione metabolism
- MBD
membrane binding domain
- MGST
microsomal glutathione-S-transferases
- mPGES-1
microsomal PGE synthase-1
- mPGES-2
microsomal PGE synthase-2
- NSAIDs
nonsteroidal anti-inflammatory drugs
- ov
ovine
- PG
prostaglandin
- PGD
prostaglandin D
- PGE
prostaglandin E
- PGF
prostaglandin F
- PGG
prostaglandin G
- PGHS
prostaglandin endoperoxide H synthase
- PGI
prostaglandin I (prostacyclin)
- PGIS
PGI (prostacyclin) synthase
- PMA
phorbol myristate acetate
- TM
transmembrane
- TbPGFS
T. brucei PGFS
- Trx
thioredoxin
- TX
thromboxane
- TXAS
TXA synthase
Biography
 William L. Smith received his B.S. in Chemistry from the University of Colorado (1967) and his Ph.D. in Biological Chemistry from the University of Michigan (1971). After postdoctoral training at the University of California, Berkeley, he spent a year as a Senior Scientist at Mead Johnson Company and then joined the faculty in Biochemistry at Michigan State University in 1975. He moved to the University of Michigan in 2003 as Minor J. Coon Professor and Chair of Biological Chemistry. His research interests are in eicosanoid biochemistry and pharmacology. Dr. Smith is an Associate Editor of the Journal of Biological Chemistry. He is the author of 200 publications and has been a recipient of both the Avanti Award (2004) and the William C. Rose Award (2006) from the American Society of Biochemistry and Molecular Biology.
William L. Smith received his B.S. in Chemistry from the University of Colorado (1967) and his Ph.D. in Biological Chemistry from the University of Michigan (1971). After postdoctoral training at the University of California, Berkeley, he spent a year as a Senior Scientist at Mead Johnson Company and then joined the faculty in Biochemistry at Michigan State University in 1975. He moved to the University of Michigan in 2003 as Minor J. Coon Professor and Chair of Biological Chemistry. His research interests are in eicosanoid biochemistry and pharmacology. Dr. Smith is an Associate Editor of the Journal of Biological Chemistry. He is the author of 200 publications and has been a recipient of both the Avanti Award (2004) and the William C. Rose Award (2006) from the American Society of Biochemistry and Molecular Biology.
 Yoshihiro Urade received a M.A. in Agriculture from Osaka Prefecture University (1978) and his Ph.D. in Medical Science from Kyoto University (1983). He spent four years as a Senior Scientist at the Hayaishi Biotransformation Transfer Project, Exploratory Research for Advanced Technology, Research Development Corporation of Japan and then joined the Roche Institute of Molecular Biology as a Visiting Professor in 1987. He moved to the International Research Laboratories, CIBA-Geigy Japan Ltd. in 1990 as Senior Scientist. In 1993 he joined the Osaka Bioscience Institute as a Vice-Head of the Molecular Behavioral Biology Department and has served as the Head of this department since1998. His research interests are in enzymology and inflammation. Dr. Urade serve on the Editorial Advisory Board of Prostaglandins, Leukotrienes, and Essential Fatty Acids and is an Associate Editor of Sleep and Biological Rhythms. He is the author of more than 270 publications. He was made an honorary citizen of New Orleans to recognize his career in Biochemistry (1997).
Yoshihiro Urade received a M.A. in Agriculture from Osaka Prefecture University (1978) and his Ph.D. in Medical Science from Kyoto University (1983). He spent four years as a Senior Scientist at the Hayaishi Biotransformation Transfer Project, Exploratory Research for Advanced Technology, Research Development Corporation of Japan and then joined the Roche Institute of Molecular Biology as a Visiting Professor in 1987. He moved to the International Research Laboratories, CIBA-Geigy Japan Ltd. in 1990 as Senior Scientist. In 1993 he joined the Osaka Bioscience Institute as a Vice-Head of the Molecular Behavioral Biology Department and has served as the Head of this department since1998. His research interests are in enzymology and inflammation. Dr. Urade serve on the Editorial Advisory Board of Prostaglandins, Leukotrienes, and Essential Fatty Acids and is an Associate Editor of Sleep and Biological Rhythms. He is the author of more than 270 publications. He was made an honorary citizen of New Orleans to recognize his career in Biochemistry (1997).
 Per-Johan Jakobsson received his Ph.D. from the Karolinska Institutet for work in the field of leukotrienes (1994). After finishing his doctoral studies he continued his training at Merck Frosst in Montreal, defining the MAPEG protein superfamily and subsequently characterizing microsomal prostaglandin E synthase-1. He received the Young Investigator Award in connection with the 6th International Conference on Eicosanoids & Other Bioactive Lipids in Cancer, Inflammation and Related Diseases in 1999. In 2000, Dr Jakobsson was appointed Associate Professor at the Karolinska Institutet and currently combines work as a clinical fellow in rheumatology with translational research on disease mechanisms in inflammation with a special focus on eicosanoid pathways and membrane proteins. He is the author of 70 publications.
Per-Johan Jakobsson received his Ph.D. from the Karolinska Institutet for work in the field of leukotrienes (1994). After finishing his doctoral studies he continued his training at Merck Frosst in Montreal, defining the MAPEG protein superfamily and subsequently characterizing microsomal prostaglandin E synthase-1. He received the Young Investigator Award in connection with the 6th International Conference on Eicosanoids & Other Bioactive Lipids in Cancer, Inflammation and Related Diseases in 1999. In 2000, Dr Jakobsson was appointed Associate Professor at the Karolinska Institutet and currently combines work as a clinical fellow in rheumatology with translational research on disease mechanisms in inflammation with a special focus on eicosanoid pathways and membrane proteins. He is the author of 70 publications.
References
- 1.Serhan C, Chiang N, Van Dyke T. Nat. Rev. Immunol. 2008;8:349. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma NP, Dong L, Yuan C, Noon KR, Smith WL. Mol. Pharmacol. 2010;77:979. doi: 10.1124/mol.109.063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groeger A, Cipollina C, Cole M, Woodcock S, Bonacci G, Rudolph T, Rudolph V, Freeman B, Schopfer F. Nat. Chem. Biol. 2010;6:433. doi: 10.1038/nchembio.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith WL, DeWitt DL, Garavito RM. Annu. Rev. Biochem. 2000;69:149. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 5.van der Donk W, Tsai A, Kulmacz R. Biochemistry. 2002;41:15451. doi: 10.1021/bi026938h. [DOI] [PubMed] [Google Scholar]
- 6.Simmons DL, Botting RM, Hla T. Pharmacol. Rev. 2004;56:387. doi: 10.1124/pr.56.3.3. [DOI] [PubMed] [Google Scholar]
- 7.Kang YJ, Mbonye UR, Delong CJ, Wada M, Smith WL. Prog. Lipid Res. 2007;46:108. doi: 10.1016/j.plipres.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider C, Pratt DA, Porter NA, Brash AR. Chem. Biol. 2007;14:473. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith WL. Trends Biochem. Sci. 2008;33:27. doi: 10.1016/j.tibs.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Rouzer CA, Marnett LJ. J. Lipid Res. 2009;50:S29. doi: 10.1194/jlr.R800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai A, Kulmacz R. Arch. Biochem. Biophys. 2010;493:103. doi: 10.1016/j.abb.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosser T, Yu Y, Fitzgerald GA. Annu. Rev. Med. 2010;61:17. doi: 10.1146/annurev-med-011209-153129. [DOI] [PubMed] [Google Scholar]
- 13.Rouzer C, Marnett L. Chem. Rev. 2003;103:2239. doi: 10.1021/cr000068x. [DOI] [PubMed] [Google Scholar]
- 14.DeWitt DL, Smith WL. Proc. Natl. Acad. Sci. U.S.A. 1988;85:1212. doi: 10.1073/pnas.85.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merlie JP, Fagan D, Mudd J, Needleman P. J. Biol. Chem. 1988;263:3550. [PubMed] [Google Scholar]
- 16.Yokoyama C, Takai T, Tanabe T. FEBS Lett. 1988;231:347. doi: 10.1016/0014-5793(88)80847-0. [DOI] [PubMed] [Google Scholar]
- 17.Roth GJ, Siok CJ, Ozols J. J. Biol. Chem. 1980;255:1301. [PubMed] [Google Scholar]
- 18.Chen Y, Bienkowski M, Marnett L. J. Biol. Chem. 1987;262:16892. [PubMed] [Google Scholar]
- 19.Sirois J, Richards JS. J. Biol. Chem. 1992;267:6382. [PubMed] [Google Scholar]
- 20.Lin HJ, Lakkides KM, Keku TO, Reddy ST, Louie AD, Kau IH, Zhou H, Gim JSY, Ma HL, Matthies CF, Dai A, Huang H-F, Materi AM, Lin JH, Frankl HD, Lee ER, Hardy SI, Herschman HR, Henderson BE, Kolonel LN, Le Marchand L, Garavito RM, Sandler RS, Haile RW, Smith WL. Cancer Epidemiol. Biomarkers Prev. 2002;11:1305. [PubMed] [Google Scholar]
- 21.Ulrich CM, Whitton J, Yu J-H, Sibert J, Sparks R, Potter JD, Bigler J. Cancer Epidemiol. Biomarkers Prev. 2005;14:616. doi: 10.1158/1055-9965.EPI-04-0510. [DOI] [PubMed] [Google Scholar]
