Summary
The Cullin 4A (Cul4A) gene is important in cell survival, development, growth, and cell cycle control and is amplified in breast and hepatocellular cancers. Recently, we reported that Cul4A plays an oncogenic role in the pathogenesis of mesothelioma. An important strategy for studying Cul4A in different tissues is targeted overexpression of this gene in vivo. Studies of Cul4A in mice have been restricted to the loss-of-function studies using Cul4A knockout mice; gain-of-function studies of Cul4A using transgenic mice have not been reported. We, therefore, generated a gain-of-function transgenic mouse model that over-expresses Cul4A in a Cre-dependent manner. Before Cre recombination, these mice express LacZ during development in most adult tissues. After Cre-mediated excision of the LacZ reporter, the transfected Cul4A gene is expressed along with a C-terminal Myc-tag in different tissues. In this study, Cre-excision was induced in mouse lungs by inhalation of an adenovirus vector encoding Cre recombinase. This mouse model provides a valuable resource for investigating the significance of Cul4A overexpression in various tissues.
Keywords: Cul4A, Cre-mediated expression, mouse model
Cullin 4A (Cul4A) belongs to the family of evolutionarily conserved cullin proteins, including seven related ones (Cul1, 2, 3, 4A, 4B, 5, and 7) in humans. Cul4A forms a part of the multifunctional ubiquitin-protein ligase E3 complex by interacting with ring finger protein (ROC1) and damaged DNA binding protein (DDB1). Through ubiquitin-mediated proteolysis, Cul4A regulates many critical processes in cells. Cul4A is a critical gene for hematopoietic cell survival and development. Overexpression of Cul4A reportedly increases cell growth and causes the disruption of the G2–M cell cycle in irradiated mammary epithelial cells. Previous studies (Kopanja et al., 2009) showed that Cul4A is amplified in breast and liver cancers, is implicated in the ubiquitination and proteolysis of tumor suppressors such as p53, and may contribute to tumorigenesis and cancer development through ubiquitination and then proteolysis of tumor suppressors.
The Cul4A gene restricts cellular repair capacity (Angers et al., 2006; Li et al., 2010). It does so by orchestrating the concerted actions of nucleotide excision repair (NER) and the DNA damage-responsive G1/S checkpoint through selective degradation of the DDB2 and XPC DNA damage sensors and the p21/CIP1/WAF1 checkpoint effector (Liu et al., 2009). We previously found that Cul4A is an oncogene for mesothelioma (Hung et al., 2009). However, the relationship between Cul4A and the occurrence and metastasis of other tumors such as mesothelioma and lung cancer is seldom reported, and the potential oncogenic role of Cul4A has been little studied.
Mouse models have been instrumental in developing our knowledge of signaling pathways in vivo and of the aberrations in these pathways that are causally associated with disease. Studies of the Cul4A gene in mice have been restricted to the loss-of-function studies using Cul4A knockout mice (Liu et al., 2009). For instance, it has been shown that Cul4A knockout mice are resistant to ultraviolet (UV)-induced skin carcinogenesis and that Cul4A restricted cellular repair capacity through selective degradation of p21, DDB2, and XPC (Liu et al., 2009). However, gain-of-function studies of Cul4A using transgenic mice have not been reported.
