Summary
Quorum sensing is a process of bacterial cell–cell communication that enables populations of cells to carry out behaviours in unison. Quorum sensing involves detection of the density-dependent accumulation of extracellular signal molecules called autoinducers that elicit population-wide changes in gene expression. In Vibrio species, CqsS is a membrane-bound histidine kinase that acts as the receptor for the CAI-1 autoinducer which is produced by the CqsA synthase. In Vibrio cholerae, CAI-1 is (S)-3-hydroxytridecan-4-one. The C170 residue of V. cholerae CqsS specifies a preference for a ligand with a 10-carbon tail length. However, a phenylalanine is present at this position in Vibrio harveyi CqsS and other homologues, suggesting that a shorter CAI-1-like molecule functions as the signal. To investigate this, we purified the V. harveyi CqsS ligand, and determined that it is (Z)-3-aminoundec-2-en-4-one (Ea-C8-CAI-1) carrying an 8-carbon tail. The V. harveyi CqsA/CqsS system is exquisitely selective for production and detection of this ligand, while the V. cholerae CqsA/CqsS counterparts show relaxed specificity in both production and detection. We isolated CqsS mutants in each species that display reversed specificity for ligands. Our analysis provides insight into how fidelity is maintained in signal transduction systems.
Introduction
Many bacteria produce and release extracellular signalling molecules called autoinducers. By monitoring the accumulation of autoinducers, bacteria track changes in population density and species complexity in the vicinity. This process, known as quorum sensing, enables groups of bacteria to synchronize gene expression and carry out collective behaviours such as bioluminescence, biofilm formation, competence and virulence factor production, which presumably are not productive when performed by a single bacterium (Fuqua and Greenberg, 2002; Pappas et al., 2004; Novick and Geisinger, 2008; Ng and Bassler, 2009; Williams and Camara, 2009).
Many Vibrio species possess two or more quorum-sensing systems that channel multiple autoinducer inputs into the output response (Bassler et al., 1994; Miller et al., 2002; Henke and Bassler, 2004). For example, the human pathogen Vibrio cholerae activates quorum sensing in response to two different autoinducers, CAI-1 and AI-2 (Chen et al., 2002; Miller et al., 2002; Higgins et al., 2007) (Fig. 1A); whereas the marine bacterium Vibrio harveyi detects three distinct autoinducers, HAI-1, AI-2 and Vh-CAI-1 (Cao and Meighen, 1989; Chen et al., 2002; Henke and Bassler, 2004) (Fig. 1A). Presumably, bacteria extract unique information from each autoinducer. Consistent with this notion, HAI-1 (Fig. 1A), identified as 3-hydroxybutanoyl homoserine lactone, is produced by the LuxM synthase and detected by the LuxN receptor. The LuxM/LuxN system is present in V. harveyi and a few other very closely related Vibrio species, and HAI-1 is suggested to be used for intra-species communication (Cao and Meighen, 1989; Bassler et al., 1993; 1994; Henke and Bassler, 2004). AI-2, identified as (2S, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran borate in Vibrios, is produced by the LuxS enzyme and detected by the LuxPQ receptor (Bassler et al., 1994; 1997; Schauder et al., 2001; Chen et al., 2002; Neiditch et al., 2005). LuxS is present in many bacterial species; thus, AI-2 is considered a signal for inter-species communication (Xavier and Bassler, 2003; Vendeville et al., 2005; Federle, 2009). CAI-1, identified in V. cholerae as (S)-3-hydroxytridecan-4-one, is produced by the CqsA synthase and detected by the CqsS receptor. The CqsA/CqsS system is conserved in many Vibrio species (Miller et al., 2002; Henke and Bassler, 2004; Higgins et al., 2007), suggesting it may be used for communication between Vibrios. The structure of the CAI-1 signal produced by V. harveyi (Vh-CAI-1) has not been examined.
Fig. 1.
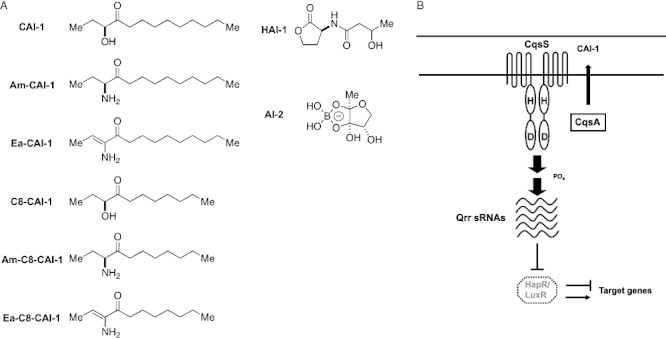
(A) Autoinducer molecules produced and detected by Vibrio species. Molecules in the left column are the CAI-1 type. HAI-1 and AI-2 are shown in the right column.B. The CqsA/CqsS quorum-sensing system of Vibrio species. Black arrows denote phosphate flow from the CqsS receptor to LuxO at low cell density. Under this condition, the Qrr sRNAs are transcribed and inhibit translation of luxR (hapR) transcripts. Thus, LuxR (HapR) proteins are not produced. At high cell density, in the presence of autoinducers, phosphate flow in the signal transduction pathway is reversed, the qrr genes are not transcribed, luxR (hapR) mRNA is translated, and LuxR (HapR) protein is produced, and it initiates the quorum-sensing response.
The information contained in the three autoinducers in V. harveyi (two in V. cholerae) is shuttled into a common phosphorelay signal transduction pathway (Fig. 1B shows the CqsA/CqsS system as an example). Collectively these autoinducers activate production of the master quorum-sensing regulator LuxR (HapR in V. cholerae). LuxR (HapR) acts as both a transcriptional activator and a transcriptional repressor. In V. harveyi, LuxR activates genes for bioluminescence and regulates at least 50 additional targets (Showalter et al., 1990; Waters and Bassler, 2006; Pompeani et al., 2008). In V. cholerae, HapR activates competence genes and HapA protease production, and represses virulence and biofilm formation (Kovacikova and Skorupski, 2002; Zhu et al., 2002; Hammer and Bassler, 2003; 2009; Zhu and Mekalanos, 2003; Blokesch and Schoolnik, 2008; Tsou et al., 2009).