- 22.Poole EM, Bigler J, Whitton J, Sibert JG, Kulmacz RJ, Potter JD, Ulrich CM. Carcinogenesis. 2007;28:1259. doi: 10.1093/carcin/bgm026. [DOI] [PubMed] [Google Scholar]
- 23.Liu W, Poole E, Ulrich C, Kulmacz R. Pharmacogenomics J. 2010 June 15; doi: 10.1038/tpj.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Ouderaa FJ, Buytenhek M, Nugteren DH, Van-Dorp DA. Biochim. Biophys. Acta. 1977;487:315. doi: 10.1016/0005-2760(77)90008-x. [DOI] [PubMed] [Google Scholar]
- 25.Otto JC, Smith WL. J. Biol. Chem. 1994;269:19868. [PubMed] [Google Scholar]
- 26.Otto JC, DeWitt DL, Smith WL. J. Biol. Chem. 1993;268:18234. [PubMed] [Google Scholar]
- 27.Otto JC. Dissertation. Michigan State University, Department of Biochemistry; East Lansing Michigan: 1994. [Google Scholar]
- 28.Picot D, Loll PJ, Garavito M. Nature. 1994;367:243. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 29.Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Nature. 1996;384:644. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 30.Wada M, Saunders TL, Morrow J, Milne GL, Walker KP, Dey SK, Brock TG, Opp MR, Aronoff DM, Smith WL. J. Biol. Chem. 2009;284:30742. doi: 10.1074/jbc.M109.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mbonye UR, Yuan C, Harris CE, Sidhu RS, Song I, Arakawa T, Smith WL. J. Biol. Chem. 2008;283:8611. doi: 10.1074/jbc.M710137200. [DOI] [PubMed] [Google Scholar]
- 32.Mutsaers JH, van-Halbeek H, Kamerling JP, Vliegenthart JF. Eur. J. Biochem. 1985;147:569. doi: 10.1111/j.0014-2956.1985.00569.x. [DOI] [PubMed] [Google Scholar]
- 33.Percival M, Bastien L, Griffin P, Kargman S, Ouellet M, O'Neill G. Prot. Exp. Purif. 1997;9:388. doi: 10.1006/prep.1996.0685. [DOI] [PubMed] [Google Scholar]
- 34.Nemeth JF, Hochgesang GP, Marnett LJ, Caprioli RM, Hochensang GP. Biochemistry. 2001;40:3109. doi: 10.1021/bi002313c. [DOI] [PubMed] [Google Scholar]
- 35.Dong L, Vecchio AJ, Sharma NP, Jurban BJ, Malkowski MG, Smith WL. J. Biol. Chem. 2011;286:19035. doi: 10.1074/jbc.M111.231969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiefer J, Pawlitz J, Moreland K, Stegeman R, Hood W, Gierse J, Stevens A, Goodwin D, Rowlinson S, Marnett L, Stallings W, Kurumbail R. Nature. 2000;405:97. doi: 10.1038/35011103. [DOI] [PubMed] [Google Scholar]
- 37.Mbonye UR, Wada M, Rieke CJ, Tang H-Y, DeWitt DL, Smith WL. J. Biol. Chem. 2006;281:35770. doi: 10.1074/jbc.M608281200. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Phipps S, Turner M, Simmons D. J. Genet. Genomics. 37:117. doi: 10.1016/S1673-8527(09)60030-8. [DOI] [PubMed] [Google Scholar]
- 39.Selinsky BS, Gupta K, Sharkey CT, Loll PJ. Biochemistry. 2001;40:5172. doi: 10.1021/bi010045s. [DOI] [PubMed] [Google Scholar]
- 40.Sidhu R, Lee J, Yuan C, Smith W. Biochemistry. 2010;49:7069. doi: 10.1021/bi1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto T, Yamamoto S, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 1974;71:3645. doi: 10.1073/pnas.71.9.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otto JC, Smith WL. J. Biol. Chem. 1996;271:9906. doi: 10.1074/jbc.271.17.9906. [DOI] [PubMed] [Google Scholar]
- 43.Spencer AG, Thuresson EA, Otto JC, Song I, Smith T, DeWitt DL, Garavito RM, Smith WL. J. Biol. Chem. 1999;274:32936. doi: 10.1074/jbc.274.46.32936. [DOI] [PubMed] [Google Scholar]
- 44.MirAfzali Z, Leipprandt JR, McCracken JL, DeWitt DL. J. Biol. Chem. 2006;281:28354. doi: 10.1074/jbc.M605206200. [DOI] [PubMed] [Google Scholar]
- 45.Wendt KU, Poralla K, Schulz GE. Science. 1997;277:1811. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 46.Bracey M, Cravatt B, Stevens R. FEBS Lett. 2004;567:159. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 47.Raykhel I, Alanen H, Salo K, Jurvansuu J, Nguyen VD, Latva-Ranta M, Ruddock L. J. Cell Biol. 2007;179:1193. doi: 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao G, Chen W, Kulmacz RJ. J. Biol. Chem. 1998;273:6801. doi: 10.1074/jbc.273.12.6801. [DOI] [PubMed] [Google Scholar]
- 49.Duggan KC, Walters MJ, Musee J, Harp JM, Kiefer JR, Oates JA, Marnett LJ. J. Biol. Chem. 2010;285:34950. doi: 10.1074/jbc.M110.162982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulmacz RJ, Lands WE. J. Biol. Chem. 1984;259:6358. [PubMed] [Google Scholar]
- 51.Kulmacz RJ, Lands WEM. J. Biol. Chem. 1985;260:12572. [PubMed] [Google Scholar]
- 52.Percival MD, Ouellet M, Vincent CJ, Yergey JA, Kennedy BP, O'Neill GP. Arch. Biochem. Biophys. 1994;315:111. doi: 10.1006/abbi.1994.1478. [DOI] [PubMed] [Google Scholar]
- 53.Yuan C, Rieke CJ, Rimon G, Wingerd BA, Smith WL. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6142. doi: 10.1073/pnas.0601805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan C, Sidhu RS, Kuklev DV, Kado Y, Wada M, Song I, Smith WL. J. Biol. Chem. 2009;284:10046. doi: 10.1074/jbc.M808634200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prusakiewicz J, Duggan K, Rouzer C, Marnett L. Biochemistry. 2009;48:7353. doi: 10.1021/bi900999z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rimon G, Sidhu RS, Lauver DA, Lee JY, Sharma NP, Yuan C, Frieler RA, Trievel RC, Lucchesi BR, Smith WL. Proc. Natl. Acad. Sci. U.S.A. 2010;107:28. doi: 10.1073/pnas.0909765106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vecchio AJ, Simmons DM, Malkowski MG. J. Biol. Chem. 2010;285:22152. doi: 10.1074/jbc.M110.119867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vecchio AJ, Malkowski MG. J. Biol. Chem. 2011;286:20736. doi: 10.1074/jbc.M111.230367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamberg M, Samuelsson B. J. Biol. Chem. 1967;242:5344. [PubMed] [Google Scholar]
- 60.Hamberg M, Samuelsson B. J. Biol. Chem. 1967;242:5336. [PubMed] [Google Scholar]
- 61.Peng S, Okeley N, Tsai A, Wu G, Kulmacz R, van der Donk W. J. Am. Chem. Soc. 2002;124:10785. doi: 10.1021/ja026880u. [DOI] [PubMed] [Google Scholar]
- 62.Shimokawa T, Kulmacz RJ, DeWitt DL, Smith WL. J. Biol. Chem. 1990;265:20073. [PubMed] [Google Scholar]
- 63.Goodwin DC, Gunther MR, Hsi LC, Crews BC, Eling TE, Mason RP, Marnett LJ. J. Biol. Chem. 1998;273:8903. doi: 10.1074/jbc.273.15.8903. [DOI] [PubMed] [Google Scholar]
- 64.Tsai A, Hsi LC, Kulmacz RJ, Palmer G, Smith WL. J. Biol. Chem. 1994;269:5085. [PubMed] [Google Scholar]
- 65.Malkowski MG, Ginell S, Smith WL, Garavito RM. Science. 2000;289:1933. doi: 10.1126/science.289.5486.1933. [DOI] [PubMed] [Google Scholar]
- 66.Malkowski MG, Thuresson ED, Lakkides KM, Rieke CJ, Micielli R, Smith WL, Garavito RM. J. Biol. Chem. 2001;276:37547. doi: 10.1074/jbc.M105982200. [DOI] [PubMed] [Google Scholar]
- 67.Thuresson ED, Lakkides KM, Rieke CJ, Sun Y, Wingerd BA, Micielli R, Mulichak AM, Malkowski MG, Garavito RM, Smith WL. J. Biol. Chem. 2001;276:10347. doi: 10.1074/jbc.M009377200. [DOI] [PubMed] [Google Scholar]
- 68.Thuresson ED, Malkowski MG, Lakkides KM, Rieke CJ, Mulichak AM, Ginell SL, Garavito RM, Smith WL. J. Biol. Chem. 2001;276:10358. doi: 10.1074/jbc.M009378200. [DOI] [PubMed] [Google Scholar]
- 69.Tsai A, Palmer G, Xiao G, Swinney DC, Kulmacz RJ. J. Biol. Chem. 1998;273:3888. doi: 10.1074/jbc.273.7.3888. [DOI] [PubMed] [Google Scholar]
- 70.Tsai A, Kulmacz RJ, Palmer G. J. Biol. Chem. 1995;270:10503. doi: 10.1074/jbc.270.18.10503. [DOI] [PubMed] [Google Scholar]
- 71.Thuresson ED, Lakkides KM, Smith WL. J. Biol. Chem. 2000;275:8501. doi: 10.1074/jbc.275.12.8501. [DOI] [PubMed] [Google Scholar]
- 72.Schneider C, Boeglin WE, Prusakiewicz JJ, Rowlinson SW, Marnett LJ, Samel N, Brash AR. J. Biol. Chem. 2002;277:13354. doi: 10.1074/jbc.M107471200. [DOI] [PubMed] [Google Scholar]
- 73.Harman CA, Rieke CJ, Garavito RM, Smith WL. J. Biol. Chem. 2004;279:42929. doi: 10.1074/jbc.M403013200. [DOI] [PubMed] [Google Scholar]
- 74.Mukherjee A, Brinkley D, Chang K, Roth J. Biochemistry. 2007;46:3975. doi: 10.1021/bi602502j. [DOI] [PubMed] [Google Scholar]
- 75.Gupta A, Mukherjee A, Matsui K, Roth J. J. Am. Chem. Soc. 2008;130:11274. doi: 10.1021/ja8042273. [DOI] [PubMed] [Google Scholar]
- 76.Liang Z, Klinman J. Curr. Opin. Struct. Biol. 2004;14:648. doi: 10.1016/j.sbi.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 77.Smith WL, Lands WEM. Biochemistry. 1972;11:3276. doi: 10.1021/bi00767a024. [DOI] [PubMed] [Google Scholar]
- 78.Ohki S, Ogino N, Yamamoto S, Hayaishi O. J. Biol. Chem. 1979;254:829. [PubMed] [Google Scholar]
- 79.Pagels WR, Sachs RJ, Marnett LJ, Dewitt DL, Day JS, Smith WL. J. Biol. Chem. 1983;258:6517. [PubMed] [Google Scholar]
- 80.Kulmacz R. Arch. Biochem. Biophys. 1986;249:273. doi: 10.1016/0003-9861(86)90003-2. [DOI] [PubMed] [Google Scholar]
- 81.Kulmacz RJ, Wang LH. J. Biol. Chem. 1995;270:24019. doi: 10.1074/jbc.270.41.24019. [DOI] [PubMed] [Google Scholar]
- 82.Landino LM, Crews BC, Timmons MD, Morrow JD, Marnett LJ. Proc. Natl. Acad. Sci. U.S.A. 1996;93:15069. doi: 10.1073/pnas.93.26.15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruf HH, Schuhn D, Nastainczyk W. FEBS Lett. 1984;165:293. doi: 10.1016/0014-5793(84)80189-1. [DOI] [PubMed] [Google Scholar]