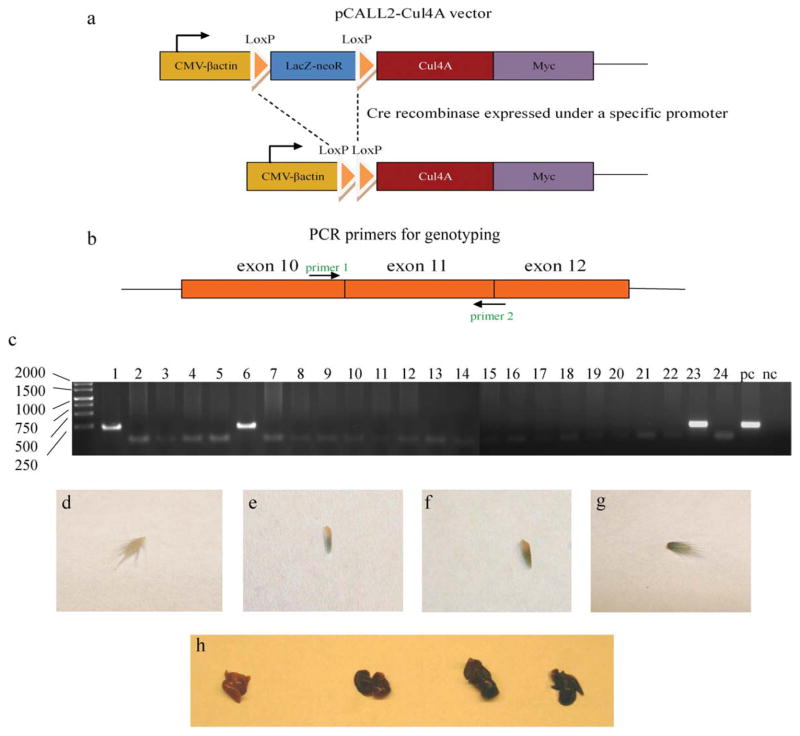
The relationship between Cul4A and cancer is not well understood. Because a transgenic mouse model is needed to explore the potential oncogenic role of Cul4A, we sought to generate one with lineage-tracing that conditionally overexpresses Cul4A protein on Cre-mediated recombination. In this mouse model, before Cre-mediated recombination, LacZ was expressed in the majority of embryonic and adult tissues that were successfully transfected with the gene vector. On Cre-excision in selected organs, LacZ expression was replaced by expression of Cul4A and a Myc-tag. In our model, Cre-excision was induced in mouse lungs by inhalation of an adenovirus vector encoding Cre recombinase (Ade-Cre) (DuPage et al., 2009). To generate this mouse model, we used the pCALL2 expression vector, which has been previously reported (Ding et al., 2002; Elghazi et al., 2008; Lobe et al., 1999; Novak et al., 2000). In our study, a Myc-tagged full-length Cul4A cDNA was subcloned downstream of the loxP-flanked LacZ/neoR. Between the loxP sites, the LacZ sequence is followed by a triple repeat of the SV40 polyadenylation signal (Zinyk et al., 1998) to ensure a transcriptional stop.
Before the Cre-mediated excision, the promoter regulated expression of the loxP-flanked LacZ/neoR. The Cre recombinase of the P1 bacteriophage catalyzes recombination between two specific 34-bp consensus sequences (loxP sites) with high specificity and efficiency (Sternberg and Hamilton, 1981). The loxP sites are palindromic, except for an 8-bp asymmetric core sequence that provides each loxP site with an orientation (Hoess et al., 1986). Introduction of Cre enzyme results in recombination between the loxP sites and excision of the intervening DNA sequence. Upon Cre-mediated excision, LacZ expression is switched to the expression of the Cul4A and a Myc-tag as a monitor of expression (Fig. 1a). The expression of Cul4A can be manipulated in an organ of interest by Cre-excision in that organ.
FIG. 1.
pCALL2-Cul4A and the three positive lines. (a) Diagram of the pCALL2-Cul4A transgene construct before and after Cre-mediated excision of the floxed LacZ-neoR sequence. (b) Design of the PCR primers for genotyping. (c) PCR data (pc, positive control; nc, negative control). The transferred gene could be amplified from lines 1, 6, and 23. (d–g) The stained tails of the transgenic mouse lines (from left to right: (d) negative control, (e) Line 6, (f) Line 1, and (g) Line 23). (h) The stained lungs of the transgenic mouse lines (from left to right: negative control, Line 6, Line 1, and Line 23).
The pCALL2 vector constructed with the Cul4A gene (We hereafter refer to the constructed vector as pCALL2-Cul4A.) was used to generate transgenic mice using embryonic stem cell (ES)-based transgenesis. By standard methods (Nagy, 1997), the pCALL2-Cul4A was transfected into ES cells. After clone selection, the chosen clones of ES cells with pCALL2-Cul4A were used to generate chimeric mice. Transgenic mice were mated with FVB/N mice to produce transgenic offspring. The pCALL2-Cul4A transgene was genotyped by polymerase chain reaction (PCR) using genomic DNA isolated from the tail. We successfully obtained three chimeric mouse lines (lines 1, 6, and 23) from 24 lines (Fig. 1c). Whole mount X-gal staining was positive in the tails (Fig. 1d–g) and the lungs (Fig. 1h) of these three mouse lines, but varied in strength. Line 6 exhibited weak LacZ expression in the staining of tail and lung, whereas Lines 1 and 23 showed similar strong LacZ expression.