Specificity in ligand–receptor interactions presumably plays a role in preventing cross-talk between related signals and eliminating noise from molecules similar to autoinducers that exist in the environment. However, some quorum-sensing systems display low signal discrimination and multiple autoinducers can activate these circuits (McClean et al., 1997; Cha et al., 1998). The CqsA/CqsS system belongs to the latter class. Specifically, spent culture fluids from a variety of Vibrio species trigger a quorum-sensing response in a V. cholerae CAI-1 reporter strain (Henke and Bassler, 2004). This finding has been interpreted to mean that CAI-1 is used for inter-Vibrio cell–cell communication.
Strikingly, however, examination of V. cholerae CqsS receptor mutants displaying altered responses to natural and synthetic CAI-1 analogues showed that the CqsS residue Cys 170 imparts a preference for a CAI-1 molecule that carries a 10-carbon hydrocarbon tail (Ng et al., 2010). Substituting residue 170 with bulky aromatic amino acids such as phenylalanine or tyrosine (C170F or C170Y) results in a receptor mutant that only recognizes a CAI-1 analogue carrying an 8-carbon tail (C8-CAI-1, Fig. 1A). Examination of conservation among different CqsS homologues shows that residue 170 is substituted with a phenylalanine in V. harveyi CqsS and other homologues. A polymorphism at this particular position in CqsS suggests that a CAI-1 molecule with a 10-carbon tail is not likely to be the autoinducer detected by all CqsS receptors.
In this study, through purification and chemical synthesis, we identify the V. harveyi CqsS ligand as (Z)-3-aminoundec-2-en-4-one (Ea-C8-CAI-1, Fig. 1A). Ea-C8-CAI-1 is also produced and detected by V. cholerae CqsA/CqsS. Although the CqsA/CqsS systems in V. harveyi and V. cholerae are highly conserved, the V. harveyi CqsA synthase is highly selective for its substrate octanoyl CoA, and the V. harveyi CqsS receptor likewise displays an exquisite specificity for its ligand Ea-C8-CAI-1. In contrast, V. cholerae CqsA has less substrate specificity, accepting both decanoyl CoA and octanoyl CoA. The V. cholerae CqsS receptor is similarly less stringent and does not discriminate well between ligands. We isolated V. cholerae and V. harveyi CqsS mutants displaying increased and decreased ligand specificity respectively. We propose that the CqsA/CqsS systems in these two organisms have diverged from a common ancestral origin, and that differences arose from selective forces that favoured decreased specificity in V. cholerae and/or increased specificity in V. harveyi.
Results
V. harveyi CqsS detects a different ligand than does V. cholerae CqsS
The CqsS receptor is conserved among many Vibrio species. Notably, the transmembrane ligand-sensing domains share more than 70% amino acid sequence identity. This high sequence identity suggests that CqsS receptors could recognize a common or a related set of signalling molecules. (S)-3-hydroxytridecan-4-one and (S)-3-aminotridecan-4-one (i.e. CAI-1 and Am-CAI-1, respectively, Fig. 1A) are two potent 13-carbon CqsS agonists produced and detected by V. cholerae (Higgins et al., 2007; Kelly et al., 2009). We previously showed that a single amino acid change in the CqsS-sensing domain results in an altered ligand specificity (Ng et al., 2010). Specifically, replacing Cys 170 with a bulky amino acid causes a loss in response to CAI-1 and an increase in sensitivity to a CAI-1 analogue carrying a shortened 8-carbon tail (C8-CAI-1, Fig. 1A) (Ng et al., 2010). We inspected all CqsS receptor homologues in sequenced Vibrios and found that many of them, including the V. harveyi CqsS, carry a phenylalanine substitution at this particular position (Fig. 2A). Therefore, we suspect that these receptors must detect molecules that are shorter than CAI-1.
Fig. 2.
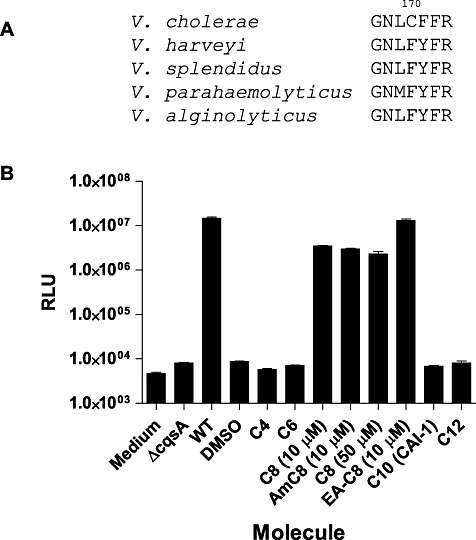
(A) Polymorphism in the CqsS receptors of Vibrio species. The C170 residue in V. cholerae is substituted with phenylalanine in multiple Vibrio species including V. harveyi.B. The V. harveyi CqsS-dependent quorum-sensing response to various CAI-1 type molecules. Activation of bioluminescence expression was measured in V. harveyi strain JMH626 in the presence of exogenously added CAI-1 type molecules. ΔcqsA and WT denote cell-free culture fluids prepared from a V. harveyi mutant lacking or containing the CqsA synthase respectively. See Fig. 1A for the structures of the different molecules. RLU denotes relative light units which is Lux/OD600.
We chose V. harveyi as the model to examine the molecules detected by these naturally occurring polymorphic CqsS receptors. To do this, we measured light production in a V. harveyi reporter strain that responds exclusively to exogenously supplied CqsS ligands (JMH626, see Experimental procedures). Sterile medium and spent culture fluid from a ΔcqsA strain do not induce light production in JMH626. However, addition of 10% (v/v) spent culture fluid prepared from wild-type V. harveyi induces light production 3000-fold in JMH626 (Fig. 2B). Presumably, this is due to production of the endogenous CqsS ligand. We tested purified CAI-1 and CAI-1 analogues carrying different hydrocarbon tail lengths for stimulation of light production in this assay. CAI-1 analogues with 4-, 6-, 10- and 12-carbon tails did not stimulate light production (Fig. 2B). However, C8-CAI-1 (10 µM) and Am-C8-CAI-1 (10 µM) induce an approximately 400-fold increases in light production (Fig. 2B). Increasing the concentration to 50 µM did not further activate light production (Fig. 2B). Combining C8-CAI-1 with spent culture medium from a V. harveyiΔcqsA strain also did not enhance induction (data not shown). These results suggest that V. harveyi CqsS does not detect the V. cholerae CAI-1 molecule (10-carbon tail) as an agonist, and that C8-CAI-1 is only a weak agonist for V. harveyi CqsS since it cannot induce maximal light production even at very high concentrations. We therefore reason that V. harveyi must produce a CAI-1 like molecule that likely has an 8-carbon tail with additional modification(s) that do not exist in CAI-1 or in C8-CAI-1.