- 84.Lambeir AM, Markey CM, Dunford HB, Marnett LJ. J. Biol. Chem. 1985;260:14894. [PubMed] [Google Scholar]
- 85.Kulmacz RJ, Tsai AL, Palmer G. J. Biol. Chem. 1987;262:10524. [PubMed] [Google Scholar]
- 86.Dietz R, Nastainczyk W, Ruf HH. Eur. J. Biochem. 1988;171:321. doi: 10.1111/j.1432-1033.1988.tb13793.x. [DOI] [PubMed] [Google Scholar]
- 87.Karthein R, Dietz R, Nastainczyk W, Ruf HH. Eur. J. Biochem. 1988;171:313. doi: 10.1111/j.1432-1033.1988.tb13792.x. [DOI] [PubMed] [Google Scholar]
- 88.Rogge C, Liu W, Wu G, Wang L, Kulmacz R, Tsai A. Biochemistry. 2004;43:1560. doi: 10.1021/bi035717o. [DOI] [PubMed] [Google Scholar]
- 89.Tsai A, Wei C, Baek HK, Kulmacz RJ, Van Wart HE. J. Biol. Chem. 1997;272:8885. doi: 10.1074/jbc.272.14.8885. [DOI] [PubMed] [Google Scholar]
- 90.Liu J, Seibold SA, Rieke CJ, Song I, Cukier RI, Smith WL. J. Biol. Chem. 2007;282:18233. doi: 10.1074/jbc.M701235200. [DOI] [PubMed] [Google Scholar]
- 91.Lassmann G, Odenwaller R, Curtis JF, DeGray JA, Mason RP, Marnett LJ, Eling TE. J. Biol. Chem. 1991;266:20045. [PubMed] [Google Scholar]
- 92.Capdevila JH, Morrow JD, Belosludtsev YY, Beauchamp DR, DuBois RN, Falck JR. Biochemistry. 1995;34:3325. doi: 10.1021/bi00010a023. [DOI] [PubMed] [Google Scholar]
- 93.Lu G, Tsai A-L, Van Wart HE, Kulmacz RJ. J. Biol. Chem. 1999;274:16162. doi: 10.1074/jbc.274.23.16162. [DOI] [PubMed] [Google Scholar]
- 94.Kulmacz R. Biochem. Biophys. Res. Commun. 2005;338:25. doi: 10.1016/j.bbrc.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 95.Swinney DC, Mak AY, Barnett J, Ramesha CS. J. Biol. Chem. 1997;272:12393. doi: 10.1074/jbc.272.19.12393. [DOI] [PubMed] [Google Scholar]
- 96.Chen W, Pawelek TR, Kulmacz RJ. J. Biol. Chem. 1999;274:20301. doi: 10.1074/jbc.274.29.20301. [DOI] [PubMed] [Google Scholar]
- 97.Shitashige M, Morita I, Murota S. Biochim. Biophys. Acta. 1998;1389:57. doi: 10.1016/s0005-2760(97)00129-x. [DOI] [PubMed] [Google Scholar]
- 98.Wu G, Wei C, Kulmacz RJ, Osawa Y, Tsai A-L. J. Biol. Chem. 1999;274:9231. doi: 10.1074/jbc.274.14.9231. [DOI] [PubMed] [Google Scholar]
- 99.Song I, Ball TM, Smith WL. Biochem. Biophys. Res. Commun. 2001;289:869. doi: 10.1006/bbrc.2001.6071. [DOI] [PubMed] [Google Scholar]
- 100.Wu G, Kulmacz R, Tsai A. Biochemistry. 2003;42:13772. doi: 10.1021/bi035415m. [DOI] [PubMed] [Google Scholar]
- 101.Wu G, Vuletich JL, Kulmacz RJ, Osawa Y, Tsai A-L. J. Biol. Chem. 2001;276:19879. doi: 10.1074/jbc.M100628200. [DOI] [PubMed] [Google Scholar]
- 102.Grosser T, Fries S, FitzGerald GA. J. Clin. Invest. 2006;116:4. doi: 10.1172/JCI27291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wada M, DeLong CJ, Hong Y, Rieke CJ, Song I, Sidhu S, Warnock S, Schmaie AH, Yokoyama C, Smyth EM, Wilson SJ, FitzGerald GA, Garavito RM, Sui DX, Regan JW, Smith WL. J. Biol. Chem. 2007;282:22254. doi: 10.1074/jbc.M703169200. [DOI] [PubMed] [Google Scholar]
- 104.Laneuville O, Breuer DK, Dewitt DL, Hla T, Funk CD, Smith WL. J. Pharm. Exp. Ther. 1994;271:927. [PubMed] [Google Scholar]
- 105.Walker M, Kurumbail R, Kiefer J, Moreland K, Koboldt C, Isakson P, Seibert K, Gierse J. Biochem. J. 2001;357:709. doi: 10.1042/0264-6021:3570709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Copeland RA, Williams JM, Giannaras J, Nurnberg S, Covington M, Pinto D, Pick S, Trzaskos JM. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11202. doi: 10.1073/pnas.91.23.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rogge C, Ho B, Liu W, Kulmacz R, Tsai A. Biochemistry. 2006;45:523. doi: 10.1021/bi051235w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rogge C, Liu W, Kulmacz R, Tsai A. J. Inorg. Chem. 2009;103:912. doi: 10.1016/j.jinorgbio.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu G, Tsai A, Kulmacz R. Biochemistry. 2009;48:11902. doi: 10.1021/bi901600f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kulmacz RJ. J. Biol. Chem. 1989;264:14136. [PubMed] [Google Scholar]
- 111.Gupta K, Selinsky B, Kaub C, Katz A, Loll P. J. Mol. Biol. 2004;335:503. doi: 10.1016/j.jmb.2003.10.073. [DOI] [PubMed] [Google Scholar]
- 112.Mathias RA, Vergara C, Gao L, Rafaels N, Hand T, Campbell M, Bickel C, Ivester P, Sergeant S, Barnes KC, Chilton FH. J. Lipid Res. 51:2766. doi: 10.1194/jlr.M008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simopoulos AP. Exp. Biol. Med. 2010;235:785. doi: 10.1258/ebm.2010.009298. [DOI] [PubMed] [Google Scholar]
- 114.Langenbach R, Loftin CD, Lee C, Tiano H. Ann. N.Y. Acad. Sci. 1999;889:52. doi: 10.1111/j.1749-6632.1999.tb08723.x. [DOI] [PubMed] [Google Scholar]
- 115.Smith WL, Langenbach R. J. Clin. Invest. 2001;107:1491. doi: 10.1172/JCI13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Choi S-H, Langenbach R, Bosetti F. FASEB J. 2008;22:1491. doi: 10.1096/fj.07-9411com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu Y, Fan J, Hui Y, Rouzer CA, Marnett LJ, Klein-Szanto AJ, FitzGerald GA, Funk CD. J. Biol. Chem. 2007;282:1498. doi: 10.1074/jbc.M609930200. [DOI] [PubMed] [Google Scholar]
- 118.Tanabe T, Tohnai N. Prostaglandins Other Lipid Mediators. 2002:68–69. 95. doi: 10.1016/s0090-6980(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 119.Kang Y-J, Wingerd BA, Arakawa T, Smith WL. J. Immunol. 2006;177:8111. doi: 10.4049/jimmunol.177.11.8111. [DOI] [PubMed] [Google Scholar]
- 120.Smith WL. In: Handbook of Eicosanoids: Prostaglandins and Related Lipids. Part A. Willis AL, editor. I. CRC Press; Boca Raton, FL: 1987. [Google Scholar]
- 121.Chulada PC, Thompson MB, Mahler JF, Doyle CM, Gaul BW, Lee C, Tiano HF, Morham SG, Smithies O, Langenbach R. Cancer Res. 2000;60:4705. [PubMed] [Google Scholar]
- 122.DeWitt D, Meade E. Arch. Biochem. Biophys. 1993;306:94. doi: 10.1006/abbi.1993.1485. [DOI] [PubMed] [Google Scholar]
- 123.Evett GE, Xie W, Chipman JG, Robertson DL, Simmons DL. Arch. Biochem. Biophys. 1993;306:169. doi: 10.1006/abbi.1993.1496. [DOI] [PubMed] [Google Scholar]
- 124.Kujubu DA, Fletcher BS, Varnum BC, Lim RW, Herschman HR. J. Biol. Chem. 1991;266:12866. [PubMed] [Google Scholar]
- 125.Xie W, Herschman HR. J. Biol. Chem. 1996;271:31742. doi: 10.1074/jbc.271.49.31742. [DOI] [PubMed] [Google Scholar]
- 126.Goerig M, Habenicht AJR, Zeh W, Salbach P, Kommerell B, Rothe DER, Nastainczyk W, Glomset JA. J. Biol. Chem. 1988;263:19384. [PubMed] [Google Scholar]
- 127.Domin J, Rozengurt E. J. Biol. Chem. 1993;268:8927. [PubMed] [Google Scholar]
- 128.Lin AH, Bienkowski MJ, Gorman RR. J. Biol. Chem. 1989;264:17379. [PubMed] [Google Scholar]
- 129.Murakami M, Naraba H, Tanioka T, Semmyo N, Nakatani Y, Kojima F, Ikeda T, Fueki M, Ueno A, Oh-ishi S, Kudo I. J. Biol. Chem. 2000;275:32783. doi: 10.1074/jbc.M003505200. [DOI] [PubMed] [Google Scholar]
- 130.Naraba H, Murakami M, Matsumoto H, Shimbara S, Ueno A, Kudo I, Oh-ishi S. J. Immunol. 1998;160:2974. [PubMed] [Google Scholar]
- 131.Murakami M, Kambe T, Shimbara S, Kudo I. J. Biol. Chem. 1999;274:3103. doi: 10.1074/jbc.274.5.3103. [DOI] [PubMed] [Google Scholar]
- 132.Dennis EA. J. Biol. Chem. 1994;269:13057. [PubMed] [Google Scholar]
- 133.Satake Y, Diaz BL, Balestrieri B, Lam BK, Kanaoka Y, Grusby MJ, Arm JP. J. Biol. Chem. 2004;279:16488. doi: 10.1074/jbc.M313748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ni Z, Okeley NM, Smart BP, Gelb MH. J. Biol. Chem. 2006;281:16245. doi: 10.1074/jbc.M513874200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reddy ST, Herschman HR. J. Biol. Chem. 1994;269:15473. [PubMed] [Google Scholar]
- 136.Bhattacharyya DK, Lecomte M, Dunn J, Morgans DJ, Smith WL. Arch. Biochem. Biophys. 1995;317:19. doi: 10.1006/abbi.1995.1130. [DOI] [PubMed] [Google Scholar]
- 137.Regier MK, DeWitt DL, Schindler MS, Smith WL. Arch. Biochem. Biophys. 1993;301:439. doi: 10.1006/abbi.1993.1168. [DOI] [PubMed] [Google Scholar]
- 138.Spencer AG, Woods JW, Arakawa T, Singer II, Smith WL. J. Biol. Chem. 1998;273:9886. doi: 10.1074/jbc.273.16.9886. [DOI] [PubMed] [Google Scholar]
- 139.Morita I, Takahashi R, Saito Y, Murota S. J. Biol. Chem. 1983;258:10197. [PubMed] [Google Scholar]
- 140.Lands W, Libelt B, Morris A, Kramer N, Prewitt T, Bowen P, Schmeisser D, Davidson M, Burns J. Biochim. Biophys. Acta. 1992;1180:147. doi: 10.1016/0925-4439(92)90063-s. [DOI] [PubMed] [Google Scholar]
- 141.Marszalek J, Lodish H. Annu. Rev. Cell Dev. Biol. 2005;21:633. doi: 10.1146/annurev.cellbio.21.122303.120624. [DOI] [PubMed] [Google Scholar]
- 142.Fischer S, Weber P. Nature. 1984;307:165. doi: 10.1038/307165a0. [DOI] [PubMed] [Google Scholar]
- 143.Fischer S, Weber P. Biomed. Mass. Spectrom. 1985;12:470. doi: 10.1002/bms.1200120905. [DOI] [PubMed] [Google Scholar]
- 144.von Schacky C, Siess W, Fischer S, Weber P. J. Lipid Res. 1985;26:457. [PubMed] [Google Scholar]
- 145.Braden G, Knapp H, Fitzgerald D, FitzGerald G. Circulation. 1990;82:178. doi: 10.1161/01.cir.82.1.178. [DOI] [PubMed] [Google Scholar]
- 146.Kulmacz RJ, Pendleton RB, Lands WEM. J. Biol. Chem. 1994;269:5527. [PubMed] [Google Scholar]
