Abstract
Toll-like receptor (TLR) signaling is linked to autophagy that facilitates elimination of intracellular pathogens. However, it is largely unknown whether autophagy controls TLR signaling. Here, we report that poly(I:C) stimulation induces selective autophagic degradation of the TLR adaptor molecule TRIF and the signaling molecule TRAF6, which is revealed by gene silencing of the ubiquitin-editing enzyme A20. This type of autophagy induced formation of autophagosomes and could be suppressed by an autophagy inhibitor and lysosomal inhibitors. However, this autophagy was not associated with canonical autophagic processes, including involvement of Beclin-1 and conversion of LC3-I to LC3-II. Through screening of TRIF-interacting ‘autophagy receptors’ in human cells, we identified that NDP52 mediated the selective autophagic degradation of TRIF and TRAF6 but not TRAF3. NDP52 was polyubiquitinated by TRAF6 and was involved in aggregation of TRAF6, which may result in the selective degradation. Intriguingly, only under the condition of A20 silencing, NDP52 could effectively suppress poly(I:C)-induced proinflammatory gene expression. Thus, this study clarifies a selective autophagic mechanism mediated by NDP52 that works downstream of TRIF–TRAF6. Furthermore, although A20 is known as a signaling fine-tuner to prevent excess TLR signaling, it paradoxically downregulates the fine-tuning effect of NDP52 on TLR signaling.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0819-y) contains supplementary material, which is available to authorized users.
Keywords: Autophagy, A20, NDP52, Signal transduction, Toll-like receptor (TLR)
Introduction
Toll-like receptors (TLRs) play a critical role in triggering of various innate immune responses associated with both physiological and pathological immunity. After recognition of pathogen-associated molecular patterns or danger-associated endogenous molecules, TLRs activate signaling through recruitment of two key adaptor molecules, termed myeloid differentiation factor 88 (MyD88) and Toll/interleukin (IL)-1 receptor homology domain-containing adaptor inducing interferon (IFN)-β (TRIF; also called TICAM-1) [1, 2]. MyD88 is a universal signaling adaptor for all TLR members except for TLR3, whereas TRIF is a unique signaling adaptor only for TLR3 and TLR4.
It is currently well understood that TLR signal transduction is largely dependent on functioning of specific E3 ubiquitin ligases. After TLR engagement, MyD88 facilitates recruitment of IRAK-4 followed by IRAK-1/2, forming the ‘Myddosome’ signaling complex [3]. This complex then interacts with the E3 ubiquitin ligase TRAF6 that functions as a signaling scaffold to catalyse Lys63-linked polyubiquitination (polyUb) of target proteins, including IRAK-1 [4], or synthesis of free Lys63-linked polyubiquitin chains [5]. These ubiquitin chains bind to TAB2 and TAB3 to activate the TAK1 complex and bind to NEMO to activate the IKK complex, both of which are crucial for NF-κB activation [1, 5, 6]. TRIF is able to directly activate TRAF6-dependent NF-κB signaling [1, 7]. In addition, TRIF activates the E3 ubiquitin ligase TRAF3 that catalyses Lys63-linked polyUb of TBK1 and IKKi/IKKε, which mediates phosphorylation of IRF3 and its nuclear translocation to produce type I IFNs [1, 7–9]. Although TRAF3 is also incorporated into the MyD88 complex [8], MyD88 activates the E3 ubiquitin ligases cIAP1/2 to facilitate TRAF3 Lys48-linked polyUb followed by proteasomal degradation, which do not work for TRIF-mediated signaling [10]. TRIF also recruits RIP1 to the C-terminal RIP homotypic interaction motif [11]. RIP1 undergoes Lys63-linked polyUb by the E3 ubiquitin ligase Peli1 to activate NF-κB signaling [12].
Excessive activation of TLR-mediated responses accumulates pathological damages, ultimately leading to the development of inflammatory disorders [13]. To obviate this, cells normally employ regulatory mechanisms for ‘fine-tuning’ of TLR signaling. The ubiquitin-editing enzyme A20 (also known as TNFAIP3) cleaves Lys63-linked polyubiquitin chains and promotes Lys48-linked polyUb [14], leading to negative regulation of TLR-mediated activation of NF-κB and/or IRF3 through regulation of its substrates, including TRAF6, TRAF3 and RIP1 [15, 16]. In addition, the E3 ubiquitin ligase Cbl-b degrades Syk-phosphorylated MyD88 and TRIF through catalysing Lys48-linked polyUb [17]. Lys48-linked polyUb is known to target substrates for proteasomal degradation [18, 19], while Lys63-linked polyUb is involved in the selective removal of target proteins by autophagy [20, 21].
Recent findings have revealed that TLRs utilize the mechanism of autophagy to eliminate intracellular pathogens [22–24]. The term autophagy is broadly used to designate the lysosomal delivery and degradation of intracellular components. The best-known form of autophagy is macroautophagy, a process of bulk lysosomal self-digestion in response to nutrient starvation and other metabolic stresses, induction of which consists of the processes of autophagosome vesicle nucleation by Beclin1 in complex with the class III phosphatidylinositol 3 (PI3) kinase VPS34, and then vesicle elongation by the covalent conjugation of the ATG proteins and LC3 to form the autophagosome [25, 26]. However, recent reports have revealed the existence of various types of autophagy that can be induced through both canonical and non-canonical processes [27–29]. Furthermore, during the process called ‘selective autophagy’, cytoplasmic constituents with Lys63-linked polyUb, including misfolded proteins, damaged organelles and intracellular pathogens, are clustered within the autophagosome by autophagy-associated ubiquitin receptors (hereafter autophagy receptors), such as sequestosome 1 (SQSTM1; also known as p62), followed by delivery to the lysosome to selectively degrade unnecessary proteins or organelles [21, 30].
In this study, we report that gene silencing of A20 revealed an unknown mechanism of selective autophagic degradation of TRIF, the process of which is promoted by the autophagy receptor nuclear dot protein (NDP) 52. NDP52 has recently been identified as an autophagy receptor recognizing ubiquitin-coated cytosolic bacteria [31]. Interestingly, under the condition of A20 silencing, NDP52 could exert an effective downregulatory effect on expression of TLR-induced proinflammatory genes. Thus, this study provides novel information regarding the selective autophagy downstream of TLRs, which has a ‘fine-tuning’ effect on TLR signaling but is normally inactivated by the fine-tuner A20.
Materials and methods
Reagents and antibodies
Synthetic poly(I:C) (polyriboinosinic polyribocytidylic acid) was obtained from Amersham. Highly purified Escherichia coli lipopolysaccharide (LPS) was as described previously [32]. Z-VAD-fmk was obtained from Zymed Laboratories. The class III PI3 kinase inhibitor 3-methyladenine (3-MA), the lysosomal thiol protease inhibitor E-64, the aspartic protease inhibitor pepstatin A, the cysteine protease leupeptin, and etoposide were purchased from Sigma-Aldrich. MG-132 was obtained from Calbiochem. The vacuolar proton-ATPase inhibitor bafilomycin A1 was obtained from Wako Chemical. Lithium dodecyl sulfate (LDS) sample buffer was obtained from Invitrogen. Sources of antibodies used are: anti-TRIF rabbit polyclonal antibody (Alexis; AL227), anti-BAG3 rabbit polyclonal antibody (Abcam; ab86298), anti-BNIP3L (NIX) rabbit polyclonal antibody (Abcam; ab8399), anti-NDP52 (CALCOCO2) rabbit polyclonal antibody (Abcam; ab68588), anti-Flag M2 monoclonal antibody (Sigma-Aldrich; F3165), anti-LC-3 monoclonal antibody (MBL; M115-2), anti-Lys63-linked polyUb rabbit monoclonal antibody Apu3 (Millipore; 05-1308), anti-Lys48-linked polyUb rabbit monoclonal antibody Apu2 (Millipore; 05-1307), FK2 anti-polyUb antibody (Nippon Biotest; MFK-004), anti-NBR1 rabbit polyclonal antibody (Cell Signaling Technology; 5202), anti-GFP rabbit monoclonal antibody (Cell Signaling Technology; 2956) and anti-Rab7 rabbit monoclonal antibody (Cell Signaling Technology; 9367). In addition, rabbit polyclonal antibodies to TRAF6 (sc-7221), TRAF3 (sc-948), RIP1 (sc-7881), SQSTM1 (sc-25575), HDAC6 (sc-11420), Beclin-1 (sc-11427), A20 (sc-22834), HA (sc-805), GAPDH (sc-25778) and β-actin (SC-8432) were obtained from Santa Cruz Biotechnology.
Cell culture
HeLa cells and human embryonic kidney (HEK) 293T cells were maintained at 37°C in a humidified atmosphere of 5% CO2 in DMEM supplemented with 10% FBS, penicillin G (100 units/ml), and streptomycin (100 μg/ml). HEK293 cells stably transfected with human TLR4, MD2 and CD14 genes (HEK293-TLR4/MD2-CD14 cells) were obtained from Invivogen and grown in DMEM supplemented with blasticidin (10 μg/ml) and hygromycin B (50 μg/ml). Cells were stimulated with LPS in antibiotic-free DMEM supplemented with 5% FBS. Human bone marrow-derived primary mononuclear cells were obtained from Lonza (2M-125C). These cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 15% FBS, penicillin G (100 units/ml) and streptomycin (100 μg/ml) for 2 h before transfection of siRNA.