In order to identify the molecule detected by the V. harveyi CqsS receptor, we overexpressed the V. harveyi cqsA gene in Escherichia coli (strain WN1327). Spent culture fluid (10% v/v) from WN1327, but not from E. coli carrying the empty vector, induced light production in JMH626 to the maximal level (data not shown). Moreover, activity could be quantitatively extracted with dichloromethane (DCM), which allowed us to separate the activity from other components. Concentration of the DCM extract to complete dryness led to a significant loss in activity; therefore in all our analyses, we avoided steps that resulted in activity decreases by monitoring the activity in each step.
A variety of reverse and normal phase HPLC methods (Supporting information) were used to isolate fractions capable of inducing maximal light production in V. harveyi JMH626. The purity of the active fractions was determined by ESI-TOF HRMS. One fraction that could induce maximal light production in JMH626 possessed a single ion peak with m/z [M+H+] of 184.1699, corresponding to a molecular formula of C11H21NO. A second fraction was weakly active and possessed a molecular ion peak with m/z [M+H+] of 187.1694, corresponding to a molecular formula of C11H22O2. Concentration of this second fraction allowed us to use NMR to determine that it contains 3-hydroxyundecan-4-one (C8-CAI-1) and its regioisomer 4-hydroxyundecan-3-one in approximately a 1:1 ratio. Through chemical synthesis and consistent with the above results, we found that C8-CAI-1 is only a weak agonist for the V. harveyi CqsS receptor. The regioisomer, 4-hydroxyundecan-3-one, is inactive (Fig. 2B and data not shown).
Direct characterization of the structure of the 184.1699 molecular ion by NMR was impossible because, as mentioned, concentration caused a complete loss in activity. We hypothesized that this molecule must have a structure similar to known CqsS agonists including CAI-1, Am-CAI-1 and C8-CAI-1. Based on its molecular formula C11H21NO, we predicted that the active molecule was likely a derivative of Am-C8-CAI-1 (Fig. 1A) possessing an additional degree of unsaturation. To identify the position of the unsaturation, we used super-critical fluid chromatography (SFC) to obtain a highly concentrated fraction of our desired molecule dissolved in a solvent suitable for direct NMR analysis (see Experimental procedures). The resulting maximally active fraction contains a molecule with a single olefinic proton by 1H-NMR (δ 5.77, q, J = 7.1 Hz) (Supporting information). Taken together with the molecular formula, the splitting pattern and chemical shift of this signal allows us to propose that the structure of the highly active V. harveyi CqsS agonist is (Z)-3-aminoundec-2-en-4-one (Ea-C8-CAI-1, Fig. 1A). We prepared Ea-C8-CAI-1 by total synthesis (Supporting information) and determined that, by 1H-NMR, the synthetic material contains the diagnostic olefinic quartet observed in the natural sample. Additionally, the synthetic compound has identical activity to the ligand-containing spent culture fluid harvested from wild-type V. harveyi; that is, they both induce maximal light production in JMH626 (Fig. 2B). We conclude the V. harveyi CqsS receptor detects Ea-C8-CAI-1 as its native ligand.
Different Vibrio species display unique production and detection profiles for the CAI-1 family of molecules
Spent culture fluids from V. harveyi and V. cholerae cross-stimulate the CqsS-dependent quorum-sensing response of one another (Henke and Bassler, 2004) (data not shown). Because V. harveyi CqsS is insensitive to CAI-1 (10-carbon tail) and quite sensitive to Ea-C8-CAI-1, we hypothesized that V. cholerae must also produce and detect Ea-C8-CAI-1. Based on the above results, we also wondered if V. cholerae produces an enamino molecule (perhaps Ea-CAI-1, 10-carbon tail, Fig. 1A) analogous to Ea-C8-CAI-1 that promotes the response we observed in V. harveyi.
To examine these possibilities, we determined how much of each CAI-1 type molecule is produced by wild-type V. harveyi and V. cholerae. Both species were grown to early stationary phase and cell-free spent culture fluids were prepared (see Experimental procedures). CAI-1 type molecules were extracted with DCM for HRMS analysis. The concentration of each CAI-1 type molecule was calculated by comparison to the intensity of each ion peak to that of d2-CAI-1 (double deuterium labelled CAI-1), which was added as an internal standard at 500 nM prior to the extraction. Correction for the different ionization potentials of each of the CAI-1 type molecules was achieved using a mixed sample containing a known amount of each of the molecules (see Experimental procedures).
In the V. cholerae spent culture fluids, we detected the molecular ions from Ea-C8-CAI-1, Ea-CAI-1 and CAI-1 at an average concentration of 19 nM, 139 nM and 222 nM respectively. We did not detect any C8-CAI-1 ion in the V. cholerae sample (Fig. 3). In the V. harveyi spent culture fluids, we only detected molecular ions from Ea-C8-CAI-1 and C8-CAI-1 at an average concentration of 50 nM and 154 nM respectively. No ion from CAI-1 or Ea-CAI-1 was detected (Fig. 3). The detection limits for Ea-C8-CAI-1, C8-CAI-1, Ea-CAI-1 and CAI-1 are 6.25 nM, 25 nM, 12.5 nM and 25 nM respectively (Supporting information).
Fig. 3.
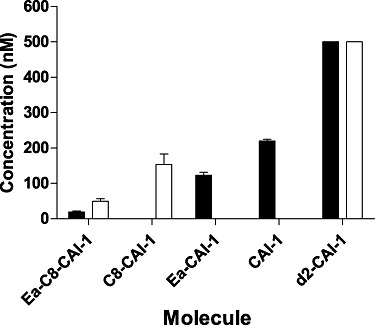
CAI-1 type molecule biosynthetic profiles for V. cholerae and V. harveyi. The concentrations of different CAI-1 type molecules present in cell-free culture fluids prepared from V. cholerae (black bars) and V. harveyi (white bars) were determined by HRMS analyses using d2-CAI-1 as the internal reference (final set of bars). Results from three independent replicates were averaged. Error bars denote standard deviations. See Fig. 1A for the structures of the different molecules.