- 147.Liu W, Cao D, Oh SF, Serhan CN, Kulmacz RJ. FASEB J. 2006;20:1097. doi: 10.1096/fj.05-5273com. [DOI] [PubMed] [Google Scholar]
- 148.Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS, Kaufmann C, Morgado-Diaz JA, Bozza PT, Viola JPB. Cancer Res. 2008;68:1732. doi: 10.1158/0008-5472.CAN-07-1999. [DOI] [PubMed] [Google Scholar]
- 149.Matsuoka T, Hirata M, Tanaka H, Takahashi Y, Murata T, Kabashima K, Sugimoto Y, Kobayashi T, Ushikubi F, Aze Y, Eguchi N, Urade Y, Yoshida N, Kimura K, Mizoguchi A, Honda Y, Nagai H, Narumiya S. Science. 2000;287:2013. doi: 10.1126/science.287.5460.2013. [DOI] [PubMed] [Google Scholar]
- 150.Lewis RA, Soter NA, Diamond PT, Austen KF, Oates JA, Roberts LJ., 2nd J. Immunol. 1982;129:1627. [PubMed] [Google Scholar]
- 151.Peters SP, Schleimer RP, Kagey-Sobotka A, Naclerio RM, MacGlashan DW, Jr., Schulman ES, Adkinson NF, Jr., Lichtenstein LM. Transactions of the Association of American Physicians. 1982;95:221. [PubMed] [Google Scholar]
- 152.Tanaka K, Ogawa K, Sugamura K, Nakamura M, Takano S, Nagata K. J. Immunol. 2000;164:2277. doi: 10.4049/jimmunol.164.5.2277. [DOI] [PubMed] [Google Scholar]
- 153.Ueno R, Ishikawa Y, Nakayama T, Hayaishi O. Biochem. Biophys. Res. Commun. 1982;109:576. doi: 10.1016/0006-291x(82)91760-0. [DOI] [PubMed] [Google Scholar]
- 154.Urade Y, Ujihara M, Horiguchi Y, Igarashi M, Nagata A, Ikai K, Hayaishi O. J. Biol. Chem. 1990;265:371. [PubMed] [Google Scholar]
- 155.Mohri I, Eguchi N, Suzuki K, Urade Y, Taniike M. Glia. 2003;42:263. doi: 10.1002/glia.10183. [DOI] [PubMed] [Google Scholar]
- 156.Urade Y, Kitahama K, Ohishi H, Kaneko T, Mizuno N, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9070. doi: 10.1073/pnas.90.19.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gerena RL, Irikura D, Urade Y, Eguchi N, Chapman DA, Killian GJ. Biol. Reprod. 1998;58:826. doi: 10.1095/biolreprod58.3.826. [DOI] [PubMed] [Google Scholar]
- 158.Tokugawa Y, Kunishige I, Kubota Y, Shimoya K, Nobunaga T, Kimura T, Saji F, Murata Y, Eguchi N, Oda H, Urade Y, Hayaishi O. Biol. Reprod. 1998;58:600. doi: 10.1095/biolreprod58.2.600. [DOI] [PubMed] [Google Scholar]
- 159.Eguchi Y, Eguchi N, Oda H, Seiki K, Kijima Y, Matsu-ura Y, Urade Y, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14689. doi: 10.1073/pnas.94.26.14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Christ-Hazelhof E, Nugteren DH. Biochim. Biophys. Acta. 1979;572:43. doi: 10.1016/0005-2760(79)90198-x. [DOI] [PubMed] [Google Scholar]
- 161.Urade Y, Fujimoto N, Hayaishi O. J. Biol. Chem. 1985;260:12410. [PubMed] [Google Scholar]
- 162.Urade Y, Fujimoto N, Ujihara M, Hayaishi O. J. Biol. Chem. 1987;262:3820. [PubMed] [Google Scholar]
- 163.Kanaoka Y, Ago H, Inagaki E, Nanayama T, Miyano M, Kikuno R, Fujii Y, Eguchi N, Toh H, Urade Y, Hayaishi O. Cell. 1997;90:1085. doi: 10.1016/s0092-8674(00)80374-8. [DOI] [PubMed] [Google Scholar]
- 164.Kanaoka Y, Fujimori K, Kikuno R, Sakaguchi Y, Urade Y, Hayaishi O. Eur. J. Biochem. 2000;267:3315. doi: 10.1046/j.1432-1327.2000.01362.x. [DOI] [PubMed] [Google Scholar]
- 165.Thomson AM, Meyer DJ, Hayes JD. Biochem. J. 1998;(Pt 2):333, 317. doi: 10.1042/bj3330317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Inoue T, Irikura D, Okazaki N, Kinugasa S, Matsumura H, Uodome N, Yamamoto M, Kumasaka T, Miyano M, Kai Y, Urade Y. Nat. Struct. Biol. 2003;10:291. doi: 10.1038/nsb907. [DOI] [PubMed] [Google Scholar]
- 167.Shimizu T, Yamamoto S, Hayaishi O. J. Biol. Chem. 1979;254:5222. [PubMed] [Google Scholar]
- 168.Urade Y, Tanaka T, Eguchi N, Kikuchi M, Kimura H, Toh H, Hayaishi O. J. Biol. Chem. 1995;270:1422. doi: 10.1074/jbc.270.3.1422. [DOI] [PubMed] [Google Scholar]
- 169.Urade Y, Nagata A, Suzuki Y, Fujii Y, Hayaishi O. J. Biol. Chem. 1989;264:1041. [PubMed] [Google Scholar]
- 170.White DM, Mikol DD, Espinosa R, Weimer B, Le Beau MM, Stefansson K. J. Biol. Chem. 1992;267:23202. [PubMed] [Google Scholar]
- 171.Nagata A, Suzuki Y, Igarashi M, Eguchi N, Toh H, Urade Y, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4020. doi: 10.1073/pnas.88.9.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Irikura D, Inui T, Beuckmann CT, Aritake K, Schreiber G, Miyano M, Inoue T, Urade Y. J. Biochem. (Tokyo) 2007;141:173. doi: 10.1093/jb/mvm016. [DOI] [PubMed] [Google Scholar]
- 173.Fujimori K, Inui T, Uodome N, Kadoyama K, Aritake K, Urade Y. Gene. 2006;375:14. doi: 10.1016/j.gene.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 174.Pervaiz S, Brew K. FASEB J. 1987;1:209. doi: 10.1096/fasebj.1.3.3622999. [DOI] [PubMed] [Google Scholar]
- 175.Hoffmann A, Conradt HS, Gross G, Nimtz M, Lottspeich F, Wurster U. J. Neurochem. 1993;61:451. doi: 10.1111/j.1471-4159.1993.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 176.Clausen J. Proc. Soc. Exp. Biol. Med. 1961;107:170. doi: 10.3181/00379727-107-26569. [DOI] [PubMed] [Google Scholar]
- 177.Watanabe K, Urade Y, Mader M, Murphy C, Hayaishi O. Biochem. Biophys. Res. Commun. 1994;203:1110. doi: 10.1006/bbrc.1994.2297. [DOI] [PubMed] [Google Scholar]
- 178.Mohri I, Taniike M, Taniguchi H, Kanekiyo T, Aritake K, Inui T, Fukumoto N, Eguchi N, Kushi A, Sasai H, Kanaoka Y, Ozono K, Narumiya S, Suzuki K, Urade Y. J. Neurosci. 2006;26:4383. doi: 10.1523/JNEUROSCI.4531-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Aritake K, Kado Y, Inoue T, Miyano M, Urade Y. J. Biol. Chem. 2006;281:15277. doi: 10.1074/jbc.M506431200. [DOI] [PubMed] [Google Scholar]
- 180.Eguchi N, Minami T, Shirafuji N, Kanaoka Y, Tanaka T, Nagata A, Yoshida N, Urade Y, Ito S, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 1999;96:726. doi: 10.1073/pnas.96.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pinzar E, Kanaoka Y, Inui T, Eguchi N, Urade Y, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4903. doi: 10.1073/pnas.090093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Urade Y, Eguchi N. Prostaglandins Other Lipid Mediators. 2002;68-69:375. doi: 10.1016/s0090-6980(02)00042-4. [DOI] [PubMed] [Google Scholar]
- 183.Urade Y, Hayaishi O. Biochim. Biophys. Acta. 2000;1482:259. doi: 10.1016/s0167-4838(00)00161-8. [DOI] [PubMed] [Google Scholar]
- 184.Song WL, Wang M, Ricciotti E, Fries S, Yu Y, Grosser T, Reilly M, Lawson JA, FitzGerald GA. J. Biol. Chem. 2008;283:1179. doi: 10.1074/jbc.M706839200. [DOI] [PubMed] [Google Scholar]
- 185.Beuckmann CT, Fujimori K, Urade Y, Hayaishi O. Neurochem. Res. 2000;25:733. doi: 10.1023/a:1007579507804. [DOI] [PubMed] [Google Scholar]
- 186.Ujihara M, Tsuchida S, Satoh K, Sato K, Urade Y. Arch. Biochem. Biophys. 1988;264:428. doi: 10.1016/0003-9861(88)90308-6. [DOI] [PubMed] [Google Scholar]
- 187.Herve M, Angeli V, Pinzar E, Wintjens R, Faveeuw C, Narumiya S, Capron A, Urade Y, Capron M, Riveau G, Trottein F. European journal of immunology. 2003;33:2764. doi: 10.1002/eji.200324143. [DOI] [PubMed] [Google Scholar]
- 188.Sommer A, Rickert R, Fischer P, Steinhart H, Walter RD, Liebau E. Infect. Immun. 2003;71:3603. doi: 10.1128/IAI.71.6.3603-3606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Joachim A, Ruttkowski B. Exp. Parasitol. 2011;127:604. doi: 10.1016/j.exppara.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 190.Kubata BK, Duszenko M, Martin KS, Urade Y. Trends Parasitol. 2007;23:325. doi: 10.1016/j.pt.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 191.Daiyasu H, Watanabe K, Toh H. Biochem. Biophys. Res. Commun. 2008;369:281. doi: 10.1016/j.bbrc.2008.02.088. [DOI] [PubMed] [Google Scholar]
- 192.Takahashi S, Tsurumura T, Aritake K, Furubayashi N, Sato M, Yamanaka M, Hirota E, Sano S, Kobayashi T, Tanaka T, Inaka K, Tanaka H, Urade Y. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:846. doi: 10.1107/S1744309110020828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tanaka H, Tsurumura T, Aritake K, Furubayashi N, Takahashi S, Yamanaka M, Hirota E, Sano S, Sato M, Kobayashi T, Tanaka T, Inaka K, Urade Y. J. Synchrotron Radiat. 2011;18:88. doi: 10.1107/S0909049510037076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pinzar E, Miyano M, Kanaoka Y, Urade Y, Hayaishi O. J. Biol. Chem. 2000;275:31239. doi: 10.1074/jbc.M000750200. [DOI] [PubMed] [Google Scholar]
- 195.Uchida Y, Urade Y, Mori S, Kohzuma T. J. Inorg. Chem. 2010;104:331. doi: 10.1016/j.jinorgbio.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 196.Yamaguchi N, Naiki T, Kohzuma T, Takada T, Sakata F, Mori M. Theor. Chem. Acc. 2011;128:191. [Google Scholar]
- 197.Shimamoto S, Yoshida T, Inui T, Gohda K, Kobayashi Y, Fujimori K, Tsurumura T, Aritake K, Urade Y, Ohkubo T. J. Biol. Chem. 2007;282:31373. doi: 10.1074/jbc.M700123200. [DOI] [PubMed] [Google Scholar]
- 198.Miyamoto Y, Nishimura S, Inoue K, Shimamoto S, Yoshida T, Fukuhara A, Yamada M, Urade Y, Yagi N, Ohkubo T, Inui T. J. Struct. Biol. 2010;169:209. doi: 10.1016/j.jsb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 199.Inoue K, Yagi N, Urade Y, Inui T. J. Biochem. 2009;145:169. doi: 10.1093/jb/mvn154. [DOI] [PubMed] [Google Scholar]
