Abstract
Diversity Arrays Technology (DArT) provides a high-throughput whole-genome genotyping platform for the detection and scoring of hundreds of polymorphic loci without any need for prior sequence information. The work presented here details the development and performance of a DArT genotyping array for apple. This is the first paper on DArT in horticultural trees. Genetic mapping of DArT markers in two mapping populations and their integration with other marker types showed that DArT is a powerful high-throughput method for obtaining accurate and reproducible marker data, despite the low cost per data point. This method appears to be suitable for aligning the genetic maps of different segregating populations. The standard complexity reduction method, based on the methylation-sensitive PstI restriction enzyme, resulted in a high frequency of markers, although there was 52–54% redundancy due to the repeated sampling of highly similar sequences. Sequencing of the marker clones showed that they are significantly enriched for low-copy, genic regions. The genome coverage using the standard method was 55–76%. For improved genome coverage, an alternative complexity reduction method was examined, which resulted in less redundancy and additional segregating markers. The DArT markers proved to be of high quality and were very suitable for genetic mapping at low cost for the apple, providing moderate genome coverage.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-011-9579-5) contains supplementary material, which is available to authorized users.
Keywords: Apple, DArT, Genetic mapping, Molecular markers, Diversity
Introduction
The genus Malus has been a focus of molecular studies since the mid-1980s, when Weeden and Lamb (1985) used isozymes as a means of discriminating between different apple cultivars. As new marker types have been developed, scientists have readily adopted them into their studies of apple genetics, progressing from rapid amplification of polymorphic DNA (RAPD) (Koller et al. 1993), through restriction fragment length polymorphism (RFLP) and isozymes (Maliepaard et al. 1998), to amplified fragment length polymorphism (AFLP) and simple sequence repeat (SSR) (Guilford et al. 1997, Hokanson et al. 1998) and on to targeted markers (e.g., Broothaerts 2003; Calenge et al. 2005; Chagné et al. 2007) and single nucleotide polymorphism (SNP) arrays (Micheletti et al. 2011). The progress in the molecular characterisation of the Malus genome has recently gained further momentum with whole-genome sequencing (Velasco et al. 2010).
Molecular markers have been applied widely to evolutionary and pedigree studies in apple, including both wild Malus species (Richards et al. 2009) and domestic cultivars (e.g., Cabe et al. 2005; Evans et al. 2010). Additionally, markers developed for the apple have been applied fairly widely to other pome species, and vice versa, notably pear (Pyrus spp., Yamamoto et al. 2001; Hemmat et al. 2003; Dondini et al. 2004), quince (Cydonia oblonga; Yamamoto et al. 2004) and loquat (Eriobotrya japonica; Gisbert et al. 2009; He et al. 2010). Simultaneously, linkage maps have been constructed for a number of domestic cultivars, and several map alignments have been reported (Maliepaard et al. 1998; N’Diaye et al. 2008; Patocchi et al. 2009; Van Dyk et al. 2010). The construction of linkage maps has facilitated the identification of molecular markers associated with numerous phenotypic traits. Among the traits examined to date are resistance to apple scab caused by the fungus Venturia inaequalis (Koller et al. 1994; Calenge et al. 2004; Bus et al. 2005a, b; Soriano et al. 2009), fire blight caused by the bacterium Erwinia amylovora (Peil et al. 2007), columnar growth habit (Moriya et al. 2009), several fruit quality traits (e.g., King et al. 2001; Liebhard et al. 2003a; Costa et al. 2005, 2008, 2010; Kenis et al. 2008) and chilling requirement (Van Dyk et al. 2010). The identification of such molecular markers is essential for marker-assisted selection in apple breeding programs (Gianfranceschi et al. 1996; Liebhard et al. 2003b; Gardiner et al. 2007; Zhu and Barrett 2008). In recent years, genomic methods have been embraced by apple researchers. The enhanced ability to study gene expression has resulted in new understandings of developmental processes (Ban et al. 2007; Espley et al. 2009). Transcription analyses of apple fruit development using cDNA microarrays (Soglio et al. 2009) and plant physiological responses to pathogens (Norelli et al. 2009) has facilitated the development of new molecular markers (e.g., Chagné et al. 2008; Igarashi et al. 2008). The further development of high-throughput genetic technologies will continue to expand the ability of scientists to investigate the details of the genetics of apple and its relatives (Shulaev et al. 2008).