We systematically analysed the relative activities of all of the CAI-1-related molecules observed in our HRMS analysis (i.e. C8-CAI-1, Ea-C8-CAI-1, CAI-1 and Ea-CAI-1) on V. harveyi and V. cholerae (Fig. 4, Table 1). As in the above experiments, we used JMH626 for the V. harveyi test. For V. cholerae, we used a mutant (WN1102, see Experimental procedures) that carries the V. harveyi luxCDABE operon (luciferase) and responds exclusively to exogenously supplied CqsS ligands. The V. harveyi wild-type CqsS receptor is highly sensitive to synthetic Ea-C8-CAI-1 (EC50 = 24 nM, EC50 is defined as the concentration of autoinducer that induces light production to half the maximum level) but only responds weakly to C8-CAI-1 (EC50 = 5 µM). The V. harveyi CqsS receptor is insensitive to Ea-CAI-1 and CAI-1 (Fig. 4A, Table 1). In contrast, the V. cholerae CqsS receptor responds to a wider array of molecules including Ea-C8-CAI-1 (EC50 = 36 nM), Ea-CAI-1 (EC50 = 4 nM) and CAI-1 (EC50 = 62 nM), but is only weakly sensitive to C8-CAI-1 (EC50 = 1.2 µM) (Fig. 4B and Table 1). We also tested Am-CAI-1 and Am-C8-CAI-1; their potencies are comparable to the CAI-1 and C8-CAI-1 counterparts in each CqsS receptor (data not shown).
Fig. 4.
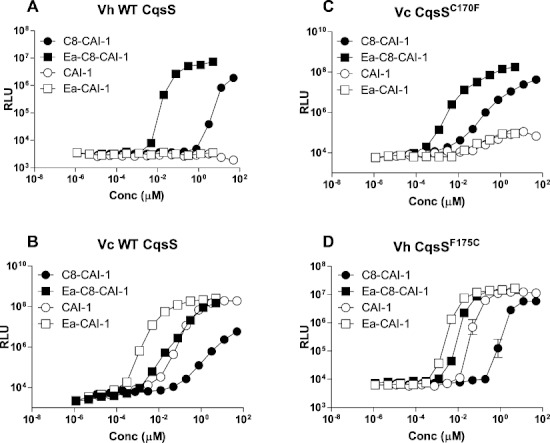
Response of V. cholerae and V. harveyi wild-type CqsS and CqsS mutants to different CAI-1 type molecules. The CqsS-dependent quorum-sensing responses to different CAI-1 type molecules were measured by assaying activation of bioluminescence expression in wild-type V. harveyi (A), wild-type V. cholerae (B), V. cholerae carrying CqsS C170F (C) and V. harveyi carrying CqsS F175C (D). C8-CAI-1, closed circles; Ea-C8-CAI-1, closed squares; CAI-1, open circles, Ea-CAI-1, open squares. See Fig. 1A for the structures of the different molecules. Representative data from at least three independent experiments are shown. See Table 1 for EC50 values for each receptor/ligand combination. RLU denotes relative light units.
Table 1.
CqsS receptor sensitivities to CAI-1 type molecules.a
| EC50 (nM) | ||||
|---|---|---|---|---|
| CqsS | C8-CAI-1 | Ea-C8-CAI-1 | CAI-1 | Ea-CAI-1 |
| Vc WT | 1175 ± 300 | 36 ± 8 | 62 ± 12 | 4 ± 3 |
| Vc C170F | 268 ± 91 | 3 ± 0.4 | NA | NA |
| Vh WT | 5294 ± 967 | 24 ± 7 | NA | NA |
| Vh F175C | 3177 ± 219 | 12 ± 3 | 36 ± 8 | 6 ± 4 |
Numbers are shown as mean ± standard error of mean.
NA, not applicable.
Based on these results, we propose that, in the growth conditions used on this study, wild-type V. harveyi only detects Ea-C8-CAI-1 through CqsS to activate quorum sensing. Although C8-CAI-1 is a weak agonist, the EC50 value is significantly larger than the concentration of C8-CAI-1 released by V. harveyi. Using the same reasoning, V. cholerae likely detects three molecules, Ea-CAI-1, CAI-1 and Ea-C8-CAI-1, but not C8-CAI-1. Therefore, V. harveyi CqsS has a stringent specificity for its ligand, while V. cholerae CqsS is somewhat promiscuous. In addition, our results show that there is a common molecule, Ea-C8-CAI-1, produced and detected efficiently by both V. cholerae and V. harveyi CqsS (Fig. 4 and Table 1). We propose that Ea-C8-CAI-1 is the chemical signal that is used for inter-Vibrio cell–cell communication.
CqsA substrate specificity correlates with the CAI-1 biosynthetic profile
In another study, we showed that V. cholerae CAI-1 is derived from the precursor Ea-CAI-1, which is synthesized by V. cholerae CqsA using S-adenosylmethionine (SAM) and decanoyl coenzyme A (C10-CoA) as substrates (Y. Wei et al., 2011). Likewise, in V. harveyi, C8-CAI-1 is derived from Ea-C8-CAI-1, which is synthesized by CqsA from SAM and octanoyl CoA (C8-CoA) (Supporting information). However, these findings do not account for all the CAI-1 type molecules we detect in cell-free culture fluids. Specifically, V. cholerae must also produce Ea-C8-CAI-1. We wondered therefore if similar to their cognate CqsS receptors, V. harveyi and V. cholerae CqsA enzymes possess high and low substrate specificity respectively. To examine this possibility, the two CqsA enzymes were purified and used in a coupled-enzyme assay (Experimental procedures) to determine the kinetic parameters with respect to processing C8-CoA or C10-CoA as substrates.
Vibrio cholerae CqsA uses C10-CoA and C8-CoA to make Ea-CAI-1 and Ea-C8-CAI-1 respectively (Table 2). However, as indicated by the differences in both Km and kcat, V. cholerae CqsA prefers C10-CoA over C8-CoA as substrate (four- to sixfold). In contrast, the V. harveyi CqsA enzyme only uses C8-CoA to produce Ea-C8-CAI-1 and is unable to use C10-CoA as a substrate (Table 2). Thus, the unique preferences of the CqsA enzymes for their acyl CoA substrates match the in vivo CAI-1 production profiles displayed by V. cholerae and V. harveyi. Interestingly, the CqsA substrate preferences also highly correlate with the CqsS ligand selectivities. We propose that the CqsA enzymes and the CqsS receptors coevolved to ensure productive ligand–receptor interactions and high-fidelity signalling.
Table 2.