- 200.Kumasaka T, Aritake K, Ago H, Irikura D, Tsurumura T, Yamamoto M, Miyano M, Urade Y, Hayaishi O. J. Biol. Chem. 2009;284:22344. doi: 10.1074/jbc.M109.018341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhou Y, Shaw N, Li Y, Zhao Y, Zhang R, Liu ZJ. FASEB J. 2010;24:4668. doi: 10.1096/fj.10-164863. [DOI] [PubMed] [Google Scholar]
- 202.Jemth P, Mannervik B. J. Biol. Chem. 2000;275:8618. doi: 10.1074/jbc.275.12.8618. [DOI] [PubMed] [Google Scholar]
- 203.Islam F, Watanabe Y, Morii H, Hayaishi O. Arch. Biochem. Biophys. 1991;289:161. doi: 10.1016/0003-9861(91)90456-s. [DOI] [PubMed] [Google Scholar]
- 204.Matsushita N, Aritake K, Takada A, Hizue M, Hayashi K, Mitsui K, Hayashi M, Hirotsu I, Kimura Y, Tani T, Nakajima H. Jpn. J. Pharmacol. 1998;78:11. doi: 10.1254/jjp.78.11. [DOI] [PubMed] [Google Scholar]
- 205.Christ A, Labzin L, Bourne G, Fukunishi H, Weber J, Sweet M, Smythe M, Flanagan J. J. Med. Chem. 2010;53:5536. doi: 10.1021/jm100194a. [DOI] [PubMed] [Google Scholar]
- 206.Hohwy M, Spadola L, Lundquist B, Hawtin P, Dahmén J, Groth-Clausen I, Nilsson E, Persdotter S, von Wachenfeldt K, Folmer R, Edman K. J. Med. Chem. 2008;51:2178. doi: 10.1021/jm701509k. [DOI] [PubMed] [Google Scholar]
- 207.Irikura D, Aritake K, Nagata N, Maruyama T, Shimamoto S, Urade Y. J. Biol. Chem. 2009;284:7623. doi: 10.1074/jbc.M808593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jones RL, Giembycz MA, Woodward DF. Br. J. Pharmacol. 2009;158:104. doi: 10.1111/j.1476-5381.2009.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hirata M, Kakizuka A, Aizawa M, Ushikubi F, Narumiya S. Proc. Natl. Acad. Sci. U. S. A. 1994;91:11192. doi: 10.1073/pnas.91.23.11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hirai H, Tanaka K, Yoshie O, Ogawa K, Kenmotsu K, Takamori Y, Ichimasa M, Sugamura K, Nakamura M, Takano S, Nagata K. J. Exp. Med. 2001;193:255. doi: 10.1084/jem.193.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mizoguchi A, Eguchi N, Kimura K, Kiyohara Y, Qu WM, Huang ZL, Mochizuki T, Lazarus M, Kobayashi T, Kaneko T, Narumiya S, Urade Y, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11674. doi: 10.1073/pnas.201398898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Qu W-M, Huang Z-L, Xu X-H, Aritake K, Eguchi N, Nambu F, Narumiya S, Urade Y, Hayaishi O. Proc. Nat. Acad. Sci. U.S.A. 2006;103:17949. doi: 10.1073/pnas.0608581103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chevalier E, Stock J, Fisher T, Dupont M, Fric M, Fargeau H, Leport M, Soler S, Fabien S, Pruniaux MP, Fink M, Bertrand CP, McNeish J, Li B. J. Immunol. 2005;175:2056. doi: 10.4049/jimmunol.175.4.2056. [DOI] [PubMed] [Google Scholar]
- 214.Nagata K, Hirai H. Prostaglandins, Leukotrienes Essent. Fatty Acids. 2003;69:169. doi: 10.1016/s0952-3278(03)00078-4. [DOI] [PubMed] [Google Scholar]
- 215.Smith WL, Marnett LJ, DeWitt DL. Pharmacol. Ther. 1991;49:153. doi: 10.1016/0163-7258(91)90054-p. [DOI] [PubMed] [Google Scholar]
- 216.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. Cell. 1995;83:803. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 217.Kliewer SA, Lenhard JM, Willson TM, Patel I, Morris DC, Lehmann JM. Cell. 1995;83:813. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 218.Bell-Parikh LC, Ide T, Lawson JA, McNamara P, Reilly M, FitzGerald GA. J. Clin. Invest. 2003;112:945. doi: 10.1172/JCI18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Powell WS. J. Clin. Invest. 2003;112:828. doi: 10.1172/JCI19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ujihara M, Urade Y, Eguchi N, Hayashi H, Ikai K, Hayaishi O. Arch. Biochem. Biophys. 1988;260:521. doi: 10.1016/0003-9861(88)90477-8. [DOI] [PubMed] [Google Scholar]
- 221.Shimura C, Satoh T, Igawa K, Aritake K, Urade Y, Nakamura M, Yokozeki H. Am. J. Pathol. 2010;176:227. doi: 10.2353/ajpath.2010.090111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ujihara M, Horiguchi Y, Ikai K, Urade Y. J. Invest. Dermatol. 1988;90:448. doi: 10.1111/1523-1747.ep12460883. [DOI] [PubMed] [Google Scholar]
- 223.Urade Y, Ujihara M, Horiguchi Y, Ikai K, Hayaishi O. J. Immunol. 1989;143:2982. [PubMed] [Google Scholar]
- 224.Mahmud I, Ueda N, Yamaguchi H, Yamashita R, Yamamoto S, Kanaoka Y, Urade Y, Hayaishi O. J. Biol. Chem. 1997;272:28263. doi: 10.1074/jbc.272.45.28263. [DOI] [PubMed] [Google Scholar]
- 225.Hyo S, Kawata R, Kadoyama K, Eguchi N, Kubota T, Takenaka H, Urade Y. Arch. Otolaryngol. Head Neck Surg. 2007;133:693. doi: 10.1001/archotol.133.7.693. [DOI] [PubMed] [Google Scholar]
- 226.Liu M, Eguchi N, Yamasaki Y, Urade Y, Hattori N, Urabe T. Neuroscience. 2009;163:296. doi: 10.1016/j.neuroscience.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 227.Mohri I, Kadoyama K, Kanekiyo T, Sato Y, Kagitani-Shimono K, Saito Y, Suzuki K, Kudo T, Takeda M, Urade Y, Murayama S, Taniike M. J. Neuropathol. Exp. Neurol. 2007;66:469. doi: 10.1097/01.jnen.0000240472.43038.27. [DOI] [PubMed] [Google Scholar]
- 228.Okano M, Fujiwara T, Sugata Y, Gotoh D, Masaoka Y, Sogo M, Tanimoto W, Yamamoto M, Matsumoto R, Eguchi N, Kiniwa M, Isik AU, Urade Y, Nishizaki K. Am. J. Rhinol. 2006;20:342. doi: 10.2500/ajr.2006.20.2865. [DOI] [PubMed] [Google Scholar]
- 229.Okano M, Fujiwara T, Yamamoto M, Sugata Y, Matsumoto R, Fukushima K, Yoshino T, Shimizu K, Eguchi N, Kiniwa M, Urade Y, Nishizaki K. Clin. Exp. Allergy. 2006;36:1028. doi: 10.1111/j.1365-2222.2006.02528.x. [DOI] [PubMed] [Google Scholar]
- 230.Okinaga T, Mohri I, Fujimura H, Imai K, Ono J, Urade Y, Taniike M. Acta Neuropathol. (Berl) 2002;104:377. doi: 10.1007/s00401-002-0567-z. [DOI] [PubMed] [Google Scholar]
- 231.Mohri I, Aritake K, Taniguchi H, Sato Y, Kamauchi S, Nagata N, Maruyama T, Taniike M, Urade Y. Am. J. Pathol. 2009;174:1735. doi: 10.2353/ajpath.2009.080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Redensek A, Rathore KI, Berard JL, Lopez-Vales R, Swayne LA, Bennett SA, Mohri I, Taniike M, Urade Y, David S. Glia. 2011;59:603. doi: 10.1002/glia.21128. [DOI] [PubMed] [Google Scholar]
- 233.Urade Y, Eguchi N, Hayaishi O. In: Lipocalins. Akerstrom B, Flower D, Salier JP, editors. Landes Bioscience/Eurekah: Georgetown; Texas, U.S.A.: 2006. [Google Scholar]
- 234.Urade Y, Hayaishi O. Future Neurol. 2010;5:363. [Google Scholar]
- 235.Hayaishi O, Urade Y, Eguchi N, Huang ZL. Arch. Ital. Biol. 2004;142:533. [PubMed] [Google Scholar]
- 236.Takemiya T, Matsumura K, Sugiura H, Yasuda S, Uematsu S, Akira S, Yamagata K. Neurochem. Int. 2011;58:489. doi: 10.1016/j.neuint.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 237.Ohinata K, Takagi K, Biyajima K, Fujiwara Y, Fukumoto S, Eguchi N, Urade Y, Asakawa A, Fujimiya M, Inui A, Yoshikawa M. FEBS Lett. 2008;582:679. doi: 10.1016/j.febslet.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 238.Taniike M, Mohri I, Eguchi N, Beuckmann CT, Suzuki K, Urade Y. J. Neurosci. 2002;22:4885. doi: 10.1523/JNEUROSCI.22-12-04885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chabas D, Baranzini SE, Mitchell D, Bernard CC, Rittling SR, Denhardt DT, Sobel RA, Lock C, Karpuj M, Pedotti R, Heller R, Oksenberg JR, Steinman L. Science. 2001;294:1731. doi: 10.1126/science.1062960. [DOI] [PubMed] [Google Scholar]
- 240.Kagitani-Shimono K, Mohri I, Oda H, Ozono K, Suzuki K, Urade Y, Taniike M. Neuropathol. Appl. Neurobiol. 2006;32:64. doi: 10.1111/j.1365-2990.2005.00690.x. [DOI] [PubMed] [Google Scholar]
- 241.Myerowitz R, Lawson D, Mizukami H, Mi Y, Tifft CJ, Proia RL. Hum. Mol. Genet. 2002;11:1343. doi: 10.1093/hmg/11.11.1343. [DOI] [PubMed] [Google Scholar]
- 242.Mohri I, Taniike M, Okazaki I, Kagitani-Shimono K, Aritake K, Kanekiyo T, Yagi T, Takikita S, Kim HS, Urade Y, Suzuki K. J. Neurochem. 2006;97:641. doi: 10.1111/j.1471-4159.2006.03753.x. [DOI] [PubMed] [Google Scholar]
- 243.Saleem S, Shah ZA, Urade Y, Dore S. Neuroscience. 2009;160:248. doi: 10.1016/j.neuroscience.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Taniguchi H, Mohri I, Okabe-Arahori H, Aritake K, Wada K, Kanekiyo T, Narumiya S, Nakayama M, Ozono K, Urade Y, Taniike M. J. Neurosci. 2007;27:4303. doi: 10.1523/JNEUROSCI.0321-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Taniguchi H, Mohri I, Okabe-Arahori H, Kanekiyo T, Kagitani-Shimono K, Wada K, Urade Y, Nakayama M, Ozono K, Taniike M. Neurosci. Lett. 2007;420:39. doi: 10.1016/j.neulet.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 246.Tachibana M, Fex J, Urade Y, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7677. doi: 10.1073/pnas.84.21.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Beuckmann CT, Gordon WC, Kanaoka Y, Eguchi N, Marcheselli VL, Gerashchenko DY, Urade Y, Hayaishi O, Bazan NG. J. Neurosci. 1996;16:6119. doi: 10.1523/JNEUROSCI.16-19-06119.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Malki S, Nef S, Notarnicola C, Thevenet L, Gasca S, Mejean C, Berta P, Poulat F, Boizet-Bonhoure B. EMBO J. 2005;24:1798. doi: 10.1038/sj.emboj.7600660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Moniot B, Declosmenil F, Barrionuevo F, Scherer G, Aritake K, Malki S, Marzi L, Cohen-Solal A, Georg I, Klattig J, Englert C, Kim Y, Capel B, Eguchi N, Urade Y, Boizet-Bonhoure B, Poulat F. Development (Cambridge, England) 2009;136:1813. doi: 10.1242/dev.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Moniot B, Boizet-Bonhoure B, Poulat F. Sex. Dev. 2008;2:96. doi: 10.1159/000129694. [DOI] [PubMed] [Google Scholar]