Since the proof-of-concept paper (Jaccoud et al. 2001), Diversity Arrays Technology (DarT) has been developed as an inexpensive whole-genome profiling technique for many organisms, especially plants. The website www.diversityarrays.com has a current list of organisms for which arrays are available (>50). DArT, in its current implementation, is a hybridisation-based genome profiling technology that does not require sequence information and uses microarrays to identify and type DNA polymorphisms. As the DArT markers are typed in parallel, it is possible to identify hundreds or even thousands of polymorphic markers in a single experiment (Wittenberg et al. 2009). This highly parallel assay results in a reduction of the price per data point to around US $0.01 in organisms with well-developed arrays. The DArT assay primarily detects dominant markers, mostly resulting from single nucleotide polymorphisms and indels in restriction sites and differences in methylation of restriction sites. When methylation-sensitive restriction enzymes (like PstI; see Gruenbaum et al. 1981) were used in the large genomes of cereals (Wenzl et al. 2004, 2006; Akbari et al. 2006), DArT markers were located preferentially at the gene-rich, subtelomeric regions of the chromosomes. The use of methylation-sensitive enzymes may provide also an insight into epigenetic variation (Wenzl et al. 2004).
Interestingly, while DArT has performed well in over 50 crops, there are no reported applications of DArT in horticultural trees. As DArT has been applied successfully to complex amphidiploid genomes like wheat (Akbari et al. 2006), oat (Tinker et al. 2009) and sugarcane (Heller-Uszynska et al. 2010), application to the duplicated genomes of the pome fruits such as apple (Velasco et al. 2010), pear and loquat should be promising too. Here, we present the development and validation of DArT for apple using a complexity reduction method similar to the one used for the cereal genomes. We compare this with a second complexity reduction method that is similar to the method used for the fungus Mycosphaerella graminicola (Wittenberg et al. 2009), and we discuss the characteristics of the detected markers. We combined DArT markers with other marker types in genetic linkage maps, providing insight into the coverage of the DArT markers in the apple genome. We used as a starting point the progeny and genetic linkage map of Prima × Fiesta, which was the first genetic linkage map for apple covering all 17 chromosomes (Maliepaard et al. 1998). In addition, we used a more recent progeny of other parents for genetic mapping. Furthermore, we evaluated the performance of DArT in a genetic diversity analysis of 44 diverse apple accessions and a set of Australian breeding lines.
Materials and methods
Plant material
For the building of the DArT libraries, care was taken to represent a wide genetic diversity, including several major founders in apple breeding worldwide, founders of more local breeding programs, modern cultivars and some very recent selections from ongoing breeding programs. Forty-four accessions of Malus were used for the library development (Online Resource 1a).
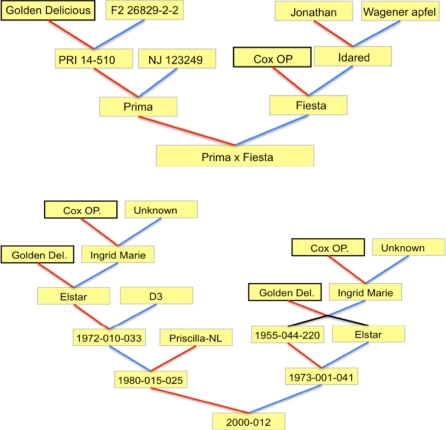
For mapping, two populations were used. The first, Prima × Fiesta, was used to examine the reliability of the DArT data by evaluating the ease by which the DArT markers were integrated in an existing, well-established linkage map. This population consists of 156 individuals (Maliepaard et al. 1998), of which a subset of 121 individuals were DArT genotyped. The second population, 2000–2012 (Soriano et al. 2009), was used to demonstrate the ease by which DArT markers allow for the alignment between mapping populations. In addition, this population was used to compare two DArT complexity reduction methods with respect to the number of markers and genome coverage. It comprises 894 individuals, 399 of which were used in the current study. The parentages of both populations are presented in Fig. 1.