DNA constructs
Constructs encoding N-terminally Flag-tagged TRIF (Flag-TRIF) and N-terminally hemagglutinin (HA)-tagged NDP52 (HA-NDP52) were generated by amplifying cDNA of THP-1 cells and by subcloning them into pcDNA3.1/V5-His (Invitrogen). SKICH domain-deleted HA-NDP52 (HA-NDP52 ΔSKICH), LIM-like domain-deleted HA-NDP52 (HA-NDP52 ΔLIM-L), and both SKICH and LIM-like domain-deleted HA-NDP52 (HA-NDP52 CC) were generated using a QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Expression plasmids for N-terminally Flag-tagged MyD88 and N-terminally HA-tagged SQSTM1 were as described previously [33]. Expression plasmids of N-terminally Flag-tagged TRAF6 and C-terminally Flag-tagged TRAF3 were kind gifts from Dr. H. Nakano (Juntendo University, Tokyo, Japan) and Dr. M. Matsumoto (Hokkaido University, Sapporo, Japan), respectively. The expression plasmid of GFP-tagged LC3 [30] was kindly provided by Prof. T. Johansen (University of Tromsö, Tromsö, Norway). Transfection of plasmids into HeLa cells and HEK293T cells was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
siRNA and gene silencing
ON-TARGET plus SMARTpool small interference RNA (siRNA) against human NDP52 (CALCOCO2, L-010637-00-0005), human A20 (TNFAIP3, L-009919-00-0005) and human Beclin-1 (BECN1, L-010552-00-0005) were purchased from Dharmacon as well as the control ON-TARGET plus non-targeting siRNA pool (D-001810-01). ON-TARGET plus SMARTpool siRNA consist of four distinct RNA oligoduplexes per target gene. The ON-TARGET plus non-targeting siRNA pool also consists of four distinct control RNA oligoduplexes. For transfection of siRNA, HeLa cells and HEK293T cells were washed once with Opti-MEM I medium (Invitrogen), and then transfection of siRNA (100 nM) was performed with Lipofectamine RNAi MAX reagent (Invitrogen) as instructed by the manufacturer (1 μl per 20 pmol of siRNA being used). After 12 h of incubation, culture media were changed to DMEM supplemented with 5% FBS, and incubation was continued for a further 12 h. Human bone marrow-derived primary mononuclear cells (4 × 106) were transfected with siRNA (30 nM) using the Nucleofector 2b electroporator (Amaxa) and the accessory program Y-001. Then, the cells were maintained in IMDM supplemented with 50 ng/ml M-CSF, 15% FBS, penicillin G and streptomycin for 48 h. For prolonged gene silencing, HeLa cells seeded on 6-well plates were transfected with siRNAs using Lipofectamine 2000 reagent. After 12 h of incubation, culture media were changed to DMEM supplemented with 10% FBS, and incubation was continued for a further 24 h. Then the cells were split 1–2 into another 6-well plate and cultured for 12 h. These procedures were further repeated twice. After 144 h (6 days), cells were used for the experiments.
RNA isolation and qRT-PCR
Total RNA was prepared from cells using a GenElute mammalian total RNA miniprep kit (Sigma-Aldrich). One μg of total RNA was reverse-transcribed using ReverTraAce reverse transcriptase (TOYOBO) with both an oligo21dT primer and random hexamer primers. qRT-PCR was performed using SYBR Premix Ex Taq (TaKaRa) on a thermal cycler dice real-time system TP800 (TaKaRa), according to the manufacturer’s instructions. All of the primer sets used in this study were obtained from TaKaRa. We confirmed that there was no critical difference between the values normalized to the levels of each of four different house-keeping genes, ACTB, GAPDH, HPRT1 and PPIA. Results shown were normalized to the level of ACTB and are representative of three independent experiments.
Immunoprecipitation
For immunoprecipitation (IP) of Flag-tagged proteins, HEK293T cells seeded on 10-cm culture dishes or 6-well plates were lysed with 880 or 250 μl of the lysis buffer [33] at 4°C for 15 min. After clarification by centrifugation at 15,000g for 10 min, cell lysates were immunoprecipitated using 75 or 25 μl anti-Flag M2 agarose (Sigma-Aldrich) for 1 h at 4°C on a rotating platform. For IP of HA epitope tag, HEK293T cells seeded on 6-well plates were lysed with 250 μl of lysis buffer. After clarification, cell lysates were immunoprecipitated using 25 μl EZview Red Anti-HA Affinity Gel (Sigma-Aldrich). The beads were washed four times with 1 ml lysis buffer, boiled with SDS sample buffer containing 2-mercaptoethanol, and subjected to immunoblotting (IB) using the indicated antibodies. Results are representative of three independent experiments.
Immunoblotting
HeLa cells and HEK293T cells seeded on 6-well plates were lysed with 300 μl of the lysis buffer [33] at 4°C for 15 min. Lysates were boiled with SDS sample buffer. In some experiments, HEK293T cells seeded on 6-well plates were lysed with 300 μl of LDS Sample Buffer at 4°C for 15 min, and then the lysates were boiled for 5 min. Lysates were separated on 10–20% gradient SDS-PAGE and transferred to Immobilon-P transfer membranes (Millipore). The membranes were blocked in 5% skim milk in PBS. Immunoreactive bands were detected using the antibodies described above. The ECL Plus Western Blotting detection system (Amersham Bioscience) was used to visualize the blots on an ECL minicamera (Amersham Bioscences) with the instant black and white film FP-3000B (Fuji Films). Results are representative of three independent experiments. Densitometric analyses were performed as described previously [33].
Luciferase reporter gene assay
HEK293T cells seeded on 24-well plates were transfected with either 50 ng of an NF-κB-driven firefly luciferase reporter plasmid (pNF-κB-Luc; Stratagene) or an IRF3-driven firefly luciferase reporter plasmid (p561-luc, kindly provided by Prof. X. Li, Cleveland Clinic Foundation, Ohio, USA) together with 5 ng of a construct directing expression of Renilla luciferase under the control of a constitutively active thymidine kinase promoter (pRL-TK; Promega) and different expression vectors for 24 h. HEK293-TLR4/MD2-CD14 cells seeded on 24-well plates were transfected with either pNF-κB-Luc or p561-luc together with pRL-TK for 18 h and were then treated with 100 ng/ml LPS for 6 h. The cells were lysed and luciferase activity was measured as described previously [32].
Immunofluorescent microscopy and image analysis
For immunofluorescence microscopy, cells were seeded on Lab-Tek chamber 8-well permanox slides (Nunc) and fixed at −20°C with methanol for 20 min. Double-immunostaining was then carried out using the primary antibody and the secondary Alexa488-conjugated antibody (Invitrogen) and then with the tertiary antibody and the fourth Alexa564-conjugated antibody (Invitrogen). Stained cells were embedded in Mowiol 4-88 (Calbiochem) in the presence of the Prolong Gold Antifade reagent (Invitrogen). Fluorescent images were obtained as described previously [33]. Cell quantification was carried out by counting cells on five merged images of microscopic fields including at least 30 TRAF6-positive cells scored. Results are representative of three separate experiments and expressed as the mean ± SD (n = 5).
Electron microscopic analysis
For electron microscopy, human bone marrow-derived primary mononuclear cells or HeLa cells were fixed with 2% glutaraldehyde in a 0.1-M phosphate buffer (pH 7.4) at 4°C. They were post-fixed with an aqueous solution of 2% OsO4. Then, the specimens were dehydrated in a graded ethanol and embedded in Epon 812. Ultrathin sections were stained with uranyl acetate and lead citrate for observation under a JEM1200EX electron microscope (Jeol). The area of every autophagic vacuole (including autophagosome) and total cytoplasmic area were calculated on the enlarged photographs with the use of a planimeter (Planix). For each cell, the autophagic area was calculated by expressing the total area of autophagic vacuoles as a percentage of the cytoplasmic area.
Statistical analysis
Data are expressed as mean ± SD (n = 3). P values were calculated by Student’s t test and one-way analysis of variance and were considered significant at levels of <0.05 or <0.01.
Results
Gene silencing of A20 reveals a mechanism of degradation of TRIF and TRAF6
Human epithelial HeLa cells are generally thought to hyporespond to stimuli of TLRs because of low expression levels of TLRs [34, 35]. HeLa cells could weakly respond to the TLR3-stimulant ligand poly(I:C) to induce mRNA expression of IL-6 and IL-8 (Fig. 1a). Expression of A20 mRNA was also increased after poly(I:C) stimulation (Fig. 1a). Given that A20 is a potent suppressor of TLR signaling, we examined whether A20 has regulatory effects on poly(I:C)-induced responses. Silencing of A20 by short-term transfection of siRNA revealed that the responsiveness of HeLa cells to poly(I:C) was restricted by A20 (Fig. 1a). The level of TRIF protein was found to be decreased after poly(I:C) stimulation under the condition of A20 silencing (Fig. 1b) although TRIF mRNA was unchanged (Fig. 1a). In addition, among three major signaling mediators downstream of TRIF (TRAF6, TRAF3 and RIP1), only TRAF6 was clearly decreased after poly(I:C) stimulation (Fig. 1b). At least expression level of GAPDH protein was not decreased (Fig. 1b). These observations were also found in HeLa cells under the condition of long-term silencing of A20 (Fig. 1c). Also, the poly(I:C)-induced decrease of TRIF and TRAF6 was observed in human bone marrow-derived primary mononuclear cells transfected with A20 siRNA (Fig. 1d).
Fig. 1.