- 251.Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. J. Biol. Chem. 2007;282:10553. doi: 10.1074/jbc.M609578200. [DOI] [PubMed] [Google Scholar]
- 252.Han F, Takeda K, Ishikawa K, Ono M, Date F, Yokoyama S, Furuyama K, Shinozawa Y, Urade Y, Shibahara S. Biochem. Biophys. Res. Commun. 2009;385:449. doi: 10.1016/j.bbrc.2009.05.092. [DOI] [PubMed] [Google Scholar]
- 253.Tokudome S, Sano M, Shinmura K, Matsuhashi T, Morizane S, Moriyama H, Tamaki K, Hayashida K, Nakanishi H, Yoshikawa N, Shimizu N, Endo J, Katayama T, Murata M, Yuasa S, Kaneda R, Tomita K, Eguchi N, Urade Y, Asano K, Utsunomiya Y, Suzuki T, Taguchi R, Tanaka H, Fukuda K. J. Clin. Invest. 2009;119:1477. doi: 10.1172/JCI37413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Miyagi M, Miwa Y, Takahashi-Yanaga F, Morimoto S, Sasaguri T. Arterioscler., Thromb., Vasc. Biol. 2005;25:970. doi: 10.1161/01.ATV.0000159702.68591.0d. [DOI] [PubMed] [Google Scholar]
- 255.Cipollone F, Fazia M, Iezzi A, Ciabattoni G, Pini B, Cuccurullo C, Ucchino S, Spigonardo F, De Luca M, Prontera C, Chiarelli F, Cuccurullo F, Mezzetti A. Arterioscler., Thromb., Vasc. Biol. 2004;24:1259. doi: 10.1161/01.ATV.0000133192.39901.be. [DOI] [PubMed] [Google Scholar]
- 256.Miwa Y, Takiuchi S, Kamide K, Yoshii M, Horio T, Tanaka C, Banno M, Miyata T, Sasaguri T, Kawano Y. Biochem. Biophys. Res. Commun. 2004;322:428. doi: 10.1016/j.bbrc.2004.07.143. [DOI] [PubMed] [Google Scholar]
- 257.Nagata N, Fujimori K, Okazaki I, Oda H, Eguchi N, Uehara Y, Urade Y. FEBS J. 2009;276:7146. doi: 10.1111/j.1742-4658.2009.07426.x. [DOI] [PubMed] [Google Scholar]
- 258.Jowsey IR, Murdock PR, Moore GB, Murphy GJ, Smith SA, Hayes JD. Prostaglandins Other Lipid Mediators. 2003;70:267. doi: 10.1016/s0090-6980(02)00134-x. [DOI] [PubMed] [Google Scholar]
- 259.Quinkler M, Bujalska IJ, Tomlinson JW, Smith DM, Stewart PM. Gene. 2006;380:137. doi: 10.1016/j.gene.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 260.Xie Y, Kang X, Ackerman W. E. t., Belury MA, Koster C, Rovin BH, Landon MB, Kniss DA. Diabetes Obes. Metab. 2006;8:83. doi: 10.1111/j.1463-1326.2005.00472.x. [DOI] [PubMed] [Google Scholar]
- 261.Fujimori K, Aritake K, Urade Y. J. Biol. Chem. 2007;282:18458. doi: 10.1074/jbc.M701141200. [DOI] [PubMed] [Google Scholar]
- 262.Ragolia L, Palaia T, Hall CE, Maesaka JK, Eguchi N, Urade Y. J. Biol. Chem. 2005;280:29946. doi: 10.1074/jbc.M502927200. [DOI] [PubMed] [Google Scholar]
- 263.Tanaka R, Miwa Y, Mou K, Tomikawa M, Eguchi N, Urade Y, Takahashi-Yanaga F, Morimoto S, Wake N, Sasaguri T. Biochem. Biophys. Res. Commun. 2009;378:851. doi: 10.1016/j.bbrc.2008.11.152. [DOI] [PubMed] [Google Scholar]
- 264.Tanaka T, Urade Y, Kimura H, Eguchi N, Nishikawa A, Hayaishi O. J. Biol. Chem. 1997;272:15789. doi: 10.1074/jbc.272.25.15789. [DOI] [PubMed] [Google Scholar]
- 265.Gallant MA, Samadfam R, Hackett JA, Antoniou J, Parent JL, de Brum-Fernandes AJ. J. Bone Miner. Res. 2005;20:672. doi: 10.1359/JBMR.041211. [DOI] [PubMed] [Google Scholar]
- 266.Gallant MA, Chamoux E, Bisson M, Wolsen C, Parent JL, Roux S, de Brum-Fernandes AJ. J. Rheumatol. 2010;37:644. doi: 10.3899/jrheum.090622. [DOI] [PubMed] [Google Scholar]
- 267.Joo M, Kwon M, Sadikot RT, Kingsley PJ, Marnett LJ, Blackwell TS, Peebles RS, Urade Y, Christman JW. J. Immunol. 2007;179:2565. doi: 10.4049/jimmunol.179.4.2565. [DOI] [PubMed] [Google Scholar]
- 268.Hokari R, Nagata N, Kurihara C, Watanabe C, Komoto S, Okada Y, Kawaguchi A, Nagao S, Hibi T, Nagata K, Urade Y, Miura S. J. Pathol. 2009;219:417. doi: 10.1002/path.2615. [DOI] [PubMed] [Google Scholar]
- 269.Hokari R, Kurihara C, Nagata N, Aritake K, Okada Y, Watanabe C, Komoto S, Nakamura M, Kawaguchi A, Nagao S, Urade Y, Miura S. Am. J. Physiol. 2010;300:G401. doi: 10.1152/ajpgi.00351.2010. [DOI] [PubMed] [Google Scholar]
- 270.Jordan W, Tumani H, Cohrs S, Eggert S, Rodenbeck A, Brunner E, Ruther E, Hajak G. Sleep. 2004;27:867. doi: 10.1093/sleep/27.5.867. [DOI] [PubMed] [Google Scholar]
- 271.Jordan W, Tumani H, Cohrs S, Rodenbeck A, Ruther E, Bechthold J, Mayer G. J. Neurol. 2005;252:1372. doi: 10.1007/s00415-005-0870-4. [DOI] [PubMed] [Google Scholar]
- 272.Bassetti CL, Hersberger M, Baumann CR. J. Neurol. 2006;253:1030. doi: 10.1007/s00415-006-0153-8. [DOI] [PubMed] [Google Scholar]
- 273.Barcelo A, de la Pena M, Barbe F, Pierola J, Bosch M, Agusti AG. Sleep Med. 2007;8:509. doi: 10.1016/j.sleep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 274.Inoue T, Eguchi Y, Matsumoto T, Kijima Y, Kato Y, Ozaki Y, Waseda K, Oda H, Seiki K, Node K, Urade Y. Atherosclerosis. 2008;201:385. doi: 10.1016/j.atherosclerosis.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 275.Miwa Y, Oda H, Shiina Y, Shikata K, Tsushima M, Nakano S, Maruyama T, Kyotani S, Eguchi N, Urade Y, Takahashi-Yanaga F, Morimoto S, Sasaguri T. Hypertens. Res. 2008;31:1931. doi: 10.1291/hypres.31.1931. [DOI] [PubMed] [Google Scholar]
- 276.Uehara Y, Makino H, Seiki K, Urade Y. Nephrol. Dial. Transplant. 2009;24:475. doi: 10.1093/ndt/gfn515. [DOI] [PubMed] [Google Scholar]
- 277.Beuckmann CT, Aoyagi M, Okazaki I, Hiroike T, Toh H, Hayaishi O, Urade Y. Biochemistry. 1999;38:8006. doi: 10.1021/bi990261p. [DOI] [PubMed] [Google Scholar]
- 278.Kanekiyo T, Ban T, Aritake K, Huang Z-L, Qu W-M, Okazaki I, Mohri I, Murayama S, Ozono K, Taniike M, Goto Y, Urade Y. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6412. doi: 10.1073/pnas.0701585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Li W, Malpica-Llanos TM, Gundry R, Cotter RJ, Sacktor N, McArthur J, Nath A. Neurology. 2008;70:1753. doi: 10.1212/01.wnl.0000282761.19578.35. [DOI] [PubMed] [Google Scholar]
- 280.Ogino N, Miyamoto T, Yamamoto S, Hayaishi O. J. Biol. Chem. 1977;252:890. [PubMed] [Google Scholar]
- 281.Tanaka Y, Ward S, Smith W. J. Biol. Chem. 1987;262:1374. [PubMed] [Google Scholar]
- 282.Jakobsson P, Thoren S, Morgenstern R, Samuelsson B. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7220. doi: 10.1073/pnas.96.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Tanioka T, Nakatani Y, Semmyo N, Murakami M, Kudo I. J. Biol. Chem. 2000;275:32775. doi: 10.1074/jbc.M003504200. [DOI] [PubMed] [Google Scholar]
- 284.Tanikawa N, Ohmiya Y, Ohkubo H, Hashimoto K, Kangawa K, Kojima M, Ito S, Watanabe K. Biochem. Biophys. Res. Commun. 2002;291:884. doi: 10.1006/bbrc.2002.6531. [DOI] [PubMed] [Google Scholar]
- 285.Ogorochi T, Ujihara M, Narumiya S. J. Neurochem. 1987;48:900. doi: 10.1111/j.1471-4159.1987.tb05602.x. [DOI] [PubMed] [Google Scholar]
- 286.Samuelsson B, Morgenstern R, Jakobsson P-J. Pharmacol. Rev. 2007;59:207. doi: 10.1124/pr.59.3.1. [DOI] [PubMed] [Google Scholar]
- 287.Hara S, Kamei D, Sasaki Y, Tanemoto A, Nakatani Y, Murakami M. Biochimie. 2010;92:651. doi: 10.1016/j.biochi.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 288.Korotkova M, Jakobsson P-J. Frontiers in Pharmacology. 2011;2:1. doi: 10.3389/fphar.2010.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Wang M, Song WL, Cheng Y, Fitzgerald GA. J. Intern. Med. 2008;263:500. doi: 10.1111/j.1365-2796.2008.01938.x. [DOI] [PubMed] [Google Scholar]
- 290.Nakanishi M, Gokhale V, Meuillet EJ, Rosenberg DW. Biochimie. 2010;92:660. doi: 10.1016/j.biochi.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Radmark O, Samuelsson B. J. Intern. Med. 2010;268:5. doi: 10.1111/j.1365-2796.2010.02246.x. [DOI] [PubMed] [Google Scholar]
- 292.Hofacker A, Coste O, Nguyen H-V, Marian C, Scholich K, Geisslinger G. J. Neurosci. 2005;25:9005. doi: 10.1523/JNEUROSCI.2190-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Jania LA, Chandrasekharan S, Backlund MG, Foley NA, Snouwaert J, Wang IM, Clark P, Audoly LP, Koller BH. Prostaglandins Other Lipid Mediators. 2009;88:73. doi: 10.1016/j.prostaglandins.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lovgren AK, Kovarova M, Koller BH. Mol. Cell. Biol. 2007;27:4416. doi: 10.1128/MCB.02314-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bresell A, Weinander R, Lundqvist G, Raza H, Shimoji M, Sun T-H, Balk L, Wiklund R, Eriksson J, Jansson C, Persson B, Jakobsson P-J, Morgenstern R. FEBS J. 2005;272:1688. doi: 10.1111/j.1742-4658.2005.04596.x. [DOI] [PubMed] [Google Scholar]
- 296.Jakobsson PJ, Morgenstern R, Mancini J, Ford-Hutchinson A, Persson B. Protein Sci. 1999;8:689. doi: 10.1110/ps.8.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Jakobsson PJ, Mancini JA, Ford-Hutchinson AW. J. Biol. Chem. 1996;271:22203. doi: 10.1074/jbc.271.36.22203. [DOI] [PubMed] [Google Scholar]