Kinetic parameters for V. cholerae and V. harveyi CqsA enzymes.
| Enzyme Substrate | Vh CqsA C8 CoA | Vh CqsA C10 CoA | Vc CqsA C10 CoA | Vc CqsA C8 CoA |
|---|---|---|---|---|
| Km (µM) | 14.1 ± 1.3 | ND | 13.0 ± 3.1 | 73 ± 4.9 |
| Vmax (µM min−1) | 0.64 ± 0.02 | ND | 1.2 ± 0.15 | 1.4 ± 0.05 |
| kcat (min−1) | 10.7 ± 0.4 | ND | 19.5 ± 2.5 | 4.5 ± 0.17 |
ND, not detected.
CqsS amino acid residues responsible for ligand specificity
The V. harveyi receptor shows a strong preference for CAI-1 molecules carrying 8-carbon tails (Fig. 4A, Table 1). In addition, the presence of the enamino group in C8-Ea-CAI-1 is crucial for its agonist activity for the V. harveyi receptor (Fig. 4A, Table 1). In contrast, V. cholerae CqsS is promiscuous and detects both 8-carbon and 10-carbon tailed molecules. The presence of the enamino group is not obligatory for agonist activity in V. cholerae (Fig. 4B, Table 1). To understand ligand preference, we used mutagenesis to pinpoint residues that are essential for detection of the different features of the ligands (i.e. chain length and presence of the enamino group).
First, we focus on chain length discrimination. We have previously performed an analysis on V. cholerae CqsS with two molecules, CAI-1 and C8-CAI-1(Ng et al., 2010). We now expand this analysis using all four molecules described here (i.e. CAI-1, C8-CAI-1, Ea-CAI-1 and Ea-C8-CAI-1), and both V. cholerae and V. harveyi CqsS receptors are studied. As mentioned, V. cholerae CqsS prefers CAI-1 over C8-CAI-1 due to the presence of Cys 170 (Ng et al., 2010). V. cholerae CqsS prefers Ea-CAI-1 over Ea-C8-CAI-1 (Fig. 4B). Consistent with this finding, V. cholerae CqsSC170F only detects Ea-C8-CAI-1 and C8-CAI-1, but not CAI-1 and Ea-CAI-1 (Fig. 4C, Table 1). Likewise, we mutated the phenylalanine (F175) in the corresponding position in the V. harveyi CqsS receptor to cysteine. We found that the CqsSF175C mutant exhibits relaxed specificity and detects both Ea-CAI-1 and CAI-1 as agonists (Fig. 4D, Table 1). In addition, and similar to wild-type V. cholerae CqsS, the V. harveyi CqsSF175C mutant also detects Ea-C8-CAI-1 and is only weakly sensitive to C8-CAI-1 (Fig. 4D, Table 1). Moreover, the V. harveyi CqsSF175C mutant is most sensitive to Ea-CAI-1. Thus, in terms of the chain length preference, the presence of a cysteine residue at position 170 decreases the overall specificity of the CqsS receptor, allowing detection of ligands with both 8- and 10-carbon tails. The presence of phenylalanine at this position increases the CqsS specificity for ligands that carry an 8-carbon tail.
Unexpectedly, detection of the enamino group also depends on the chain length of the molecule. Specifically, the enamino group is critical for activity only when it is present in a molecule carrying an 8-carbon tail (Fig. 4, Table 1). For example, there is a 10-fold difference in EC50 between CAI-1 and Ea-CAI-1 in V. cholerae CqsS and in the V. harveyi CqsSF175C mutant. In contrast, there are 30-fold and 200-fold differences in EC50 between C8-CAI-1 and Ea-C8-CAI-1 in these two receptors (Table 1). Moreover, unlike CAI-1, C8-CAI-1 does not elicit a maximum quorum-sensing response in any CqsS receptor tested (Fig. 4, Table 1). These results suggest that C8-CAI-1 lacking the enamino group cannot properly interact with either the V. cholerae or the V. harveyi CqsS receptor. We have not been able to isolate any mutant that eliminates the requirement for the enamino group in Ea-C8-CAI-1. Thus, the molecular determinant for this preference remains unclear. One possibly is that the amino group forms a hydrogen bond with a backbone carbonyl in the receptor.
Discussion
The CqsA/CqsS quorum-sensing system responds to molecules made by a variety of Vibrio species (Henke and Bassler, 2004), yet the identities of the active signalling molecules have not been well characterized. The first CAI-1 signal examined, that of V. cholerae, is (S)-3-hydroxytridecan-4-one (Higgins et al., 2007) (CAI-1, Fig. 1A). Here, we show that CAI-1 cannot be detected by the V. harveyi CqsS receptor due to an incompatibility between the 10-carbon tail in CAI-1 and the presence of a bulky F175 residue in the V. harveyi CqsS receptor. Using purification and total in vitro synthesis, we identified the ligand for the V. harveyi CqsS to be (Z)-3-aminoundec-2-en-4-one (Ea-C8-CAI-1, Fig. 1A). Ea-C8-CAI-1 fulfils the requirements for being an inter-Vibrio quorum-sensing signal because it is produced and detected by multiple Vibrio species (Figs 3 and 4, Table 1).
Although we focused in this study on V. harveyi and V. cholerae as test cases, the CqsS and CqsA sequences from most other Vibrio species can be readily categorized into one of the two classes represented by these examples. The V. cholerae system has relaxed specificity in both substrate selection by CqsA and ligand detection by CqsS (Figs 3 and 4, Tables 1 and 2). V. cholerae produces and detects three different molecules: CAI-1, Ea-CAI-1 and Ea-C8-CAI-1. In addition, the absence of an enamino group in CAI-1 does not significantly hamper its agonist activity. In contrast, the V. harveyi system is stringent in both CqsA-dependent production and CqsS-directed detection of the ligand (Figs 3 and 4, Tables 1 and 2). V. harveyi produces C8-CAI-1 and Ea-C8-CAI-1, but only Ea-C8-CAI-1 is detected. The presence of the enamino group is critical for ligand activity in V. harveyi (Figs 3 and 4, Tables 1 and 2). Based on these results, we suggest that CqsA/CqsS systems similar to the V. harveyi system are stringent, in which the unique substrate for CqsA is C8-CoA and the unique signal for CqsS is Ea-C8-CAI-1. Consistent with this prediction, we found that only Ea-C8-CAI-1 and C8-CAI-1, but not CAI-1 and Ea-CAI-1, are present in spent culture fluids prepared from several of these species (e.g. Vibrio parahaemolyticus, Vibrio alginolyticus, Vibrio anguillarum and Vibrio furnissii, data not shown). In contrast, CqsA/CqsS systems similar to the V. cholerae system are relaxed (e.g. Vibrio mimicus and some sequenced natural Vibrio isolates). Both C8-CoA and C10-CoA can be used as substrates for CqsA, and multiple signals including CAI-1, Ea-CAI-1 and Ea-C8-CAI-1 can be detected by CqsS. Ea-C8-CAI-1 links the two types of systems as it is produced and detected by both.