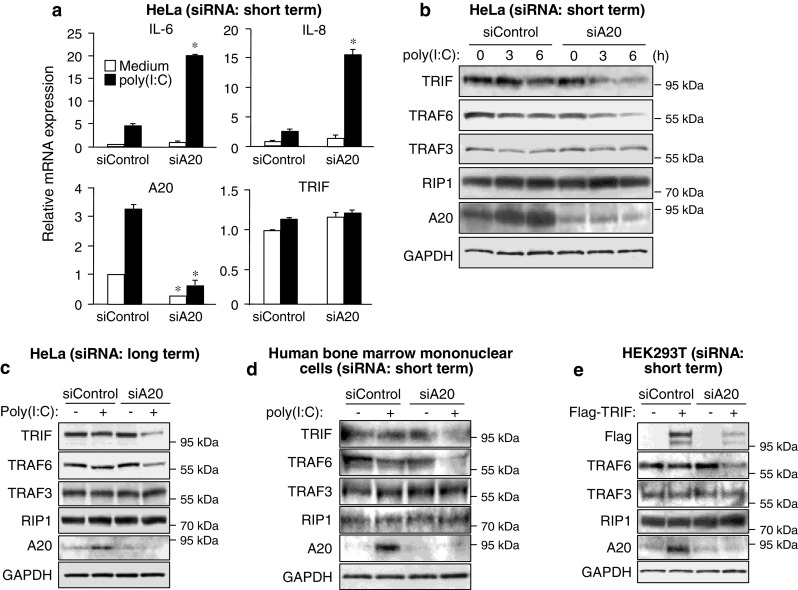
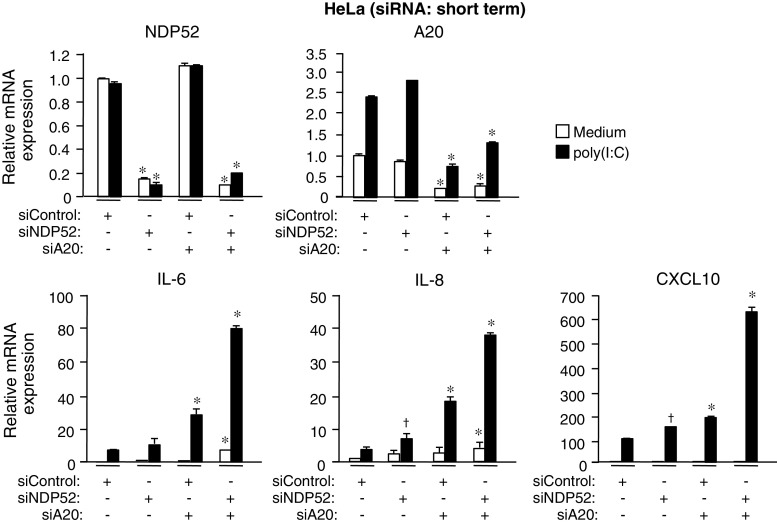
A20 conceals the mechanism of degradation of TRIF and TRAF6 by autophagy. a HeLa cells were transfected with control siRNA or A20 siRNA for 24 h. Cells were then stimulated with 50 μg/ml poly (I:C) for 6 h. The mRNA levels of IL-6, IL-8, A20 and TRIF were determined by qRT-PCR. Data are expressed as the mean fold induction ±SD (n = 3) relative to control levels (Control siRNA, Medium), for representative of three independent experiments. *P < 0.01, for comparison with the Control siRNA group. b HeLa cells were transfected with control siRNA or A20 siRNA for 24 h. Cells were then stimulated with 50 μg/ml poly (I:C) for the indicated periods. Cell lysates were analyzed by IB with antibodies to TRIF, TRAF6, TRAF3, RIP1, A20 and GAPDH. c HeLa cells were repetitively transfected with control siRNA or A20 siRNA for three times during 6 days. Cells were then stimulated with 50 μg/ml poly (I:C) for 6 h. Cell lysates were analyzed by IB with antibodies to TRIF, TRAF6, TRAF3, RIP1, A20 and GAPDH. d Human bone marrow-derived primary mononuclear cells were transfected with control siRNA or A20 siRNA and incubated in medium supplemented with 50 ng/ml M-CSF for 48 h. Cells were then stimulated with 50 μg/ml poly(I:C) for 6 h. Cell lysates were analyzed by IB with antibodies to TRIF, TRAF6, TRAF3, RIP1, A20 and GAPDH. e HEK293T cells were transfected with control siRNA or A20 siRNA for 24 h. Cells were further transfected with Flag-TRIF for 18 h. Cell lysates were then analyzed by IB with antibodies to Flag epitope, TRAF6, TRAF3, RIP1, A20 and GAPDH. All results are representative of three independent experiments
In contrast to endogenous TRIF, transfected TRIF automatically activates downstream signaling in a TLR stimulus-independent manner [36]. In HEK293T cells transfected with TRIF, the level of TRIF protein was found to be decreased by silencing of A20 (Fig. 1e). Accompanying this, endogenous TRAF6, but not TRAF3 and RIP1, was clearly decreased (Fig. 1e). These results indicate that A20 has a potential to conceal the selective decrease of TRIF and TRAF6 after TRIF-dependent signal transduction.
TRIF and TRAF6 are degraded by a machinery of non-canonical autophagy
We next investigated what machinery is involved in the selective decrease of TRIF and TRAF6. TRIF was previously reported to undergo proteolytic cleavage by caspases [37]. However, the poly(I:C)-induced decrease of TRIF and TRAF6 in A20-silenced HeLa cells was not affected by treatment with the pan-caspase inhibitor Z-VAD-fmk (Fig. 2a). Generally, eukaryotic cells utilize the ubiquitin-26S proteasome system and autophagy as the major protein degradation pathways. Importantly, treatment of A20-silenced HeLa cells with the autophagy inhibitor 3-MA, but not the 26S proteasome inhibitor MG-132, could inhibit the decrease of TRIF and TRAF6 (Fig. 2b). In HEK293T cells, 3-MA treatment obviously increased the level of overexpressed TRIF and endogenous TRAF6, whereas MG-132 conversely decreased these levels (Fig. 2c). As inhibition of the proteasome was reported to promote autophagy [38], activated TRIF and TRAF6 may be preferentially degraded by autophagy. Furthermore, the decrease of TRIF and TRAF6 in A20-silenced HeLa cells was also suppressed by the lysosomal protease inhibitors E-64, pepstatin A and leupeptin, and the lysosomal vacuolar H+-ATPase inhibitor bafilomycin A1 (Fig. 2d). We also investigated whether poly(I:C) stimulation induces formation of autophagic vacuoles in A20-silenced human bone marrow mononuclear cells. The autophagy-inducer etoposide clearly promoted formation of autophagic vacuoles in human bone marrow mononuclear cells (Supplementary material, Fig. S1). Some of autophagic vacuoles were double-membrane structures indicative of autophagosomes (Supplementary material, Fig. S1). In A20-silenced human bone marrow mononuclear cells, poly(I:C) increased formation of autophagic vacuoles (Fig. 2e). Poly(I:C)-induced formation of autophagic vacuoles were also observed in A20-silenced HeLa cells (data not shown). These results indicate that TRIF and TRAF6 are degraded within lysosomes through autophagic processes.
Fig. 2.
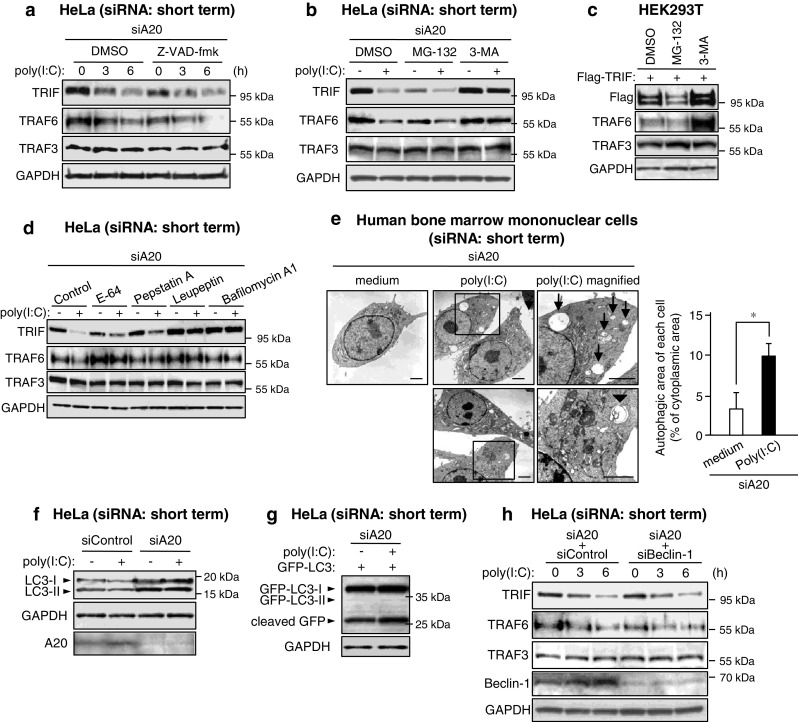
TRIF and TRAF6 are degraded through non-canonical autophagy. a HeLa cells were transfected with A20 siRNA for 24 h. Cells were pretreated with or without 25 μM Z-VAD-fmk for 1 h and then stimulated with 50 μg/ml poly (I:C) for the indicated periods. Cell lysates were analyzed by IB with antibodies to TRIF, TRAF6, TRAF3 and GAPDH. b HeLa cells were transfected with A20 siRNA for 24 h. Cells were pretreated with either 20 μM MG-132 or 10 mM 3-MA for 3 h and then stimulated with 50 μg/ml poly (I:C) for 6 h. Cell lysates were analyzed by IB with antibodies to TRIF, TRAF6, TRAF3 and GAPDH. c HEK293T cells were transfected with Flag-TRIF for 18 h. Cells were then treated with either 20 μM MG-132 or 10 mM 3-MA for 6 h. Cell lysates were analyzed by IB with antibodies to Flag epitope, TRAF6, TRAF3 and GAPDH. d HeLa cells were transfected with A20 siRNA for 24 h. Cells were pretreated with 30 μM E-64, 30 μM Pepstatin A, 30 μM Leupeptin or 100 nM Bafilomycin A1 for 4 h. Then cells were stimulated with 50 μg/ml poly (I:C) for 6 h. Cell lysates were analyzed by IB with antibodies to TRIF, TRAF6, TRAF3 and GAPDH. e Human bone marrow-derived primary mononuclear cells were transfected with A20 siRNA and maintained in medium supplemented with 50 ng/ml M-CSF for 48 h. Cells were stimulated with 50 μg/ml poly(I:C) for 6 h. Then the cells were assessed by electron microscopy. Arrows autophagic vacuoles. Arrowheads autophagosomes. The magnified photo represents the area indicated by the square. For each cell, the autophagic area was calculated by expressing the total area of autophagic vacuoles as a percentage of the cytoplasmic area. Scale bar 2 μm. Original magnification, ×3,000. Error bars ±SD (n = 5). *P ≤ 0.01. f HeLa cells were transfected with control siRNA or A20 siRNA for 24 h. Cells were then stimulated with 50 μg/ml poly (I:C) for 6 h. Cell lysates were analyzed by IB with antibodies to LC3, GAPDH and A20. g HeLa cells were cotransfected with A20 siRNA and GFP-LC3 for 24 h. Cells were then stimulated with 50 μg/ml poly (I:C) for 6 h. Cell lysates were analyzed by IB with antibodies to GFP and GAPDH. h HeLa cells were transfected with A20 siRNA together with control siRNA or Beclin-1 siRNA for 24 h. Cells were then stimulated with 50 μg/ml poly (I:C) for the indicated periods. Cell lysates were analyzed by IB with antibodies to TRIF, TRAF6, TRAF3, Beclin-1 and GAPDH
We further investigated the involvement of canonical autophagic processes in the decrease of TRIF and TRAF6. Generally, conversion of normal LC3 (LC3-I) to the phosphatidylethanolamine-conjugated form (LC3-II) is thought of as a hallmark response of canonical autophagy flux [39]. In HeLa cells, we could observe conversion of LC3-I to LC3-II after treatment of cells with bafilomycin A1 (Supplementary material, Fig. S2). However, poly(I:C) stimulation did not affect LC3 conversion at all (Fig. 2f). Furthermore, in A20-silenced HeLa cells, basal LC3 conversion was slightly promoted, but poly(I:C) did not promote the conversion (Fig. 2f). Also, poly(I:C) stimulation did not affect transfected GFP-tagged LC3 (GFP-LC3) in HeLa cells (Fig. 2g). We next tested involvement of general inducers of canonical autophagy. Silencing of Beclin-1 by siRNA did not affect the decrease of TRIF and TRAF6 after poly(I:C) stimulation in A20-silenced HeLa cells (Fig. 2h). Also, neither silencing of ATG5 nor of ATG7 affected these decreases (data not shown). Thus, our results collectively suggest that TRIF and TRAF6 are selectively degraded by non-canonical autophagy that is thought to be normally inactivated by A20.