Transgene expression was assessed by X-gal staining in the brain, heart, lung, stomach, gut, liver, pancreas, spleen, kidney, and muscle of transgenic mice from the chimeric mouse lines 1 and 23. These lines showed similar stronger LacZ (X-gal) expression in most organs when compared with negative controls (The X-gal staining data for lung, brain, stomach, gut, muscle, pancreas, and kidney are shown in Fig. 2. Data for heart, liver, and spleen are not shown.). However, LacZ expression varied in different organs and tissues. In the brain, LacZ expression was higher in the white matter than in the gray matter. In the lung, the bronchi and alveoli showed the same level of LacZ expression. Overall, LacZ expression was strongest in the gastrointestinal organs, especially in mucous membrane, although it was weak in the chorion and smooth muscle layers and the glands of mucous membrane in the gastrointestinal organs (Fig. 2f,h). In the kidney, expression was higher in the medulla than in the cortex (Fig. 2n). In the pancreas, LacZ expression was apparent, especially in the pancreas islets (Fig. 2l). In the spleen, expression of LacZ was homogeneous (data not shown). Lower levels of LacZ expression were observed in the muscle (Fig. 2j) and heart (data not shown), and none was observed in the liver (data not shown). The fact that high levels of the transgene were expressed in the stomach, gut, lung, brain, spleen, and pancreas suggests that, in this mouse model, studies of Cul4A should be feasible in these organs. The lower levels of LacZ expression in the muscle and heart and the lack of expression in the liver suggest that studies of Cul4A would not be feasible in those organs.
FIG. 2.
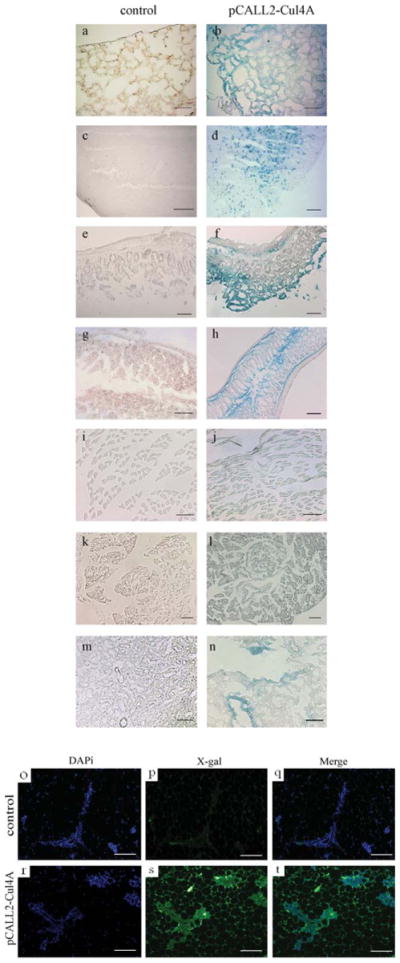
Transgene expression assessed by X-gal staining in several organs of transgenic mice from lines 1 and 23 (a–n), and immunofluorescence staining for anti-LacZ (X-gal) in mammary glands from Line 23 (o–t). In (a–n), the transgene line (pCALL2-Cul4A) is shown on the right, and the negative control is shown on the left. (a,b) Lung; (c,d) Brain; (e,f) Stomach; (g,h) Gut; (i,j) Muscle; (k,l) Pancreas; and (m,n) Kidney. (o–t) Immunofluorescence staining for anti-LacZ in mammary gland sections from control mice (top panels) and pCALL2-Cul4A mice (bottom panels). Scale bar, 100 μm for (a–h,m–t) and 50 μm for (i–l).
We also did immunofluorescence staining for anti-LacZ (X-gal) in mammary glands. The data showed strong LacZ expression in the chimeric mouse lines 1 and 23 and a remarkable difference between control and pCALL2-Cul4A mouse lines 1 (data not shown) and 23 (Fig. 2o–t). This result suggested that the pCALL2 vector highly expressed in mammary glands of the chimeric mouse lines and that studies of Cul4A should be feasible in mammary glands in this mouse model.