- 298.Jakobsson PJ, Mancini JA, Riendeau D, FordHutchinson AW. J. Biol. Chem. 1997;272:22934. doi: 10.1074/jbc.272.36.22934. [DOI] [PubMed] [Google Scholar]
- 299.Murakami M, Nakashima K, Kamei D, Masuda S, Ishikawa Y, Ishii T, Ohmiya Y, Watanabe K, Kudo I. J. Biol. Chem. 2003;278:37937. doi: 10.1074/jbc.M305108200. [DOI] [PubMed] [Google Scholar]
- 300.Thoren S, Weinander R, Saha S, Jegerschold C, Pettersson PL, Samuelsson B, Hebert H, Hamberg M, Morgenstern R, Jakobsson P-J. J. Biol. Chem. 2003;278:22199. doi: 10.1074/jbc.M303227200. [DOI] [PubMed] [Google Scholar]
- 301.Ouellet M, Falgueyret JP, Ear PH, Pen A, Mancini JA, Riendeau D, Percival MD. Protein Exp. Purif. 2002;26:489. doi: 10.1016/s1046-5928(02)00566-1. [DOI] [PubMed] [Google Scholar]
- 302.Watanabe K, Kurihara K, Suzuki T. Biochim. Biophys. Acta. 1999;1439:406. doi: 10.1016/s1388-1981(99)00084-0. [DOI] [PubMed] [Google Scholar]
- 303.Kozak KR, Crews BC, Morrow JD, Wang LH, Ma YH, Weinander R, Jakobsson PJ, Marnett LJ. J. Biol. Chem. 2002;277:44877. doi: 10.1074/jbc.M206788200. [DOI] [PubMed] [Google Scholar]
- 304.Guindon J, Hohmann AG. Br. J. Pharmacol. 2008;153:1341. doi: 10.1038/bjp.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Holm PJ, Bhakat P, Jegerschold C, Gyobu N, Mitsuoka K, Fujiyoshi Y, Morgenstern R, Hebert H. J. Mol. Biol. 2006;360:934. doi: 10.1016/j.jmb.2006.05.056. [DOI] [PubMed] [Google Scholar]
- 306.Ferguson AD, McKeever BM, Xu S, Wisniewski D, Miller DK, Yamin TT, Spencer RH, Chu L, Ujjainwalla F, Cunningham BR, Evans JF, Becker JW. Science. 2007;317:510. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 307.Martinez Molina D, Wetterholm A, Kohl A, McCarthy AA, Niegowski D, Ohlson E, Hammarberg T, Eshaghi S, Haeggstrom JZ, Nordlund P. Nature. 2007;448:613. doi: 10.1038/nature06009. [DOI] [PubMed] [Google Scholar]
- 308.Ago H, Kanaoka Y, Irikura D, Lam BK, Shimamura T, Austen KF, Miyano M. Nature. 2007;448:609. doi: 10.1038/nature05936. [DOI] [PubMed] [Google Scholar]
- 309.Jegerschold C, Pawelzik SC, Purhonen P, Bhakat P, Gheorghe KR, Gyobu N, Mitsuoka K, Morgenstern R, Jakobsson PJ, Hebert H. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11110. doi: 10.1073/pnas.0802894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Weaver AJ, Sullivan WP, Felts SJ, Owen BA, Toft DO. J. Biol. Chem. 2000;275:23045. doi: 10.1074/jbc.M003410200. [DOI] [PubMed] [Google Scholar]
- 311.Yamada T, Komoto J, Watanabe K, Ohmiya Y, Takusagawa F. J. Mol. Biol. 2005;348:1163. doi: 10.1016/j.jmb.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 312.Watanabe K, Ohkubo H, Niwa H, Tanikawa N, Koda N, Ito S, Ohmiya Y. Biochem. Biophys. Res. Commun. 2003;306:577. doi: 10.1016/s0006-291x(03)01025-8. [DOI] [PubMed] [Google Scholar]
- 313.Hebert H, Jegerschold C. Curr. Opin. Struct. Biol. 2007;17:396. doi: 10.1016/j.sbi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 314.Martinez Molina D, Eshaghi S, Nordlund P. Curr. Opin. Struct. Biol. 2008;18:442. doi: 10.1016/j.sbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 315.Rinaldo-Matthis A, Wetterholm A, Martinez Molina D, Holm J, Niegowski D, Ohlson E, Nordlund P, Morgenstern R, Haeggstrom JZ. J. Biol. Chem. 2010;285:40771. doi: 10.1074/jbc.M110.105940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Saino H, Ukita Y, Ago H, Irikura D, Nisawa A, Ueno G, Yamamoto M, Kanaoka Y, Lam BK, Austen KF, Miyano M. J. Biol. Chem. 2011;286:16392. doi: 10.1074/jbc.M110.150177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hammarberg T, Hamberg M, Wetterholm A, Hansson H, Samuelsson B, Haeggstrom JZ. J. Biol. Chem. 2009;284:301. doi: 10.1074/jbc.M808365200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Prage EB, Pawelzik SC, Busenlehner LS, Kim K, Morgenstern R, Jakobsson PJ, Armstrong RN. Biochemistry. 2011 doi: 10.1021/bi2010448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Koeberle A, Werz O. Curr. Med. Chem. 2009;16:4274. doi: 10.2174/092986709789578178. [DOI] [PubMed] [Google Scholar]
- 320.Xu D, Rowland SE, Clark P, Giroux A, Cote B, Guiral S, Salem M, Ducharme Y, Friesen RW, Methot N, Mancini J, Audoly L, Riendeau D. J. Pharmacol. Exp. Therap. 2008;326:754. doi: 10.1124/jpet.108.138776. [DOI] [PubMed] [Google Scholar]
- 321.Pawelzik SC, Uda NR, Spahiu L, Jegerschold C, Stenberg P, Hebert H, Morgenstern R, Jakobsson PJ. J. Biol. Chem. 2010;285:29254. doi: 10.1074/jbc.M110.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Scholich K, Geisslinger G. Trends Pharmacol. Sci. 2006;27:399. doi: 10.1016/j.tips.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 323.Facemire CS, Griffiths R, Audoly LP, Koller BH, Coffman TM. Hypertension. 2010;55:531. doi: 10.1161/HYPERTENSIONAHA.109.145631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Degousee N, Fazel S, Angoulvant D, Stefanski E, Pawelzik S-C, Korotkova M, Arab S, Liu P, Lindsay TF, Zhuo S, Butany J, Li R-K, Audoly L, Schmidt R, Angioni C, Geisslinger G, Jakobsson P-J, Rubin BB. Circulation. 2008;117:1701. doi: 10.1161/CIRCULATIONAHA.107.749739. [DOI] [PubMed] [Google Scholar]
- 325.Kihara Y, Matsushita T, Kita Y, Uematsu S, Akira S, Kira J, Ishii S, Shimizu T. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21807. doi: 10.1073/pnas.0906891106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Chaudhry UA, Zhuang H, Crain BJ, Dore S. Alzheimers Dement. 2008;4:6. doi: 10.1016/j.jalz.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Ikeda-Matsuo Y, Tanji H, Ota A, Hirayama Y, Uematsu S, Akira S, Sasaki Y. Br. J. Pharmacol. 2010;160:847. doi: 10.1111/j.1476-5381.2010.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Pecchi E, Dallaporta M, Jean A, Thirion S, Troadec J. Physiol. Behav. 2009;97:279. doi: 10.1016/j.physbeh.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 329.Herlenius E. Respir. Physiol. Neurobiol. 2011 doi: 10.1016/j.resp.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 330.Watanabe K. Prostaglandins Other Lipid Mediators. 2002;68-69:401. doi: 10.1016/s0090-6980(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 331.Liston TE, Roberts LJ., 2nd Proc. Natl. Acad. Sci. U.S.A. 1985;82:6030. doi: 10.1073/pnas.82.18.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Penning TM, Drury JE. Arch. Biochem. Biophys. 2007;464:241. doi: 10.1016/j.abb.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Jez JM, Flynn TG, Penning TM. Biochem. Pharmacol. 1997;54:639. doi: 10.1016/s0006-2952(97)84253-0. [DOI] [PubMed] [Google Scholar]
- 334.Kabututu Z, Martin SK, Nozaki T, Kawazu S, Okada T, Munday CJ, Duszenko M, Lazarus M, Thuita LW, Urade Y, Kubata BK. Int. J. Parasitol. 2003;33:221. doi: 10.1016/s0020-7519(02)00254-0. [DOI] [PubMed] [Google Scholar]
- 335.Moriuchi H, Koda N, Okuda-Ashitaka E, Daiyasu H, Ogasawara K, Toh H, Ito S, Woodward DF, Watanabe K. J. Biol. Chem. 2008;283:792. doi: 10.1074/jbc.M705638200. [DOI] [PubMed] [Google Scholar]
- 336.Watanabe K, Yoshida R, Shimizu T, Hayaishi O. J. Biol. Chem. 1985;260:7035. [PubMed] [Google Scholar]
- 337.Watanabe K, Fujii Y, Nakayama K, Ohkubo H, Kuramitsu S, Kagamiyama H, Nakanishi S, Hayaishi O. Proc. Natl. Acad. Sci. U.S.A. 1988;85:11. doi: 10.1073/pnas.85.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Hayashi H, Fujii Y, Watanabe K, Urade Y, Hayaishi O. J. Biol. Chem. 1989;264:1036. [PubMed] [Google Scholar]
- 339.Hayaishi O, Watanabe K, Fujii Y, Nakayama K, Ohkubo H, Kuramitsu S, Kagamiyama H, Nakanishi S. Progress in clinical and biological research. 1988;274:577. [PubMed] [Google Scholar]
- 340.Urade Y, Watanabe K, Eguchi N, Fujii Y, Hayaishi O. J. Biol. Chem. 1990;265:12029. [PubMed] [Google Scholar]
- 341.Chen LY, Watanabe K, Hayaishi O. Arch. Biochem. Biophys. 1992;296:17. doi: 10.1016/0003-9861(92)90539-9. [DOI] [PubMed] [Google Scholar]
- 342.Suzuki T, Fujii Y, Miyano M, Chen LY, Takahashi T, Watanabe K. J. Biol. Chem. 1999;274:241. doi: 10.1074/jbc.274.1.241. [DOI] [PubMed] [Google Scholar]
- 343.Koda N, Tsutsui Y, Niwa H, Ito S, Woodward DF, Watanabe K. Arch. Biochem. Biophys. 2004;424:128. doi: 10.1016/j.abb.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 344.Suzuki-Yamamoto T, Nishizawa M, Fukui M, Okuda-Ashitaka E, Nakajima T, Ito S, Watanabe K. FEBS Lett. 1999;462:335. doi: 10.1016/s0014-5793(99)01551-3. [DOI] [PubMed] [Google Scholar]
- 345.Wintergalen N, Thole HH, Galla HJ, Schlegel W. Eur. J. Biochem. 1995;234:264. doi: 10.1111/j.1432-1033.1995.264_c.x. [DOI] [PubMed] [Google Scholar]
- 346.Kabututu Z, Manin M, Pointud J-C, Maruyama T, Nagata N, Lambert S, Lefrancois- Martinez A-M, Martinez A, Urade Y. J. Biochem. 2009;145:161. doi: 10.1093/jb/mvn152. [DOI] [PubMed] [Google Scholar]
- 347.Nakashima K, Ueno N, Kamei D, Tanioka T, Nakatani Y, Murakami M, Kudo I. Biochim. Biophys. Acta. 2003;1633:96. doi: 10.1016/s1388-1981(03)00092-1. [DOI] [PubMed] [Google Scholar]
- 348.Ueno N, Takegoshi Y, Kamei D, Kudo I, Murakami M. Biochem. Biophys. Res. Commun. 2005;338:70. doi: 10.1016/j.bbrc.2005.08.152. [DOI] [PubMed] [Google Scholar]