Recruitment of autophagy receptors to TRIF
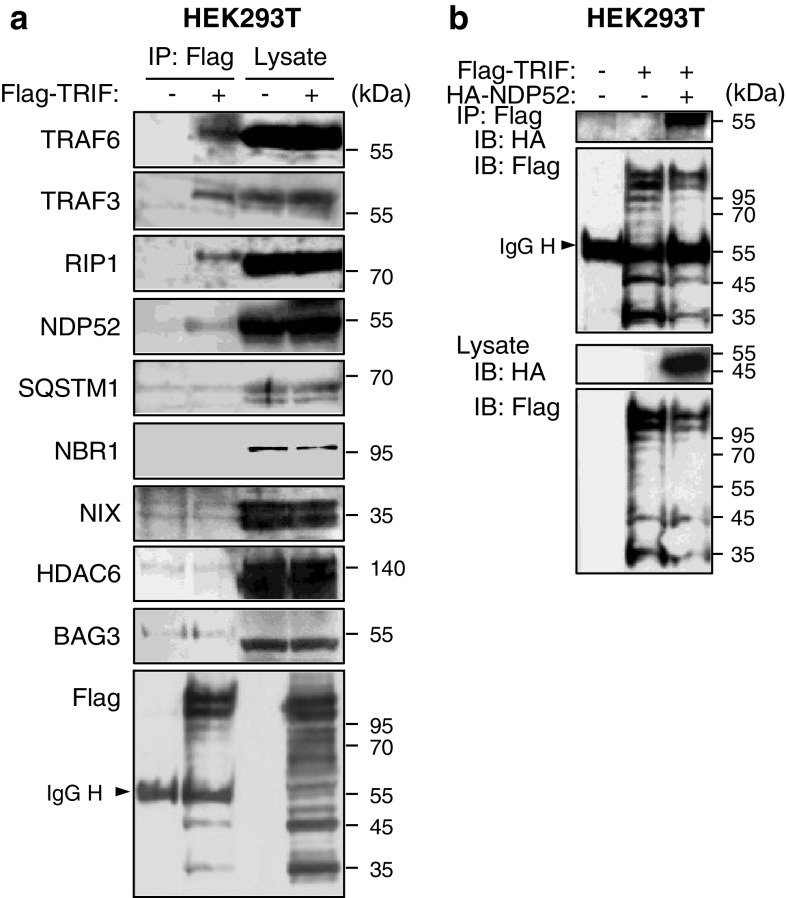
To our knowledge, any mechanisms for selective autophagic degradation of TRIF have not been clarified. However, given that selective autophagic degradation of proteins is recently suggested to be mediated by autophagy receptors [21], it is possible that the degradation of TRIF is mediated by specific autophagy receptors. To test whether or not TRIF interacts with autophagy receptors, we immunoprecipitated the Flag-tagged version of TRIF expressed in HEK293T cells followed by immunoblot analyses of endogenous co-immunoprecipitated molecules. In this experiment, TRAF6, TRAF3 and RIP1 could be detectable (Fig. 3a), indicating that immunoprecipitated TRIF forms signaling complexes. We therefore screened the established autophagy receptors, SQSTM1 [40–42], NBR1 [43], NIX/BNIP3L [44], HDAC6 [45], BAG3 [46] and NDP52 [31]. As a result, interestingly, only NDP52 was detectable (Fig. 3a). Moreover, in HEK293T cells, we could observe interaction of TRIF with NDP52 by cotransfection of differently tagged versions followed by IP and IB (Fig. 3b). Thus, activated TRIF recruits the autophagy receptor NDP52, which may mediate the selective degradation of activated TRIF.
Fig. 3.
Recruitment of autophagy receptors to TRIF. a HEK293T cells were transfected with Flag-TRIF or empty vector for 24 h. Then, IP with anti-Flag agarose was carried out with clarified cell lysates, followed by IB with indicated antibodies. b HEK293T cells were transfected with Flag-TRIF and HA-NDP52 for 24 h. IP with anti-Flag-agarose was carried out with clarified cell lysates, followed by IB with antibodies to HA epitope and Flag epitope. All results are representative of three independent experiments
NDP52 selectively degrades TRIF and TRAF6 through autophagy
We next investigated whether NDP52 is indeed involved in TRIF degradation after poly(I:C) stimulation in A20-silenced HeLa cells. Importantly, silencing of NDP52 by siRNA significantly restored poly(I:C)-induced degradation of TRIF (Fig. 4a). Furthermore, overexpression of TRIF with NDP52 in HEK293T cells revealed that NDP52 could degrade TRIF, and the degradation was restored by treatment with 3-MA (Fig. 4b).
Fig. 4.
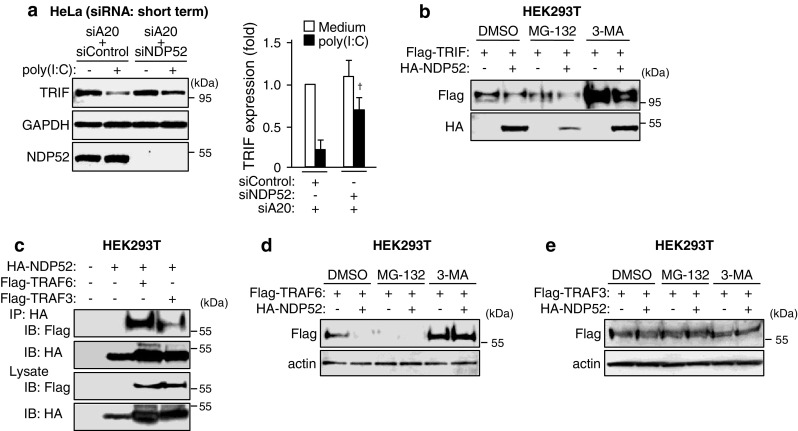
NDP52 degrades TRIF and TRAF6 through selective autophagy. a HeLa cells were transfected with A20 siRNA together with control siRNA or NDP52 siRNA for 24 h. Cells were then stimulated with 50 μg/ml poly (I:C) for 6 h. Cell lysates were analyzed by IB with antibodies to TRIF, GAPDH and NDP52. Densitometric quantification was performed on all of the immunoblot bands obtained by three independent experiments. Data are expressed as the mean fold increase ±SD relative to the control level (A20 siRNA + Control siRNA, Medium). † P < 0.05, for comparison with the value of ‘A20 siRNA + Control siRNA, poly (I:C)’. b HEK293T cells were transfected with Flag-TRIF and HA-NDP52 for 18 h. Cells were then treated with either 20 μM MG-132 or 10 mM 3-MA for 6 h. Cell lysates were analyzed by IB with antibodies to Flag epitope and HA epitope. c HEK293T cells were transfected with HA-NDP52 together with Flag-TRAF6 or Flag-TRAF3 for 24 h. IP with anti-HA affinity gel was carried out with clarified cell lysates, followed by IB with antibodies to Flag epitope and HA epitope. d HEK293T cells were transfected with Flag-TRAF6 and HA-NDP52 for 18 h. Cells were then treated with either 20 μM MG-132 or 10 mM 3-MA for 6 h. Cell lysates were analyzed by IB with antibodies to Flag epitope and β-actin. e HEK293T cells were transfected with Flag-TRAF3 and HA-NDP52 for 18 h. Cells were then treated with either 20 μM MG-132 or 10 mM 3-MA for 6 h. Cell lysates were analyzed by IB with antibodies to Flag epitope and β-actin. All results are representative of three independent experiments
There is some doubt about the degradation of TRIF because overexpressed or activated TRIF forms inclusion bodies [47]. Inclusion bodies are generally resistant to mild detergents, and it is therefore possible that the TRIF degradation was only attributable to formation of insoluble inclusions. We therefore prepared cell lysates with the strong detergent LDS, which is able to prevent generation of insoluble protein precipitants. The results with Triton X-100 and LDS were almost identical (Supplementary material, Fig. S3).
As shown in Fig. 1b, the TRAF family member TRAF6, but not TRAF3, is selectively degraded after poly(I:C) stimulation in A20-silenced HeLa cells. Unexpectedly, in HEK293T cells, we could observe interaction of NDP52 with both TRAF6 and TRAF3 by cotransfection of differently tagged versions followed by IP and IB (Fig. 4c). Overexpression of TRAF6 with NDP52 in HEK293T cells revealed that NDP52 could degrade TRAF6, and the degradation was restored by treatment with 3-MA (Fig. 4d). However, in contrast, NDP52 hardly affected TRAF3 (Fig. 4e). Thus, NDP52 indeed mediates selective degradation of TRIF and TRAF6.