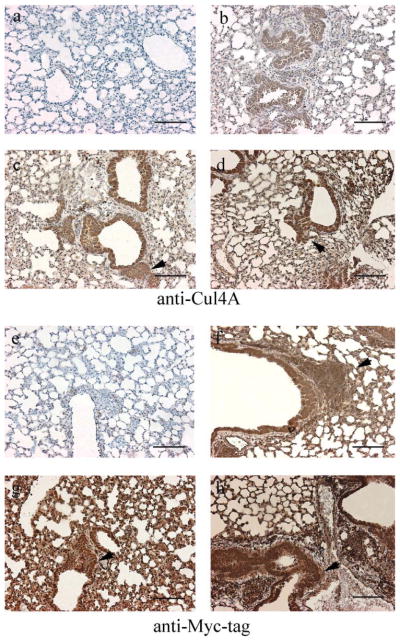
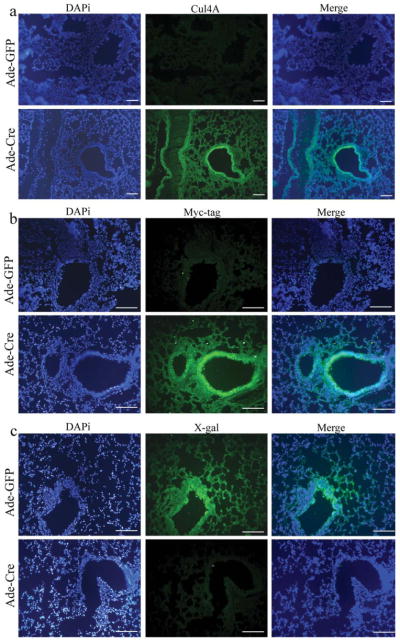
To assess Cul4A expression, Cre excision was induced in the lungs of transgenic mice. Half of the transgenic mice inhaled Ade-Cre. Simultaneously, the other half inhaled Ade-GFP without Cre. Six weeks later, lungs from these mice were dissected. To determine the expression of Cul4A and Myc-tag with or without Cre-excision, lung sections from pCALL2-Cul4A mouse lines 1, 6, and 23 with Ade-GFP or Ade-Cre were immunostained with anti-Cul4A and anti-Myc-tag antibodies, respectively. The lungs of mice with Ade-Cre were positive for both anti-Cul4A and anti-Myc-tag, whereas the lungs of mice without Cre-excision were negative for both. Interestingly, we noticed some potential hyperplasia regions in the mouse lungs after Ade-Cre inhalation (Fig. 3, the potential hyperplasia regions were marked by arrows.). Possibly, this is due to the oncogenic functions of the Cul4A protein. Further studies with longer induction periods will be needed to study these effects. Among the three lines, immunostaining was weakest in Line 6 (Fig. 3b,f). This result was similar to that of the whole mount X-gal staining (Fig. 1d–h). The lung sections from pCALL2-Cul4A mouse lines 1, 6, and 23 with Ade-GFP and Ade-Cre were also immunofluorescence stained for anti-Cul4A, anti-Myc-tag, and anti-LacZ. The results for Cul4A and Myc-tag were the same as those for immunostaining—the lungs of mice with Ade-Cre were positive, and the lungs of mice with Ade-GFP were negative. Immunofluorescence staining for LacZ was negative in lungs with Cre-excision and positive in lungs without Cre-excision (Fig. 4a–c for mouse line 23; data not shown for mouse lines 1 and 6). Taken together, all of these results confirmed that Cre-excision was successful. In mice that inhaled Ade-Cre, the LacZ gene was excised; therefore, these lungs stained positive for anti-Cul4A and anti-Myc-tag antibody and negative for anti-LacZ antibody. We used the same virus particle of Ade-Cre but with GFP (instead of Cre) as control. In mice that inhaled Ade-GFP, the pCALL2-Cul4A gene could not be expressed because the LacZ and the triple repeat of the SV40 sequences were not excised. Consequently, LacZ continued to be expressed instead of Cul4A and Myc-tag.
FIG. 3.
Immunostaining in lung sections from pCALL2-Cul4A mice with Ade-GFP (a,e) or Ade-Cre (b–d, f–h). (a–d) Anti-Cul4A; (e–h) Anti-Myc-tag; (b,f) Line 6; (c,g) Line 1; and (d,h) Line 23. The potential hyperplasia regions were marked by arrows in c, d, f, g, and h. Scale bar, 100 μm.
FIG. 4.
Immunofluorescence staining in lung sections from pCALL2-Cul4A mice with Ade-GFP (top panels) or Ade-Cre (bottom panels). (a) Anti-Cul4A. (b) Anti-Myc-tag. (c) Anti-LacZ. Scale bar, 100 μm.
In conclusion, we have produced a gain-of-function transgenic mouse model that overexpresses Cul4A in a Cre-dependent manner. We believe that these transgenic mice are valuable for further characterization of the Cul4A gene in different organs and at different developmental stages. The lineage tracing capabilities of our model are powerful tools to elucidate the importance of Cul4A in cell fate during development, as well as in the plasticity of mature cells. This mouse strain provides a new model for studying the Cul4A complex in vivo.