Ea-CAI-1 and Ea-C8-CAI-1 are both potent agonists for V. cholerae CqsS, and thus we wondered why we did not identify these enamino compounds previously (Higgins et al., 2007). We suspect that the enamino activities were destroyed during our previous sample preparation because our present results demonstrate that enamino molecules are labile following concentration or heat treatment. The procedures we used to initially isolate V. cholerae CAI-1 likely did not preserve the enamino molecules. However, because the V. cholerae CqsS receptor shows low stringency for the enamino group (Fig. 4, Table 1), we were able to identify CAI-1 as one of the V. cholerae CqsS ligands (Higgins et al., 2007). A detailed analysis of the mechanism by which Ea-CAI-1 is produced by CqsA and how Ea-CAI-1 is subsequently converted into CAI-1 is described elsewhere (Y. Wei et al., 2011).
In terms of signal specificity, the residue conferring the difference in receptor specificity is located in the final transmembrane helix of CqsS (Ng et al., 2010). Receptor specificity can be manipulated by incorporation of a single substitution at the C170 or F175 position of the CqsS receptor of V. cholerae or V. harveyi respectively. This polymorphism is particularly important for chain length preference. The presence of a small amino acid residue such as cysteine decreases the receptor selectivity, while the presence of a bulky amino acid residue such as phenylalanine increases selectivity (Fig. 4, Table 1).
The analogous mechanism underlying CqsA substrate preference is not understood. Based on the V. cholerae CqsA crystal structure, it is believed that the 10-carbon hydrocarbon chain sits in an enclosed hydrophobic pocket lined by H30, V32, F79, F257, I263, F264, C346, P348 and A349 (Jahan et al., 2009; Kelly et al., 2009). All of these residues are conserved in V. harveyi CqsA so they cannot be responsible for specifying the C8 substrate tail length preference. Additional biochemical and genetic studies are required to understand CqsA substrate specificity in V. harveyi or lack thereof in V. cholerae.
Although a few homologues of the CqsA/CqsS system are found in non-Vibrio species (see Tiaden et al., 2010a), to date, only one such system (i.e. the Legionella pneumophila LqsA/LqsS system) has been studied (Tiaden et al., 2007; 2008; 2010b; Spirig et al., 2008). An α-hydroxyketone molecule LAI-1 (3-hydroxypentadecan-4-one, that is, CAI-1 with a 12-carbon tail) is proposed to be the LqsS ligand. It is not known if the LqsA synthase uses a mechanism identical to CqsA to produce an enamino compound which is subsequently converted to LAI-1. The sequence homology is weak for the ligand–receptor interaction region in the sixth transmembrane helix of LqsS and CqsS; therefore, it is unclear how LqsS detects a ligand with a 12-carbon tail.
In Gram-negative bacteria, LuxI/LuxR quorum-sensing systems are commonly used. LuxI type proteins produce acyl homoserine lactones, which are detected by cognate cytoplasmic LuxR type receptors (Fuqua et al., 2001; Pappas et al., 2004). In Gram-positive bacteria, oligopeptides are usually detected by membrane-bound receptors for quorum sensing (Havarstein et al., 1995; Novick and Geisinger, 2008; Thoendel and Horswill, 2010). Examination of the phylogenetic relationship among all LuxI/LuxR systems leads to the conclusion that these quorum-sensing systems are ancient and thus cell–cell communication arose early in evolution (Gray and Garey, 2001; Lerat and Moran, 2004). The majority of luxI/luxR genes are located contiguously on the chromosome and therefore presumably retain their pairwise functional relationships through co-evolution as single cassettes (Gray and Garey, 2001; Lerat and Moran, 2004). The same genomic arrangement is observed for peptide-based quorum-sensing systems in Gram-positive bacteria (Pestova et al., 1996; Novick and Geisinger, 2008). That is, the gene encoding the autoinducer peptide is linked to the gene encoding the receptor. When we examine the gene organization of the different CqsA/CqsS homologues, we find that the cqsA and cqsS genes are usually adjacent in Vibrio species. In other species that possess homologous systems, such as L. pneumophila, Burkholderia xenovorans and Ralstonia eutropha, the genes that encode the synthase and the receptor are also contiguous (as in R. eutropha) or at least in close proximity with an occasional insertion of an accessory gene (e.g. in L. pneumophila and B. xenovorans). Finally, signal biosynthesis and signal detection profiles in V. cholerae and V. harveyi match closely (Figs 3 and 4), further suggesting that the cqsA and cqsS genes in each species coevolved. We therefore suggest that an ancestral CqsA/CqsS system must have diverged to give rise to the V. cholerae and V. harveyi CqsA/CqsS systems. V. cholerae and V. harveyi reside in different environmental niches. V. cholerae is a human pathogen that cycles between periods in the aquatic environment and periods inside the host; while V. harveyi is a marine bacterium that is pathogenic to many marine vertebrates and invertebrates. Although we do not know the forces that drove these two species to evolve different signalling specificities, we suspect that stringent signalling in the case of V. harveyi and promiscuous signalling in the case of V. cholerae increases the fitness of each species in its respective environmental niche.
Experimental procedures
Bacterial strains and culture conditions
All V. cholerae strains are derivatives of wild-type C6706str (Thelin and Taylor, 1996). All V. harveyi strains are derivatives of wild-type V. harveyi BB120 (Bassler et al., 1997). E. coli S17-1 pir, DH5α and Top10 were used for cloning. Strains JMH626 and WN1102 were used in bioluminescence assays (see below). JMH626 is a V. harveyiΔcqsAΔluxQΔluxN mutant that does not produce Vh-CAI-1 due to the cqsA mutation. In addition, JMH626 does not detect HAI-1 and AI-2 because it lacks both the LuxN and LuxQ receptors (Henke and Bassler, 2004). WN1102 is a V. choleraeΔcqsAΔluxQ mutant that carries the V. harveyi luxCDABE operon on a cosmid. This strain does not produce CAI-1 and it does not detect AI-2. The relevant genotypes of all plasmids and strains are provided in Supporting information. Unless specified, E. coli was grown in LB medium at 37°C with shaking, V. cholerae and V. harveyi were grown in LM medium at 30°C with shaking. Unless specified, antibiotic concentrations are as follows: ampicillin, gentamicin and kanamycin, 100 mg l−1; chloramphenicol and tetracycline, 10 mg l−1; streptomycin, 5 g l−1; polymyxin B, 50 U l−1.