NDP52 negatively regulates TLR-triggered transcriptional activities of NF-κB and IRF3
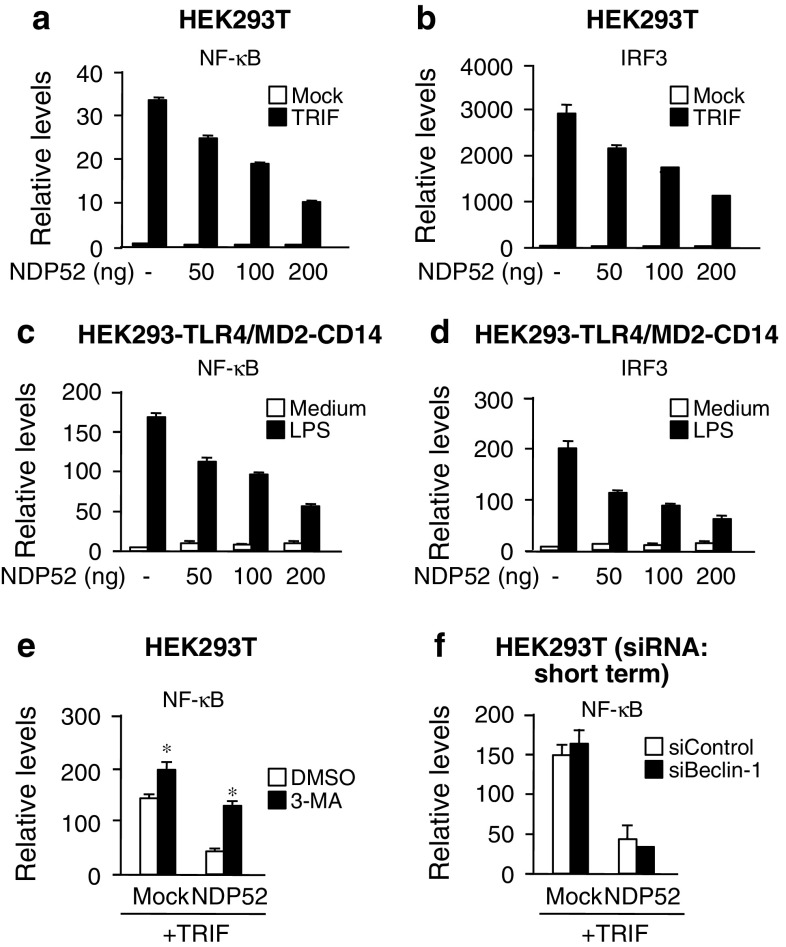
Degradation of signaling molecules is generally thought to be linked to downregulation of signal transduction. We therefore investigated whether NDP52 is able to negatively regulate TRIF-mediated signaling. In HEK293T cells, transfection of NDP52 could attenuate the TRIF overexpression-induced transcriptional activities of NF-κB and IRF3 (Fig. 5a, b). In addition, in HEK293 cells stably expressing TLR4/MD-2, transfection of NDP52 attenuated the LPS-induced activation of NF-κB and IRF3 (Fig. 5c, d). Moreover, treatment of cells with 3-MA potentiated TRIF-induced activation of NF-κB, and attenuated the negative regulatory effect of NDP52 (Fig. 5e). In addition, consistent with the result shown in Fig. 2h, silencing of Beclin-1 did not affect the negative regulatory effect of NDP52 (Fig. 5f). Thus, NDP52-mediated autophagic degradation has a potential to negatively regulate TRIF-mediated signaling.
Fig. 5.
NDP52 negatively regulates TRIF-triggered transcriptional activities of NF-κB and IRF3. a, b HEK293T cells were transfected with Flag-TRIF and pNF-κB-Luc (a) or p561-luc (b) together with indicated amounts of HA-NDP52 for 24 h. Then, luciferase activity was measured. Data are expressed as the mean ± SD (n = 3). c, d HEK293-TLR4/MD2-CD14 cells were transfected with pNF-κB-Luc (c) or p561-luc (d) together with indicated amounts of HA-NDP52 for 18 h. Cells were stimulated with 100 ng/ml E. coli LPS for 6 h. Then, luciferase activity was measured. Data are expressed as the mean ± SD (n = 3). e HEK293T cells were transfected with Flag-TRIF and pNF-κB-Luc together with HA-NDP52 or empty vector (Mock) for 18 h. Cells were treated with 10 mM 3-MA for 6 h. Then, luciferase activity was measured. Data are expressed as the mean ± SD (n = 3). *P < 0.01, for comparison with the value of DMSO. f HEK293T cells were transfected with control siRNA or Beclin-1 siRNA for 24 h. Cells were further transfected with Flag-TRIF, pNF-κB-Luc together with HA-NDP52 or empty vector (Mock) for 18 h. Then, luciferase activity was measured. Data are expressed as the mean ± SD (n = 3). All results are representative of three independent experiments
The C-terminal region of NDP52 is required for the activity of NDP52
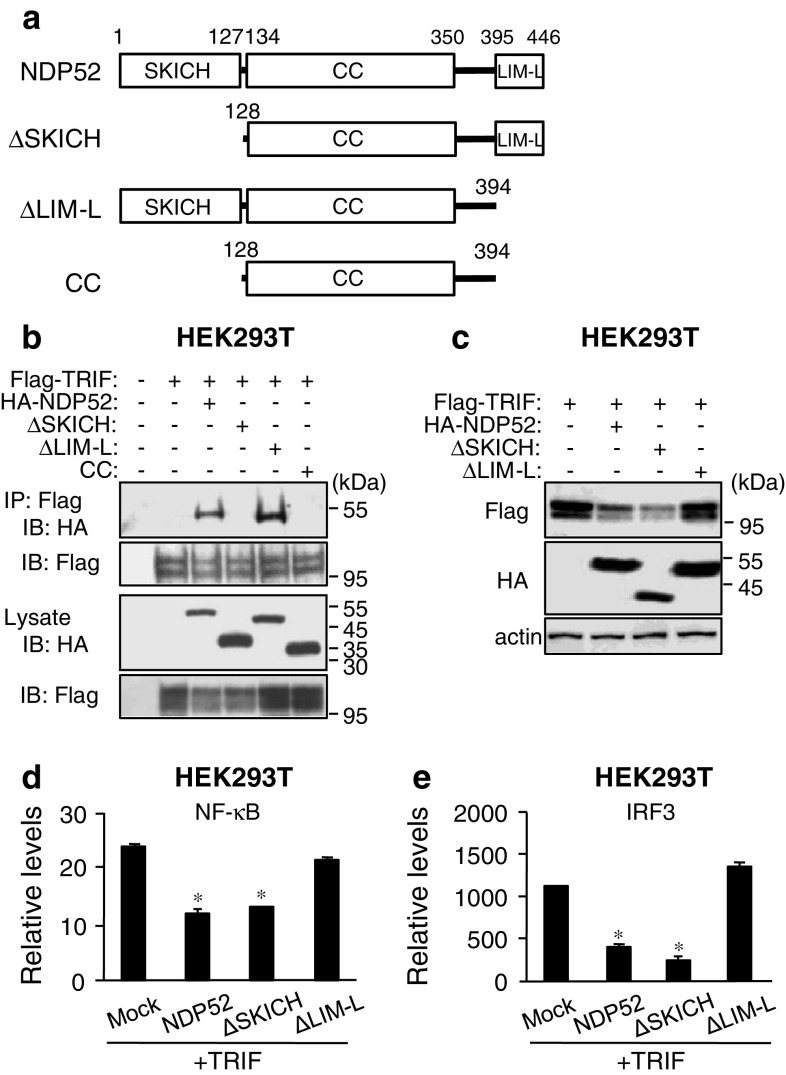
Since NDP52 is a recently identified autophagy receptor [31], its detailed function in autophagy is largely unknown. NDP52 is a mainly cytosolic protein and is ubiquitously expressed in various tissues and cells [48]. Human NDP52 was previously identified as a component of the nuclear promyelocytic leukemia body [49] and a myosin VI-interacting molecule [50]. Two vertebrate paralogs, COCOA/CALCOCO1 and TAX1BP1/CALCOCO3 (also known as T6BP), exist. Human NDP52 comprises an N-terminal skeletal muscle and kidney-enriched inositol phosphatase carboxyl homology (SKICH) domain (amino acids 1–127), an intermediate coiled-coil (CC) domain with a leucine zipper sequence (amino acids 134–350) and a C-terminal Lin11, Isl-1 and Mec-3 (LIM)-like domain (amino acids 395–446) that consists of two zinc fingers (Fig. 6a; Supplementary material, Fig. S4). All domains of NDP52 are conserved in humans, chimpanzees, bovines, dogs and chickens, whereas mouse Ndp52 has a highly mutated CC domain and completely lacks the LIM-like domain (Supplementary material, Fig. S4). Since it is suggested that the LIM-like domain of human NDP52 specifically binds to ubiquitin chains [31], the functions of NDP52 may be considerably different in humans and mice. We investigated which domain is required for the interaction and degradation. Mapping of NDP52 revealed that the SKICH domain was required for binding to TRIF (Fig. 6b). In contrast, the LIM-like domain was required for the degradation of TRIF (Fig. 6c). These results suggest that NDP52 selectively interacts with activated TRIF through the SKICH domain and subsequently recognizes their polyUb through the LIM-like domain, followed by induction of selective degradation. Although NDP52 lacking the SKICH domain (ΔSKICH) still had a degradative effect on TRIF (Fig. 6c), in which the LIM-like domain might be responsible for autophagic degradation, we could not determine the reason. Importantly, transfection of deletion mutants of NDP52 revealed that the LIM-like domain was essential for the negative regulatory effect of NDP52 on TRIF signaling (Fig. 6d, e), suggesting that the LIM-like domain-dependent protein degradation results in suppression of TRIF-dependent signaling.
Fig. 6.
The C-terminal region of NDP52 is required for degradation of TRIF. a Schematic diagram of human NDP52 and its domain-deleted mutants. b HEK293T cells were transfected with Flag-TRIF together with HA-NDP52 or HA-tagged NDP52 mutant for 24 h. IP with anti-Flag agarose was carried out with clarified cell lysates, followed by IB with antibodies to HA epitope and Flag epitope. c HEK293T cells were transfected with Flag-TRIF together with HA-NDP52 or HA-tagged NDP52 mutant for 24 h. Cell lysates were analyzed by IB with antibodies to Flag epitope, HA epitope and β-actin. d, e HEK293T cells were transfected with Flag-TRIF and pNF-κB-Luc (d) or p561-luc (e) together with HA-NDP52, HA-tagged NDP52 mutant or empty vector (Mock) for 24 h. Then luciferase activity was measured. Data are expressed as the mean ± SD (n = 3). *P < 0.01, for comparison with the value of Mock. All results are representative of three independent experiments
NDP52 interacts with and degrades MyD88
We next examined whether or not the effect of NDP52 is specific to the TRIF–TRAF6-dependent pathway because TRAF6 is also involved in the MyD88-dependent pathway. In HEK293T cells, we found that transfection of NDP52 could attenuate the MyD88 overexpression-induced transcriptional activities of NF-κB (Supplementary material, Fig. S5a). Also, in a way similar to TRIF, NDP52 could degrade overexpressed MyD88 (Supplementary material, Fig. S5b), and the degradation was restored by treatment with 3-MA (Supplementary material, Fig. S5c). We also observed interaction of MyD88 with NDP52 (Supplementary material, Fig. S5d). Also, the SKICH domain of NDP52 was required for binding to MyD88 (Supplementary material, Fig. S5e), and the LIM-like domain was required for degradation of MyD88 (data not shown). These results strongly suggest that the effect of NDP52 is exerted not only on activated TRIF but also on activated MyD88. Possibly, recruitment of NDP52 to activated TRIF and MyD88 may be mediated by TRAF6. Indeed, interaction of NDP52 with TRAF6 was observed by cotransfection of differently tagged versions followed by IP and IB (Fig. 4c). This interaction was mediated by the SKICH domain of NDP52 (data not shown). Thus, NDP52 is suggested to be recruited to TRAF6 after activation of the two TLR adaptor molecules, followed by induction of selective autophagic degradation of these TLR adaptor-associated signaling complexes.