- 349.Madore E, Harvey N, Parent J, Chapdelaine P, Arosh JA, Fortier MA. J. Biol. Chem. 2003;278:11205. doi: 10.1074/jbc.M208318200. [DOI] [PubMed] [Google Scholar]
- 350.Bresson E, Boucher-Kovalik S, Chapdelaine P, Madore E, Harvey N, Laberge PY, Leboeuf M, Fortier MA. J. Clin. Endocrinol. Metab. 2010;96:210. doi: 10.1210/jc.2010-1589. [DOI] [PubMed] [Google Scholar]
- 351.Ramana KV. Biomol. Concepts. 2011;2:103. doi: 10.1515/BMC.2011.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Fujimori K, Ueno T, Nagata N, Kashiwagi K, Aritake K, Amano F, Urade Y. J. Biol. Chem. 2010;285:8880. doi: 10.1074/jbc.M109.077164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Fujimori K, Ueno T, Amano F. Prostaglandins Other Lipid Mediators. 2010;93:52. doi: 10.1016/j.prostaglandins.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 354.Lambert-Langlais S, Pointud JC, Lefrancois-Martinez AM, Volat F, Manin M, Coudore F, Val P, Sahut-Barnola I, Ragazzon B, Louiset E, Delarue C, Lefebvre H, Urade Y, Martinez A. PloS one. 2009;4:e7309. doi: 10.1371/journal.pone.0007309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Mutayoba BM, Meyer HH, Osaso J, Gombe S. Theriogenology. 1989;32:545. doi: 10.1016/0093-691x(89)90276-8. [DOI] [PubMed] [Google Scholar]
- 356.Pentreath VW, Rees K, Owolabi OA, Philip KA, Doua F. Trans. R. Soc. Trop. Med. Hyg. 1990;84:795. doi: 10.1016/0035-9203(90)90085-s. [DOI] [PubMed] [Google Scholar]
- 357.Kubata BK, Duszenko M, Kabututu Z, Rawer M, Szallies A, Fujimori K, Inui T, Nozaki T, Yamashita K, Horii T, Urade Y, Hayaishi O. J. Exp. Med. 2000;192:1327. doi: 10.1084/jem.192.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Kilunga KB, Inoue T, Okano Y, Kabututu Z, Martin SK, Lazarus M, Duszenko M, Sumii Y, Kusakari Y, Matsumura H, Kai Y, Sugiyama S, Inaka K, Inui T, Urade Y. J. Biol. Chem. 2005;280:26371. doi: 10.1074/jbc.M413884200. [DOI] [PubMed] [Google Scholar]
- 359.Samaras N, Spithill TW. J. Biol. Chem. 1989;264:4251. [PubMed] [Google Scholar]
- 360.Kubata BK, Kabututu Z, Nozaki T, Munday CJ, Fukuzumi S, Ohkubo K, Lazarus M, Maruyama T, Martin SK, Duszenko M, Urade Y. J. Exp. Med. 2002;196:1241. doi: 10.1084/jem.20020885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Yoshikawa K, Takei S, Hasegawa-Ishii S, Chiba Y, Furukawa A, Kawamura N, Hosokawa M, Woodward DF, Watanabe K, Shimada A. Brain Res. 2011;1367:22. doi: 10.1016/j.brainres.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 362.Hoog SS, Pawlowski JE, Alzari PM, Penning TM, Lewis M. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2517. doi: 10.1073/pnas.91.7.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Wilson DK, Bohren KM, Gabbay KH, Quiocho FA. Science. 1992;257:81. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- 364.Wilson DK, Nakano T, Petrash JM, Quiocho FA. Biochemistry. 1995;34:14323. doi: 10.1021/bi00044a009. [DOI] [PubMed] [Google Scholar]
- 365.Petrash JM. Cell. Mol. Life Sci. 2004;61:737. doi: 10.1007/s00018-003-3402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Komoto J, Yamada T, Watanabe K, Takusagawa F. Biochemistry. 2004;43:2188. doi: 10.1021/bi036046x. [DOI] [PubMed] [Google Scholar]
- 367.Kubiseski TJ, Hyndman DJ, Morjana NA, Flynn TG. J. Biol. Chem. 1992;267:6510. [PubMed] [Google Scholar]
- 368.Nagata N, Kusakari Y, Fukunishi Y, Inoue T, Urade Y. FEBS J. 2011;278:1288. doi: 10.1111/j.1742-4658.2011.08049.x. [DOI] [PubMed] [Google Scholar]
- 369.Yamaguchi K, Okamoto N, Tokuoka K, Sugiyama S, Uchiyama N, Matsumura H, Inaka K, Urade Y, Inoue T. J. Synchrotron Rad. 2011;18:66. doi: 10.1107/S0909049510033595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Okamoto N, Yamaguchi K, Mizohata E, Tokuoka K, Uchiyama N, Sugiyama S, Matsumura H, Inaka K, Urade Y, Inoue T. J. Biochem. :mvr096. doi: 10.1093/jb/mvr096. [DOI] [PubMed] [Google Scholar]
- 371.Crawford K, Kaufman PL, Gabelt BT. Curr. Eye Res. 1987;6:1035. doi: 10.3109/02713688709034874. [DOI] [PubMed] [Google Scholar]
- 372.Weber PC. Contrib. Nephrol. 1980;23:83. doi: 10.1159/000390001. [DOI] [PubMed] [Google Scholar]
- 373.Sasaki S, Hozumi Y, Kondo S. Exp. Dermatol. 2005;14:323. doi: 10.1111/j.0906-6705.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 374.McCracken JA, Custer EE, Lamsa JC. Physiol. Rev. 1999;79:263. doi: 10.1152/physrev.1999.79.2.263. [DOI] [PubMed] [Google Scholar]
- 375.Sugimoto Y, Hasumoto K, Namba T, Irie A, Katsuyama M, Negishi M, Kakizuka A, Narumiya S, Ichikawa A. J. Biol. Chem. 1994;269:1356. [PubMed] [Google Scholar]
- 376.Sugimoto Y, Yamasaki A, Segi E, Tsuboi K, Aze Y, Nishimura T, Oida H, Yoshida N, Tanaka T, Katsuyama M, Hasumoto K, Murata T, Hirata M, Ushikubi F, Negishi M, Ichikawa A, Narumiya S. Science. 1997;277:681. doi: 10.1126/science.277.5326.681. [DOI] [PubMed] [Google Scholar]
- 377.Wilson SJ, Roche AM, Kostetskaia E, Smyth EM. J. Biol. Chem. 2004;279:53036. doi: 10.1074/jbc.M405002200. [DOI] [PubMed] [Google Scholar]
- 378.Lim H, Gupta RA, Ma W.-g., Paria BC, Moller DE, Morrow JD, DuBois RN, Trzaskos JM, Dey SK. Genes & Dev. 1999;13:1561. doi: 10.1101/gad.13.12.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Ali FY, Egan K, FitzGerald GA, Desvergne B, Wahli W, Bishop-Bailey D, Warner TD, Mitchell JA. Am. J. Respir. Cell Mol. Biol. 2006;34:242. doi: 10.1165/rcmb.2005-0289OC. [DOI] [PubMed] [Google Scholar]
- 380.Ullrich V, Castle L, Weber P. Biochem. Pharmacol. 1981;30:2033. doi: 10.1016/0006-2952(81)90218-5. [DOI] [PubMed] [Google Scholar]
- 381.Wada M, Yokoyama C, Hatae T, Shimonishi M, Nakamura M, Imai Y, Ullrich V, Tanabe T. J. Biochem. (Tokyo) 2004;135:455. doi: 10.1093/jb/mvh059. [DOI] [PubMed] [Google Scholar]
- 382.Schildknecht S, Ullrich V. Arch. Biochem. Biophys. 2009;484:183. doi: 10.1016/j.abb.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 383.DeWitt D, Smith W. J. Biol. Chem. 1983;258:3285. [PubMed] [Google Scholar]
- 384.Hara S, Miyata A, Yokoyama C, Inoue H, Brugger R, Lottspeich F, Ullrich V, Tanabe T. J. Biol. Chem. 1994;269:19897. [PubMed] [Google Scholar]
- 385.Miyata A, Hara S, Yokoyama C, Inoue H, Ullrich V, Tanabe T. Biochem. Biophys. Res. Commun. 1994;200:1728. doi: 10.1006/bbrc.1994.1652. [DOI] [PubMed] [Google Scholar]
- 386.Schmidt P, Youhnovski N, Daiber A, Balan A, Arsic M, Bachschmid M, Przybylski M, Ullrich V. J. Biol. Chem. 2003;278:12813. doi: 10.1074/jbc.M208080200. [DOI] [PubMed] [Google Scholar]
- 387.Hecker M, Ullrich V. J. Biol. Chem. 1989;264:141. [PubMed] [Google Scholar]
- 388.Smyth E. Clin. Lipidol. 2010;5:209. doi: 10.2217/CLP.10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Tanabe T, Ullrich V. J. Lipid Mediat. Cell Signal. 1995;12:243. doi: 10.1016/0929-7855(95)00031-k. [DOI] [PubMed] [Google Scholar]
- 390.Ullrich V. Arch. Biochem. Biophys. 2003;409:45. doi: 10.1016/s0003-9861(02)00410-1. [DOI] [PubMed] [Google Scholar]
- 391.Vermylen J, Deckmyn H. Cardiovasc. Drugs Ther. 1992;6:29. doi: 10.1007/BF00050914. [DOI] [PubMed] [Google Scholar]
- 392.Dogne J, de Leval X, Benoit P, Delarge J, Masereel B. Am. J. Respir. Med. 2002;1:11. doi: 10.1007/BF03257158. [DOI] [PubMed] [Google Scholar]
- 393.Howes L, James M, Florin T, Walker C. Expert Opin. Investig. Drugs. 2007;16:1255. doi: 10.1517/13543784.16.8.1255. [DOI] [PubMed] [Google Scholar]
- 394.Ghuysen A, Dogne J, Chiap P, Rolin S, Masereel B, Lambermont B, Kolh P, Tchana-Sato V, Hanson J, D'Orio V. Cardiovasc. Drug Rev. 2005;23:1. doi: 10.1111/j.1527-3466.2005.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 395.Wang L-H, Tsai A-L, Hsu P-Y. J. Biol. Chem. 2001;276:14737. doi: 10.1074/jbc.M009177200. [DOI] [PubMed] [Google Scholar]
- 396.Oliw E, Johnsen O. Biochim. Biophys. Acta. 1988;963:295. doi: 10.1016/0005-2760(88)90294-9. [DOI] [PubMed] [Google Scholar]
- 397.Stark K, Bylund J, Torma H, Sahlen G, Oliw E. Prostaglandins Other Lipid Mediators. 2005;75:47. doi: 10.1016/j.prostaglandins.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 398.Bylund J, Hidestrand M, Ingelman-Sundberg M, Oliw EH. J. Biol. Chem. 2000;275:21844. doi: 10.1074/jbc.M001712200. [DOI] [PubMed] [Google Scholar]
- 399.Woodward D, Protzman C, Krauss A, Williams L. Prostaglandins. 1993;46:371. doi: 10.1016/0090-6980(93)90102-d. [DOI] [PubMed] [Google Scholar]
- 400.Caggiano A, Kraig R. J. Neurochem. 1999;72:565. doi: 10.1046/j.1471-4159.1999.0720565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Halm DR, Halm ST. Am. J. Physiol. 2001;281:G984. doi: 10.1152/ajpgi.2001.281.4.G984. [DOI] [PubMed] [Google Scholar]
- 402.Woodward DF, Jones RL, Narumiya S. Pharmacol. Rev. 2011;63:471. doi: 10.1124/pr.110.003517. [DOI] [PubMed] [Google Scholar]
- 403.Inui T, Ohkubo T, Emi M, Irikura D, Hayaishi O, Urade Y. J. Biol. Chem. 2003;278:2845. doi: 10.1074/jbc.M209934200. [DOI] [PubMed] [Google Scholar]
- 404.Lazarus M, Kubata BK, Eguchi N, Fujitani Y, Urade Y, Hayaishi O. Arch. Biochem. Biophys. 2002;397:336. doi: 10.1006/abbi.2001.2614. [DOI] [PubMed] [Google Scholar]