METHODS
Construction of the Vector
The study was approved by the University of California at San Francisco (UCSF) Institutional Animal Care and Use Committee. To generate a mouse model that would conditionally overexpress Cul4A protein, we used the pCALL2 vector (Lobe et al., 1999; Novak et al., 2000). This construct contains a chicken β-actin promoter with upstream cytomegalovirus (CMV) enhancer (pCAGGS) (Niwa et al., 1991). This promoter is followed by a loxP-flanked LacZ/neoR fusion with three SV40 polyadenylation (PA) signals and the full-length Cul4A cDNA with a joint but independent sequence of Myc-tag. After the Myc-tag sequence, the construct contained a rabbit β-globin polyadenylation sequence (Fig. 1a). This allowed us to detect cells that expressed Cul4A and to perform lineage tracing experiments. We refer to the expression vector as pCALL2-Cul4A.
ES Culture
We used ES-based transgenesis to generate chimeric mice. The pCALL2-Cul4A vector was transfected by standard methods into ES cells (Nagy, 1997). After cells were cultured, ES cell clones that carried the transgene were selected through neomycin resistance. After selection, colonies were picked and replica-plated in 96-well formats. DNA was extracted from the plates for confirmatory analysis. One clone of cells was chosen, expanded, and frozen. Three clones containing the transgene were selected for generation of chimeric mice by ES cell/embryo aggregation.
Generation of Transgenic Mice Expressing Cul4A
The chosen clones with a single copy of the transgene were used for microinjection to generate transgenic mice by conventional techniques (Nagy, 1997). Transgenic mice were mated with FVB/N mice to produce transgenic offspring. The pCALL2-Cul4A transgene was genotyped by PCR using genomic DNA isolated from the tail. We used the following Cul4A primers (Fig. 1b shows the design and positions of the primers): forward: 5′-GCACTGGAGCGAGTACATCA-3′, and reverse: 5′-CACATGCTTTGCGATCAGTT-3′. A group of mice from the PCR-positive lines were fed for up to 6 months and used to generate offspring.
Tissue Preparation and LacZ Staining
Organs, including brain, lung, heart, stomach, pancreas, liver, gut, kidney, spleen, tail, and muscle, from 4-week-old mice from three different lines and a negative control group were obtained after the mice were killed using cervical dislocation. The organs were removed from the mice and fixed for 6 h in LacZ fixing solution (0.2% glutaraldehyde, 5 mM ethylene glycol tetraacetic acid (EGTA), pH 7.3, 0.1 M MgCl2 in phosphate buffered saline (PBS), pH 7.3) as described previously (Elghazi et al., 2008). Organs were washed in PBS and cryoprotected in 30% sucrose overnight. Tissues were then embedded in optimal cutting temperature (OCT) medium over dry ice. Ten-micrometer frozen sections were cut, collected onto polylysine-coated slides, and stored at −20°C. Slides were fixed in PBS containing 0.2% glutaraldehyde for 10 min, washed in LacZ wash buffer (2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet-P40 in PBS, pH 7.3) and then stained in LacZ wash buffer containing 1 mg/mL X-gal (Roche Diagnostics, Indianapolis, IN), 5 mM potassium ferrocyanide, and 5 mM ferricyanide at 37°C. After staining, slides were rinsed in LacZ wash buffer and then PBS.
Cre-Excision
The intranasal administration of the adenoviruses was performed as previously described (DuPage et al., 2009). The Ade-Cre (Ad5CMVCre) and Ade-GFP (Ad5CMVeGFP) were purchased from the Gene Transfer Vector Core of the University of Iowa, and these adenoviruses were used in several studies (Roh et al., 2006; DuPage et al., 2009). Transgenic mice were anesthetized with 2.5% Avertin via intraperitoneal injection, after which half of the mice inhaled approximately 106 or 107 particles of Ade-Cre introduced directly into the lungs. Simultaneously, the other half inhaled Ade-GFP without Cre. Six weeks later, the mice were killed, and the lungs were dissected.