NDP52 mediates aggregation of TRAF6
Autophagy receptors are known to mediate selective aggregation of targets, and this process is required for selective removal of them by autophagy [21]. We therefore tested whether NDP52 is associated with TRAF6 aggregation. Transfected TRAF6 hardly formed aggregates in HeLa cells and was diffusedly present in the cytoplasm (Fig. 7a, upper, and b). However, cotransfection of TRAF6 with NDP52 was found to be markedly increased in aggregated TRAF6 in the cytoplasm, and the TRAF6 aggregates were colocalized with NDP52 (Fig. 7a, middle, and b). NDP52-induced TRAF6 aggregates were detectable as proteins with polyUb (Fig. 7c, middle and lower, and d), although TRAF6 itself could sufficiently increase protein aggregates with polyUb even in the absence of NDP52 transfection (Fig. 7c, upper, and d). In some cells, formation of autolysosome-like vacuoles was observed (Fig. 7a, lower, and e, lower), and such vacuoles were partly colocalized with the autolysosomal marker Rab7 (Fig. 7e, lower). Rab7-positive vacuoles hardly colocalized with TRAF6 aggregations (Fig. 7e), suggesting that TRAF6 may be rapidly degraded in Rab7-positive vacuoles. Transfection of TRAF6 with NDP52 did not significantly increase the formation of Rab7-positive vacuoles (Fig. 7f). We next tested whether A20 silencing affects these observations. In A20-silenced HeLa cells, NDP52-induced TRAF6 aggregates were clearly increased (Fig. 7g, h). Aggregates of polyUb proteins were also increased and TRAF6 aggregates were colocalized with polyUb of proteins (Fig. 7i, j). Under the condition of A20 silencing, autolysosome-like vacuoles were increased and partly colocalized with Rab7 (Fig. 7k, l). These results collectively suggest that, after recruitment to TRAF6, NDP52 participates in the aggregation of the TRAF6 conjugated with polyubiquitin chains. This process may lead to degradation of TRAF6 in autolysosome although such a function of NDP52 is restricted by A20.
Fig. 7.
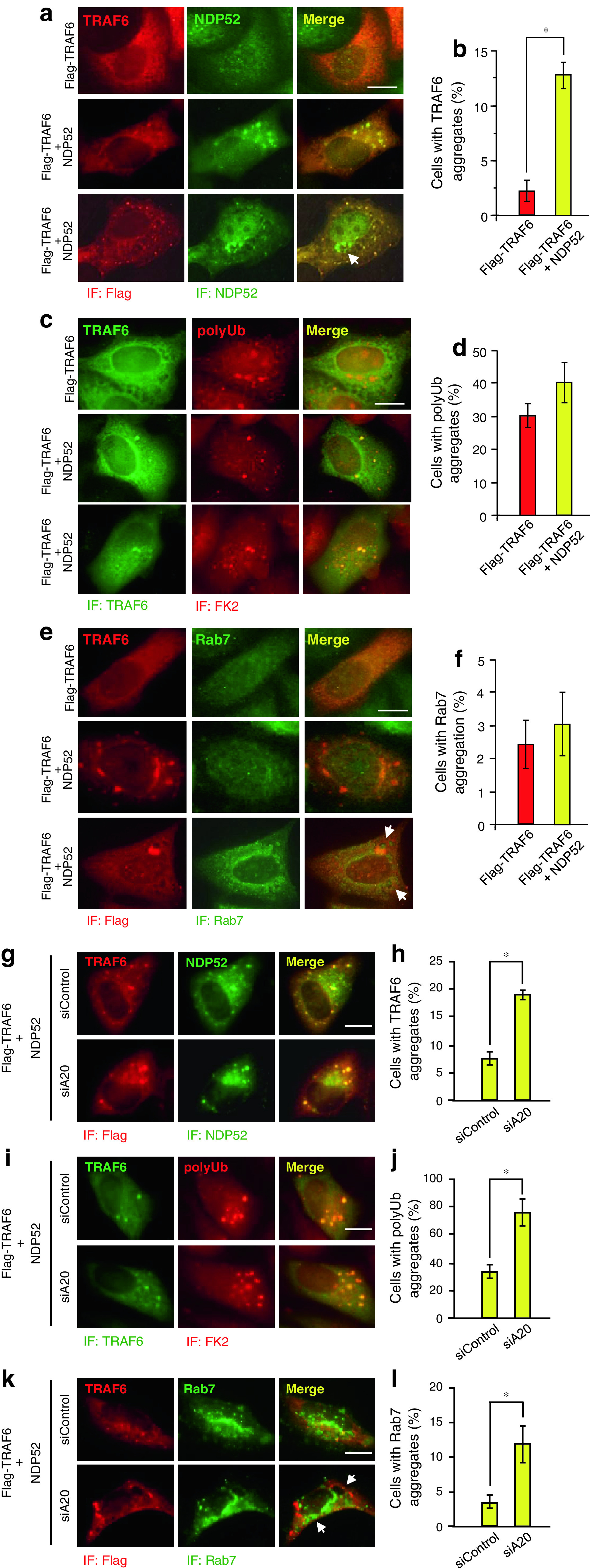
NDP52 promotes aggregation of TRAF6. a, c, e, g, i, k HeLa cells were transfected with Flag-TRAF6 and HA-NDP52 for 24 h (a, c, e). HeLa cells were transfected with control siRNA or A20 siRNA for 24 h. Then cells were further transfected with HA-NDP52 and Flag-TRAF6 for 18 h (g, i, k). Cells were fixed and stained with anti-Flag mouse monoclonal antibody and Alexa564-conjugated anti-mouse IgG antibody and then with anti-NDP52 rabbit polyclonal antibody and Alexa488-conjugated anti-rabbit IgG antibody (a, g), with anti-TRAF6 rabbit polyclonal antibody and Alexa488-conjugated anti-rabbit IgG antibody and then with anti-FK2 mouse monoclonal antibody and Alexa564-conjugated anti-mouse IgG antibody (c, i) or with anti-Flag mouse monoclonal antibody and Alexa564-conjugated anti-mouse IgG antibody and then with anti-Rab7 rabbit monoclonal antibody and Alexa488-conjugated anti-rabbit IgG antibody (e, k). Arrows autolysosome-like vesicles. Scale bar 10 μm. b, d, f, h, j, l Quantification of the percentage of cells containing TRAF6 aggregates (b, h), polyUb aggregates (d, j), and Rab7-positive vesicles (f, l) were performed by counting Flag-TRAF6-positive cells on five different images of microscopic fields including at least 30 cells. Results, shown as the mean ± SD (n = 5), are representative of three independent experiments. *P < 0.01
NDP52 undergoes Lys63- and Lys48-linked polyUb by TRAF6
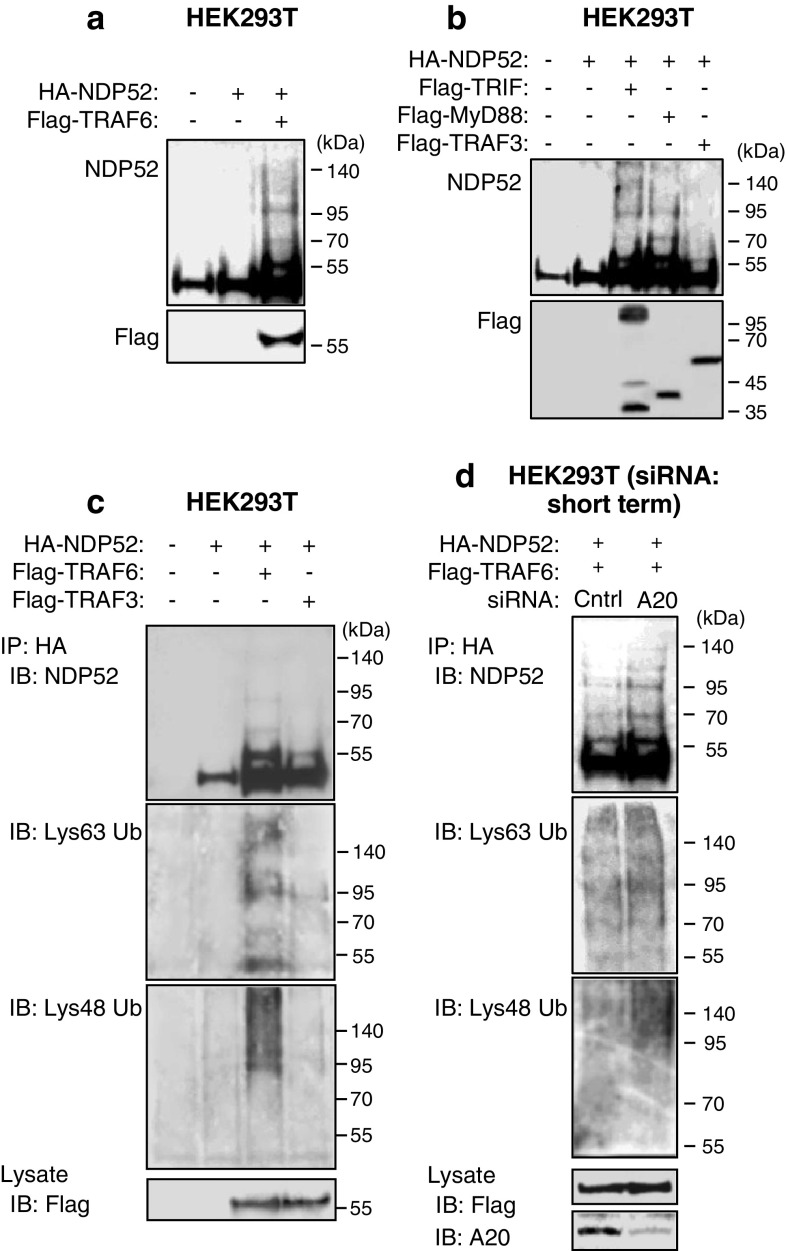
We next asked how NDP52 aggregates TRAF6. Intracellular protein aggregation is generally thought to be linked to accumulation of polyubiquitinated proteins in the cytoplasm [20]. Polymerization of the polyubiquitinated proteins results in the formation of microscopically visible structures known as inclusion bodies and aggresomes [20]. We tested whether NDP52 undergoes polyUb by TRAF6. We found that overexpression of NDP52 with TRAF6 resulted in appearance of shifted bands of NDP52 with higher molecular weights (Fig. 8a). Also, overexpression of NDP52 with TRIF or with MyD88 resulted in appearance of shifted bands (Fig. 8b). TRAF3 exerted a similar effect, but it was very weak (Fig. 8b). Importantly, an IP study revealed that TRAF6 yielded both Lys63- and Lys48-linked polyUb of NDP52 (Fig. 8c). Although TRAF3 had a similar activity, it was very weak (Fig. 8c). Of note, under the condition of A20 silencing, only Lys48-linked polyUb of NDP52 by TRAF6 were increased (Fig. 8d). Thus, NDP52 undergoes polyUb by TRAF6, which may be involved in promotion of TRAF6 aggregation.