DNA manipulation, site-directed mutagenesis and mutant construction
All DNA manipulations were performed using standard procedures (Sambrook et al., 1989). Oligonucleotide sequences used for PCR, site-directed mutagenesis and sequencing reactions will be provided upon request. Site-directed mutageneses were performed with QuikChange II XL Site-Directed Mutagenesis Kit according to the manufacturer's instructions. To examine the effects of cqsS mutations on the CAI-1 response of V. cholerae, point mutations were first constructed in pDH345 (Ng et al., 2010), and mutated cqsS alleles were subcloned into the suicide plasmid pKAS32 (Skorupski and Taylor, 1996). The mutated cqsS allele was introduced onto the V. cholerae chromosome using pKAS32 as previously described (Skorupski and Taylor, 1996). The pBB1 cosmid carrying the V. harveyi luxCDABE operon was introduced into V. cholerae strains via conjugation. To examine the effects of cqsS mutations on the CAI-1 response in V. harveyi, point mutations were first constructed in plasmid pJMH280 (Henke and Bassler, 2004), mutated cqsS alleles were subcloned into the low-copy-number cosmid pLAFR2, and the constructs were introduced into the V. harveyiΔcqsS strain via conjugation, and the mutant cqsS alleles were maintained as episomes.
Bioluminescence assay for V. cholerae and V. harveyi CqsS agonists
Culture fluids were prepared from overnight cultures unless otherwise specified. Cells were removed from the fluids by centrifugation and subsequent filtration through 0.22 µm filters. To assess CAI-1 activity in these cell-free fluids, overnight cultures of reporter strains were grown in LM medium (for V. harveyi) or LM with tetracycline (for V. cholerae carrying pBB1 and V. harveyi carrying cqsS on pLAFR2) and diluted 20-fold with sterile medium. Approximately 10–30% (v/v) of spent culture fluids were added to the diluted reporter strains. Bioluminescence and OD600 were measured in an Envison Multilabel Reader following 4 h incubation at 30°C with shaking. Sterile medium was used as the negative control. Synthetically prepared CAI-1 analogues were dissolved in DMSO and supplied at varying concentrations to the reporter strains. DMSO was used as the negative control.
Extraction and purification of V. harveyi Ea-C8-CAI-1
Escherichia coli overexpressing V. harveyi cqsA (WN1327) was grown overnight in LB with kanamycin at 30°C with shaking. The culture was diluted 1000-fold in M9 minimal salts medium (Sigma) supplemented with 0.05% (w/v) leucine, 2 mM MgCl2, 0.1 mM CaCl2, 0.01% (w/v) thiamine and 0.4% (w/v) glucose with 10 mg l−1 kanamycin. The culture was incubated overnight at 30°C with shaking. Cells were removed by centrifugation and the cleared fluid was extracted by DCM. The extract was carefully concentrated without heating to a small volume (5 ml) in a Rotovap. The concentrated extract was fractionated using HPLC (see Supporting information). Each HPLC fraction was assayed in triplicate for V. harveyi CqsS agonist activity using the reporter strain JMH626 as described above. Fractions containing activity were further characterized (Supporting information).
Purification of CqsA
The V. cholerae CqsA enzyme was purified as previously described (Kelly et al., 2009). For purification of V. harveyi CqsA, the open reading frame encoding V. harveyi CqsA was amplified by PCR and cloned into plasmid pET28B that had been previously digested with NdeI and BamHI. The resulting plasmid was transformed into E. coli BL21 Gold (DE3) resulting in strain WN1666. Strain WN1666 was grown in LB with kanamycin at 30°C with shaking until the OD600 of the culture reached ∼1.0. IPTG was added at a final concentration of 100 µM, and the culture was incubated for an additional 4 h at 30°C with shaking. Cells were harvested by centrifugation, suspended in lysis buffer (20 mM HEPES pH 8.0, 0.2 M NaCl, 5 mM β-mercaptoethanol, 100 µM pyridoxal-5′-phosphate, 1 mM MgCl2 and 20 mM imidazole), and lysed using a Cell Cracker. Soluble materials were loaded onto a HiTrap chelating column charged with nickel, the column was washed extensively with lysis buffer, and His6-tagged V. harveyi CqsA was eluted using a linear gradient of increasing concentration of imidazole dissolved in lysis buffer. Fractions containing CqsA were pooled, the buffer was exchanged, and protein was stored at −20°C in storage buffer [20 mM HEPES pH 8.0, 0.2 M NaCl, 1 mM DTT, 50% (v/v) glycerol]. Protein concentrations were determined by Bio-Rad Protein Assay Reagent.
Coupled-enzyme assay for CqsA enzyme kinetics
A coupled-enzyme assay (modified from Hunter and Ferreira, 1995; Webster et al., 2000) was used to measure the rate of CoA production by CqsA in the presence of SAM and different acyl CoA substrates. Released CoA is reacted with α-ketoglutarate and NAD+ to form succinyl CoA, NADH and CO2 when excess α-ketoglutarate dehydrogenase is supplied. The rate of NADH production is followed at 340 nm using a spectrophotometer. Briefly, the rates of CoA release from V. cholerae and V. harveyi CqsA enzymes were measured in reactions containing 20 mM potassium phosphate buffer pH 7.0, 3 mM MgCl2, 1 mM NAD+, 1 mM α-ketoglutarate, 200 µM SAM, 1 unit of α-ketoglutarate dehydrogenase (Sigma) and 60–300 nM CqsA. Decanoyl CoA or octanoyl CoA was added to the reactions at concentrations of 1–200 µM. The rate of CoA production was monitored for 30 min. Data were fitted using Graphpad Prism to obtain the kinetic parameters.