Assessment of Cul4A and Myc-Tag Expression in Transgenic Cell Lines
Lung sections from pCALL2-Cul4A mice with Ade-GFP or Ade-Cre were immunostained with anti-Cul4A and anti-Myc-tag and immunofluorescence-stained for anti-Cul4A, anti-Myc-tag, and anti-LacZ antibodies, respectively. The sections were washed with PBST (0.1% Triton X-100 in PBS) for 15 min and blocked with 2% horse serum in PBST for 2 h. The sections were then incubated with primary antibody diluted in PBST with 2% horse serum overnight at 4°C. The primary antibodies used in this study were rabbit polyclonal anti-Cul4A (1:400, ab34897, Abcam, Cambridge, MA), rabbit polyclonal anti-Myc-tag (1:20; sc-789; Santa Cruz Biotechnology, Santa Cruz, CA), and goat polyclonal anti-LacZ (1:2,500; 4600-1409; Biogenesis, Poole, UK). The sections were washed four times for 5 min each in PBST. Appropriate secondary antibody was diluted in PBST with 2% horse serum and used in the sections. For histostaining, biotinylated secondary goat anti-rabbit antibody (1:200; BA1000; Vector Laboratories, Burlingame, CA) was used for both anti-Cul4A and anti-Myc-tag primary antibodies. The tissue stainings were visualized using the Vectastain ABC-HRP kit (PK6100; Vector Laboratories). Finally, the slides were counterstained with hematoxylin. For immunofluorescence staining, fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit antibody (1:200; 711-097-003; Jackson Immuno Research Laboratories, West Grove, PA) was used for both anti-Cul4A and anti-Myc-tag primary antibodies; FITC-conjugated donkey anti-goat antibody (1:200; 705-095-003; Jackson ImmunoResearch Laboratories) was used for the anti-LacZ primary antibody. The sections were placed in dark. Two hours later, the sections were washed three times with PBS for 5 min each, counterstained with Vectashield, covered with cover-slips, and viewed using a fluorescent microscope. All sections were photographed using a Zeiss AxioCam camera with Zeiss AxioVision software.
Mammary gland sections from pCALL2-Cul4A mice and control mice were immunofluorescence-stained for anti-LacZ antibody, respectively, using the same procedure that was used for lungs.
Acknowledgments
Contract grant sponsor: National Institutes of Health, Contract grant number: 1R01 CA140654-01A1 (to L.Y.), Contract grant sponsor: Estate of Norman Mancini and the Barbara Isackson Lung Cancer Research Fund
The authors thank Dr. Corrinne Lobe (University of Toronto) for providing the pCALL2 vector. They are grateful to the support from Kazan, McClain, Abrams, Fernandez, Lyons, Greenwood, Harley & Oberman Foundation, Inc., the Estate of Robert Griffiths, the Jeffrey and Karen Peterson Family Foundation, and Paul and Michelle Zygielbaum. They thank Lorretta Chan in the UCSF Cancer Center Tissue Core for her help. They also thank Pamela Derish in the UCSF Department of Surgery for editorial assistance with the manuscript.
LITERATURE CITED
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zhang N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Ding H, Wu X, Nagy A. Mice with Cre recombinase activatable PDGF-C expression. Genesis. 2002;32:181–183. doi: 10.1002/gene.10065. [DOI] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elghazi L, Weiss AJ, Gould AP, Hegedus B, Gutmann DH, Bernal-Mizrachi E. Generation of a reporter mouse line expressing Akt and EGFP upon Cre-mediated recombination. Genesis. 2008;46:256–264. doi: 10.1002/dvg.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung MS, Mao JH, Xu Z, Yang CT, Yu JS, Harvard C, Lin YC, Bravo DT, Jablons DM, You L. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00971.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopanja D, Stoyanova T, Okur MN, Huang E, Bagchi S, Raychaudhuri P. Proliferation defects and genome instability in cells lacking Cul4A. Oncogene. 2009;28:2456–2465. doi: 10.1038/onc.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Robert EI, van Breugel PC, Strubin M, Zheng N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol. 2010;17:105–111. doi: 10.1038/nsmb.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Pengbo Zhou. Cul4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for Cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- Nagy A. Formation of mouse chimeric embryos from ES cells. Amsterdam: Taylor & Francis; 1997. p. 4. [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Roh M, Kim J, Song C, Wills M, Abdulkadir SA. Transgenic mice for Cre-inducible overexpression of the oncogenes c-MYC and Pim-1 in multiple tissues. Genesis. 2006;44:447–453. doi: 10.1002/dvg.20235. [DOI] [PubMed] [Google Scholar]
- Sternberg N, Hamilton D. Bacteriophage P1 site specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- Zinyk D, Mercer E, Harris E, Anderson D, Joyner A. Fate mapping of the mouse midbrain-hind-brain constriction using a site-specific recombination system. Curr Biol. 1998;8:665–668. doi: 10.1016/s0960-9822(98)70255-6. [DOI] [PubMed] [Google Scholar]