Fig. 8.
Lys63- and Lys48-linked polyUb of NDP52 by TRAF6 and their control by A20. a HEK293T cells were transfected with HA-NDP52 and Flag-TRAF6 for 24 h. Cell lysates were analyzed by IB with anti-NDP52 and anti-Flag antibodies. b HEK293T cells were transfected with HA-NDP52 together with Flag-TRIF, Flag-MyD88 or Flag-TRAF3 for 24 h. Cell lysates were analyzed by IB with anti-NDP52 and anti-Flag antibodies. c HEK293T cells were transfected with HA-NDP52 together with Flag-TRAF6 or Flag-TRAF3 for 24 h. Cell lysates were boiled for 5 min, rapidly cooled and then subjected to IP with anti-HA affinity gel, followed by IB with antibodies to NDP52, Lys63-linked polyUb and Lys48-linked polyUb. d HEK293T cells were transfected with control siRNA or A20 siRNA for 24 h. Then cells were further transfected with HA-NDP52 and Flag-TRAF6 for 18 h. Cell lysates were boiled and then subjected to IP with anti-HA affinity gel, followed by IB with antibodies to NDP52, Lys63-linked polyUb and Lys48-linked polyUb. All results are representative of three independent experiments
A20 controls negative regulation of proinflammatory responses by NDP52
This study suggests a possibility that the autophagy receptor NDP52 exerts a negative regulatory effect on TLR-induced proinflammatory responses through degradation of activated TLR adaptor molecules and TRAF6. However, this effect may be normally inactivated by A20. To verify this, we investigated the effect of silencing of NDP52 together with A20 by siRNA. In HeLa cells, poly(I:C) induced mRNA expression of IL-6, IL-8 and A20 and the IRF3-responsible gene CXCL10 (Fig. 9). Expression of NDP52 was not clearly affected by poly(I:C) stimulation (Fig. 9). Compared with the control cells, silencing of NDP52 hardly affected mRNA expression of IL-6, IL-8 and CXCL10 (Fig. 9). Interestingly, silencing of the combination of A20/NDP52 promoted the induction of gene expression of IL-6, IL-8 and CXCL10 (Fig. 9). Thus, A20 indeed serves as a negative regulator for the negative regulatory effect of NDP52.
Fig. 9.
A20-controlled negative regulation of poly(I:C)-induced genes by NDP52. HeLa cells were transfected with control siRNA or NDP52 siRNA together with A20 siRNA for 24 h. Then cells were stimulated with 50 μg/ml poly (I:C) for 6 h. The mRNA levels of NDP52, A20, IL-6, IL-8 and CXCL10 were determined by qRT-PCR. Data are expressed as the mean fold induction ±SD (n = 3) relative to control levels (Control siRNA, Medium), for representative of three independent experiments. *P < 0.01, and † P < 0.05, for comparison with the Control siRNA group
Discussion
The critical link between pathogen recognition of TLRs and autophagy has been shown [22–24], but it is largely unknown whether autophagy receptors have any regulatory effects on TLR signaling. In this study, we identified an unanticipated mechanism, in which the autophagy receptor NDP52 works downstream of TLR adaptors to negatively regulate TLR signaling. NDP52 seems to promote TRAF6 aggregation that is required for the selective degradation of TLR adaptor-associated signaling complex in a non-canonical autophagy-dependent manner. Thus, TRAF6 and NDP52 can cooperatively suppress TLR signal transduction. In addition, A20 downregulates the activity of NDP52 probably through suppression of polyUb by TRAF6. This may be related to the previous finding that TRAF6 induction of Lys63-linked polyUb of Beclin-1 is negatively regulated by A20 [51]. Although biological significance of the presence of such complicated mechanisms for negative regulation of TLR signaling by A20 and NDP52 is obscure, NDP52 may preferentially work to suppress excess TLR signaling in the case of the absence or an insufficient expression level of A20.
NDP52 mediates selective autophagic degradation of TRIF and TRAF6 after TLR signal transduction. This type of autophagy could be suppressed by the autophagy inhibitor 3-MA, but was not associated with ‘canonical’ autophagic mechanisms, such as conversion of LC3-I to LC3-II and involvement of Beclin-1 and ATG proteins. Given that 3-MA is an inhibitor for class III PI3 kinase, induction of autophagy found in this study is at least dependent on PI3 kinase. Although we could not determine the reason why this type of autophagy does not depend on Beclin-1 and ATG proteins, several recent reports have shown existence of non-canonical autophagy, the process of which does not require these proteins [27, 29]. Recently, such autophagy was found to be mediated by the anti-apoptotic protein Bcl-xL [28]. NDP52-mediated autophagy may also be activated by Bcl-xL.
NDP52 negatively regulates not only NF-κB activation downstream of TRAF6 but also IRF3 activation downstream of TRAF3 (Fig. 5). In addition, NDP52 can physically bind TRAF6 and TRAF3 (Fig. 4c). However, NDP52 can specifically degrade TRAF6, but not TRAF3 (Fig. 4d, e). Therefore, NDP52 has differential biological functions for binding and regulation of degradation between TRAF6 and TRAF3. TRAF6 ubiquitinates NDP52 possibly through the interaction with NDP52 (Fig. 8c), which ultimately leads to induction of TRAF6 aggregation (Fig. 7). TRIF may be involved in this process through the TRIF–TRAF6 interaction and may be selectively degraded together with TRAF6 through selective autophagy. This process may lead to suppression of TLR- and TRAF6-mediated NF-κB activation. In contrast to TRAF6, TRAF3 hardly ubiquitinates NDP52 (Fig. 8c) although NDP52 can physically bind TRAF3. A previous report indicated that NDP52 exerts different effects on TLR signaling downstream of TRAF3, in which NDP52 may work as an adaptor molecule for the TRAF3 downstream molecule TBK1 [31]. This observation suggests that TRAF3 requires NDP52 to induce appropriate activation of downstream TBK1–IRF3 signaling. The reason why NDP52 could suppress IRF3 activation may be caused by NDP52-induced decrease of TRIF. In TLR signaling, TRAF3 activation never occurs in the absence of TRIF.
A20 is known to act not only as a deubiquitinase for the targets with Lys63-linked polyUb but also as an E3 ubiquitin ligase to catalyse Lys48-linked polyUb [14]. In our study, A20 was at least involved in the regulation of Lys48-linked polyUb of NDP52, which may be dependent on suppression of TRAF6-induced Lys48-linked polyUb (Fig. 8d). However, we could not clearly see the implication of A20 in catalysis of Lys63-linked Ub of NDP52 (Fig. 8d). A recent report has shown an additional mechanism of A20 for substrate inactivation, whereby A20 disrupts the interaction of an E3 ubiquitin ligase with a substrate or of an E2 ubiquitin-conjugating enzyme with an E3 ubiquitin ligase, leading to attenuation of the activation of specific E3 ubiquitin ligases, such as TRAF6 [16]. Therefore, it is possible that this mechanism of A20 is preferentially involved in the TRAF6 regulation of NDP52 and that NDP52 is not a substrate of A20.
NDP52/CALCOCO2 and TAX1BP1/CALCOCO3 are paralogs of COCOA/CALCOCO1. These molecules share the CALCOCO domain that consists of the N-terminal SKICH domain and the subsequent CC domain. TAX1BP1 is a TRAF6-binding molecule [52] and negatively regulates TRAF6-induced NF-κB activation through assistance of the function of A20 and the E3 ubiquitin ligase Itch [16, 53]. Such a function of TAX1BP1 is intrinsically different from the function of NDP52 that is associated with selective autophagy downstream of TRAF6. Since TAX1BP1 works together with A20, it is possible that TAX1BP1 negatively regulates the function of NDP52. TAX1BP1 has C-terminal zinc fingers that contain two Pro-Pro-X-Tyr motifs, which are essential for the negative regulatory effect on NF-κB signaling [53], while NDP52 and COCOA do not have such motifs in the C-terminal ends (Supplementary material, Fig. S6). The C-terminal LIM-like domain of NDP52 mediates the recognition of ubiquitin chains [31] and is analogous to the N-terminal regions of CRP, CRIP1 and CRIP2 [49] (Supplementary material, Fig. S6). Therefore, each structure of the C-terminal end may be responsible for the different functions of TAX1BP1 and NDP52. Also, TAX1BP1 has an atypical TRAF6-binding motif, Pro-X-Glu-X-X-X-[Aromatic (Ar) or acidic (Ac) amino acid] [54], corresponding to the sequence PAERKME (597–603 amino acids) (but not a typical TRAF6-binding motif, Pro-X-Glu-X-X-(Ar/Ac) [55]). NDP52 does not have any TRAF6-binding motifs, suggesting that the interaction of NDP52 with TRAF6 may be indirect. However, the long and short of it is that TAX1BP1 and NDP52 share the function to negatively regulate TRAF6-mediated signaling. It is also possible that COCOA negatively regulates TRAF6-mediated signaling through an unknown mechanism.
Initially, it was reported that TLR4 recognition of LPS induces autophagy through TRIF but not MyD88 [24]. After that, recognition of poly(I:C) by TLR3 was shown to trigger autophagy through TRIF, while recognition of imiquimod by TLR7 induces autophagy through MyD88 [22]. It was also reported that both TRIF and MyD88 interact with Beclin1 and induce autophagy [56]. In this study, we obtained several lines of evidence that activated TRIF and MyD88 recruit the autophagy receptor NDP52 to trigger aggregation of signaling complexes involving TRAF6, in which the activity of NDP52 may be regulated by TRAF6 through polyUb (Fig. 8). NDP52 probably recognizes Lys63-linked polyUb of TRIF, MyD88 and TRAF6 through the LIM-like domain (Fig. 6) and is subsequently involved in the aggregation of them in the cytoplasm. This process does not stimulate induction of canonical autophagy flux but can selectively facilitate TLR signaling molecules to yield selective autophagic degradation probably in a non-canonical autophagy-dependent manner. The principal functions of NDP52 found in this study are summarized in Fig. 10.