Measurement of the concentration of CAI-1 type molecules in V. harveyi, V. cholerae and other Vibrio species culture fluids
Bacteria were grown in LM medium at 30°C overnight with shaking. The cultures were diluted 200-fold in M9 minimal salts medium (Sigma) supplemented with 2 mM MgCl2, 0.1 mM CaCl2, 1× MEM vitamins (Sigma), 0.3 M NaCl, 0.5% (w/v) glucose and incubated at 30°C with shaking. Cell growth was monitored at OD600. When OD600 reached ∼1.0, cells were removed by centrifugation. Pure d2-CAI-1 was added to 9 ml of collected cell-free fluids at 500 nM as an internal control. The mixtures were extracted into 500 µl of DCM. The organic layer was separated and 100 µl of this extract was diluted into 100 µl of HRMS solvent (9:1 acetonitrile : H2O with 0.1% formic acid), and this sample was injected directly for HRMS analysis. Due to variability in the aqueous/organic partitioning during extraction and the different ionization potentials of the CAI-1 type molecules, we determined a correction factor for each molecule by preparing a mixed sample containing known concentrations of each of the CAI-1 type molecules together with an internal standard (d2-CAI-1) in M9 medium. This sample was treated in an identical manner to the cell-free fluids described above.
Chemical synthesis and analytical methods
All chemical syntheses and analytical methods are provided in Supporting information.
Acknowledgments
We thank members of the Bassler laboratory for insightful discussions and suggestions. We thank Neal Byrne for assistance with the chemical separations. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health (NIH) Grant 5R01GM065859, NIH Grant 5R01AI054442, National Science Foundation (NSF) Grant MCB-0343821 to B.L.B., and an NIH postdoctoral fellowship GM082061 to W.-L.N.
Supplementary Material
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Bassler BL, Wright M, Showalter RE, Silverman MR. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M, Schoolnik GK. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J Bacteriol. 2008;190:7232–7240. doi: 10.1128/JB.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao JG, Meighen EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol Plant Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib Microbiol. 2009;16:18–32. doi: 10.1159/000219371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Gray KM, Garey JR. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology. 2001;147:2379–2387. doi: 10.1099/00221287-147-8-2379. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J Bacteriol. 2009;191:169–177. doi: 10.1128/JB.01307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- Hunter GA, Ferreira GC. A continuous spectrophotometric assay for 5-aminolevulinate synthase that utilizes substrate cycling. Anal Biochem. 1995;226:221–224. doi: 10.1006/abio.1995.1217. [DOI] [PubMed] [Google Scholar]
- Jahan N, Potter JA, Sheikh MA, Botting CH, Shirran SL, Westwood NJ, Taylor GL. Insights into the biosynthesis of the Vibrio cholerae major autoinducer CAI-1 from the crystal structure of the PLP-dependent enzyme CqsA. J Mol Biol. 2009;392:763–773. doi: 10.1016/j.jmb.2009.07.042. [DOI] [PubMed] [Google Scholar]
- Kelly RC, Bolitho ME, Higgins DA, Lu W, Ng WL, Jeffrey PD, et al. The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA. Nat Chem Biol. 2009;5:891–895. doi: 10.1038/nchembio.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G, Skorupski K. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol Microbiol. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- Lerat E, Moran NA. The evolutionary history of quorum-sensing systems in bacteria. Mol Biol Evol. 2004;21:903–913. doi: 10.1093/molbev/msh097. [DOI] [PubMed] [Google Scholar]
- McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, et al. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143(Part 12):3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Wei Y, Perez LJ, Cong J, Long T, Koch M, et al. Probing bacterial transmembrane histidine kinase receptor–ligand interactions with natural and synthetic molecules. Proc Natl Acad Sci USA. 2010;107:5575–5580. doi: 10.1073/pnas.1001392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- Pappas KM, Weingart CL, Winans SC. Chemical communication in proteobacteria: biochemical and structural studies of signal synthases and receptors required for intercellular signalling. Mol Microbiol. 2004;53:755–769. doi: 10.1111/j.1365-2958.2004.04212.x. [DOI] [PubMed] [Google Scholar]
- Pestova EV, Havarstein LS, Morrison DA. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- Pompeani AJ, Irgon JJ, Berger MF, Bulyk ML, Wingreen NS, Bassler BL. The Vibrio harveyi master quorum-sensing regulator, LuxR, a TetR-type protein is both an activator and a repressor: DNA recognition and binding specificity at target promoters. Mol Microbiol. 2008;70:76–88. doi: 10.1111/j.1365-2958.2008.06389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Showalter RE, Martin MO, Silverman MR. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J Bacteriol. 1990;172:2946–2954. doi: 10.1128/jb.172.6.2946-2954.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- Spirig T, Tiaden A, Kiefer P, Buchrieser C, Vorholt JA, Hilbi H. The Legionella autoinducer synthase LqsA produces an alpha-hydroxyketone signaling molecule. J Biol Chem. 2008;283:18113–18123. doi: 10.1074/jbc.M801929200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoendel M, Horswill AR. Biosynthesis of peptide signals in gram-positive bacteria. Adv Appl Microbiol. 2010;71:91–112. doi: 10.1016/S0065-2164(10)71004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Weber SS, Bruggemann H, Bosshard R, Buchrieser C, Hilbi H. The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol. 2007;9:2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Carranza P, Bruggemann H, Riedel K, Eberl L, et al. Synergistic contribution of the Legionella pneumophila lqs genes to pathogen–host interactions. J Bacteriol. 2008;190:7532–7547. doi: 10.1128/JB.01002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Hilbi H. Bacterial gene regulation by alpha-hydroxyketone signaling. Trends Microbiol. 2010a;18:288–297. doi: 10.1016/j.tim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Sahr T, Walti MA, Boucke K, Buchrieser C, Hilbi H. The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ Microbiol. 2010b;12:1243–1259. doi: 10.1111/j.1462-2920.2010.02167.x. [DOI] [PubMed] [Google Scholar]
- Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res. 2009;37:2747–2756. doi: 10.1093/nar/gkp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 2006;20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster SP, Alexeev D, Campopiano DJ, Watt RM, Alexeeva M, Sawyer L, Baxter RL. Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies. Biochemistry. 2000;39:516–528. doi: 10.1021/bi991620j. [DOI] [PubMed] [Google Scholar]
- Wei Y, Perez LJ, Ng WL, Semmelhack MF, Bassler BL. Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem Biol. 2011 doi: 10.1021/cb1003652. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6:191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