Fig. 10.
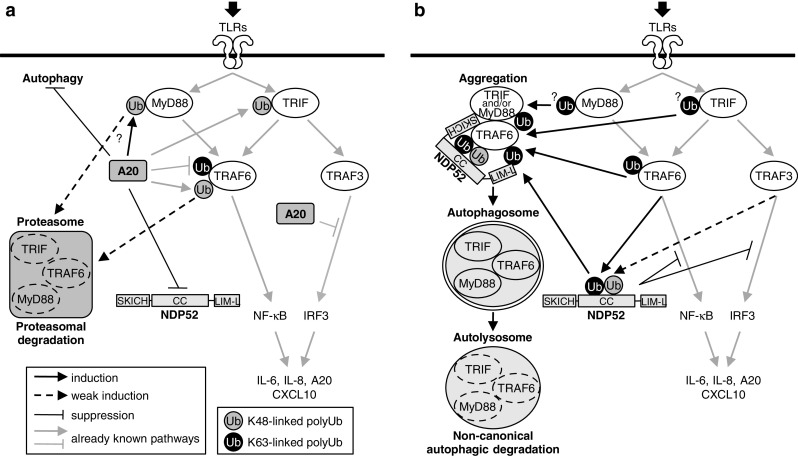
Schematic of regulation of TLR signaling by NDP52 under the control of A20. TLR triggering activates signaling pathways via two key adaptor molecules, MyD88 and TRIF. MyD88 interacts with TRAF6 that catalyses Lys63-linked polyUb of target proteins, leading to activation of downstream NF-κB signaling. TRIF is also able to activate TRAF6-dependent signaling. In addition, TRIF activates TRAF3-dependent signaling that catalyses Lys63-linked polyUb of target proteins, which mediates IRF3 signaling. a In the presence of A20, this enzyme cleaves Lys63-linked polyubiquitin chains, leading to negative regulation of TRAF6-mediated activation of NF-κB and TRAF3-mediated activation of IRF3. A20 further promotes catalyses of Lys48-linked polyUb of target proteins, including MyD88 and TRIF, which may lead to their proteasomal degradation. In addition, A20 is thought to suppress induction of autophagy [51]. Although NDP52 has a potential to be activated by TRAF6 to mediate selective autophagy, A20 normally inactivates NDP52 through deubiquitination. b In the absence of A20, NDP52 may be fully activated by TRAF6. In this case, NDP52 can mediate aggregation of the complexes of MyD88–TRAF6 and TRIF–TRAF6. This process is thought to be important to selectively degrade unnecessary signaling molecules in lysosomes via an autophagic mechanism. This type of autophagy seems to be induced through non-canonical processes. Such a function of NDP52-mediated autophagy ultimately leads to negative regulation of TLR signaling
Beyond its critical function of cell-autonomous protection from starvation, autophagy assumes the responsibility of regulation of innate immune responses [26, 57]. The most well-known function is selective elimination of intracellular pathogens, including bacteria and viruses. Recently, it is becoming clear that several autophagy receptors, including NDP52 and SQSTM1, play an important role in the selective elimination of pathogenic bacteria [21, 31, 58]. During this, autophagy seems to be useful for cytoprotection towards excess innate immune signaling that often generates a cytotoxic level of ROS [59, 60]. Our results regarding the functions of NDP52 for negative regulation of TLR signaling support this idea. On the other hand, recent findings from the analyses of gene knockout mice have revealed that certain autophagy receptors, including SQSTM1 and NBR1, are involved in T helper type 2 cell differentiation [61, 62]. Therefore, autophagy receptor-mediated negative regulation of TLR signaling may be involved in effective induction of acquired immunity. Future investigations should reveal unknown functions of autophagy receptors in innate and adaptive immunity, which may be found to be under the control of A20.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Dr. M. Matsumoto and Dr. H. Nakano for providing the expression plasmids of TRAF3 and TRAF6, respectively. We also heartily thank Prof. T. Johansen and Prof. X. Li for providing the expression plasmids of GFP-LC3 and p561-luc, respectively. We thank Dr. T. Hanaichi (Hanaichi Ultrastructure Research Institute, Okazaki, Japan) for his technical assistance for electron microscopy. This work was supported by the 2010 Science Research Promotion Fund (to T. Into) from the Promotion and Mutual Aid Corporation for Private Schools of Japan and Asahi University. This work was also supported by a Grant-in-Aid for Young Scientists (Start-up; 21890275 and B; 23792158) to M. Inomata, provided by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- CC
Coiled-coil
- GFP
Green fluorescent protein
- IFN
Interferon
- IRAK
IL-1 receptor-associated kinase
- LIM
Lin11, Isl-1 and Mec-3
- qRT-PCR
Quantitative reverse transcription-coupled PCR
- PI3
Phosphatidylinositol 3
- SKICH
Skeletal muscle and kidney-enriched inositol phosphatase carboxyl homology
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 3.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136:1019–1098. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 8.Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Hacker G, Mann M, Karin M. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 10.Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meylan E, Burns K, Hofmann K. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-κB activation. Nat Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 12.Chang M, Jin W, Sun SC. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of Toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 14.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 15.Parvatiyar K, Barber GN, Harhaj EW. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1–IKKi kinases. J Biol Chem. 2010;285:14999–15009. doi: 10.1074/jbc.M110.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat Immunol. 2010;11:734–742. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 18.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/S0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 19.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 20.Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 21.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 22.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 28.Priault M, Hue E, Marhuenda F, Pilet P, Oliver L, Vallette FM. Differential dependence on Beclin 1 for the regulation of pro-survival autophagy by Bcl-2 and Bcl-xL in HCT116 colorectal cancer cells. PLoS One. 2010;5:e8755. doi: 10.1371/journal.pone.0008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 30.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 32.Into T, Inomata M, Nakashima M, Shibata K, Hacker H, Matsushita K. Regulation of MyD88-dependent signaling events by S nitrosylation retards Toll-like receptor signal transduction and initiation of acute-phase immune responses. Mol Cell Biol. 2008;28:1338–1347. doi: 10.1128/MCB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Into T, Inomata M, Niida S, Murakami Y, Shibata K. Regulation of MyD88 aggregation and the MyD88-dependent signaling pathway by sequestosome 1 and histone deacetylase 6. J Biol Chem. 2010;285:35759–35769. doi: 10.1074/jbc.M110.126904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uehara A, Fujimoto Y, Fukase K, Takada H. Various human epithelial cells express functional Toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol Immunol. 2007;44:3100–3111. doi: 10.1016/j.molimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Wyllie DH, Kiss-Toth E, Visintin A, Smith SC, Boussouf S, Segal DM, Duff GW, Dower SK. Evidence for an accessory protein function for Toll-like receptor 1 in anti-bacterial responses. J Immunol. 2000;165:7125–7132. doi: 10.4049/jimmunol.165.12.7125. [DOI] [PubMed] [Google Scholar]
- 36.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 37.Rebsamen M, Meylan E, Curran J, Tschopp J. The antiviral adaptor proteins Cardif and Trif are processed and inactivated by caspases. Cell Death Differ. 2008;15:1804–1811. doi: 10.1038/cdd.2008.119. [DOI] [PubMed] [Google Scholar]
- 38.Wu WK, Wu YC, Yu L, Li ZJ, Sung JJ, Cho CH. Induction of autophagy by proteasome inhibitor is associated with proliferative arrest in colon cancer cells. Biochem Biophys Res Commun. 2008;374:258–263. doi: 10.1016/j.bbrc.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 39.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 41.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 43.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 44.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthias P, Yoshida M, Khochbin S. HDAC6 a new cellular stress surveillance factor. Cell Cycle. 2008;7:7–10. doi: 10.4161/cc.7.1.5186. [DOI] [PubMed] [Google Scholar]
- 46.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funami K, Sasai M, Oshiumi H, Seya T, Matsumoto M. Homo-oligomerization is essential for Toll/interleukin-1 receptor domain-containing adaptor molecule-1-mediated NF-κB and interferon regulatory factor-3 activation. J Biol Chem. 2008;283:18283–18291. doi: 10.1074/jbc.M801013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sternsdorf T, Jensen K, Zuchner D, Will H. Cellular localization, expression, and structure of the nuclear dot protein 52. J Cell Biol. 1997;138:435–448. doi: 10.1083/jcb.138.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korioth F, Gieffers C, Maul GG, Frey J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J Cell Biol. 1995;130:1–13. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morriswood B, Ryzhakov G, Puri C, Arden SD, Roberts R, Dendrou C, Kendrick-Jones J, Buss F. T6BP and NDP52 are myosin VI binding partners with potential roles in cytokine signalling and cell adhesion. J Cell Sci. 2007;120:2574–2585. doi: 10.1242/jcs.007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ling L, Goeddel DV. T6BP, a TRAF6-interacting protein involved in IL-1 signaling. Proc Natl Acad Sci USA. 2000;97:9567–9572. doi: 10.1073/pnas.170279097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, Harhaj EW. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 54.Meads MB, Li ZW, Dalton WS. A novel TNF receptor-associated factor 6 binding domain mediates NF-κB signaling by the common cytokine receptor beta subunit. J Immunol. 2010;185:1606–1615. doi: 10.4049/jimmunol.0902026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, Du K, Singh S, Pike JW, Darnay BG, Choi Y, Wu H. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 56.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–33182. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2010;7:22–26. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 60.Takeda K, Noguchi T, Naguro I, Ichijo H. Apoptosis signal-regulating kinase 1 in stress and immune response. Annu Rev Pharmacol Toxicol. 2008;48:199–225. doi: 10.1146/annurev.pharmtox.48.113006.094606. [DOI] [PubMed] [Google Scholar]
- 61.Martin P, Diaz-Meco MT, Moscat J. The signaling adapter p62 is an important mediator of T helper 2 cell function and allergic airway inflammation. EMBO J. 2006;25:3524–3533. doi: 10.1038/sj.emboj.7601250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang JQ, Liu H, Diaz-Meco MT, Moscat J. NBR1 is a new PB1 signalling adapter in Th2 differentiation and allergic airway inflammation in vivo. EMBO J. 2010;29:3421–3433. doi: 10.1038/emboj.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.