Fig. 1.
Pedigree of the two mapping populations examined. Common parents are highlighted
DNA extraction
For the development of the DArT libraries and genetic diversity studies, leaves were collected from grafted trees with a preference for younger, actively expanding leaf material. Genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB)/chloroform/isoamylalcohol protocol based on the method of Doyle and Doyle (1987). The addition of 2% w/v polyvinylpyrrolidone (PVP-40, Sigma, K value: 29–32) (Aljanabi et al. 1999; Kim et al. 1997) appeared to be essential for preventing the inhibition of restriction endonuclease digestion in many of the apple leaf samples tested. For the mapping populations, DNA was extracted according to Maliepaard et al. (1998) and Soriano et al. (2009).
Construction of DArT arrays
A crucial step in the Diversity Arrays Technology is the complexity reduction of genomic representations. In this manuscript, complexity reduction refers to the reproducible selection of a subset of DNA fragments from a whole genome. These fragments, after being cloned into E. coli vectors (TOPO) and amplified with M13 primers, were printed onto slides as probes for microarray hybridisations. The complexity reduction method used most often in DArT involves digestion with the methylation-sensitive restriction enzyme, PstI. In conjunction with digestion using this relatively rarely-cutting restriction enzyme (six bp recognition site plus methylation sensitivity; Gruenbaum et al. 1981), an enzyme with frequent cutting capabilities is used (Wenzl et al. 2004). In this study, the frequently-cutting enzymes AluI, BstNI, TaqI or MseI were used. PCR adapters were ligated to the PstI fragment ends, and the PCR-amplification was performed using primers complementary to the PstI adapters, according to Wenzl et al. (2004). Only those fragments with PstI adapters at both ends were amplified.
Initial assessment of the enzyme combinations was performed by agarose gel electrophoresis, as described by Jaccoud et al. (2001). On this basis, the genomic representations produced by the digestion with PstI, in combination with either AluI or BstNI (PstI/AluI and PstI/BstNI), were considered to be the most suitable due to the absence of visible bands in the gel smear (Kilian et al. 2005). An initial library of 768 clones was prepared for PstI/AluI and a second initial library of the same size for PstI/BstNI, using DNA from 15 diverse heritage apple varieties (Online Resource 1a). The inserts of the 2 × 768 clones were amplified and printed on glass slides to provide small arrays for testing. The 15 cultivars listed in Table 1 were hybridised in duplicate to the arrays, according to Wenzl et al. (2004). The PstI/AluI complexity reduction method was found to give a higher number of candidate polymorphic markers (16% of clones) than the PstI/BstNI method (10% of clones). The PstI/AluI library was therefore expanded by an additional 3,840 clones derived from the 15 heritage cultivars, and 9,984 clones from modern apple cultivars and breeding lines (Online Resource 1a). The total size of the expanded library for the PstI/AluI complexity reduction was 14,592 clones.
Table 1.
Number of DArT markers from the standard complexity reduction method, during successive mapping stages in the Prima × Fiesta population
| Mapping stages | Prima | Fiesta’ | Both parents | Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| No. of events | No. cumulative | No. of events | No. cumulative | No. of events | No. cumu- lative | No. of events | % events | No. cumulative | |
| Preparation data | |||||||||
| Polymorphic | 257 | 234 | 285 | 776 | |||||
| Unclear parentage | 11 | 246 | 14 | 220 | 0 | 285 | 25 | 3.2 | 751 |
| Polymorphic Information Content < 0.10 | 47 | 199 | 49 | 171 | 3 | 282 | 99 | 12.8 | 652 |
| Mapping | |||||||||
| Redundant, completely identical | 1 | 198 | 40 | 131 | 73 | 209 | 114 | 14.7 | 538 |
| Remaining ungrouped | 0 | 198 | 4 | 127 | 0 | 209 | 4 | 0.5 | 534 |
| Insufficient linkage for phase determination | 1 | 197 | 4 | 123 | 32 | 177 | 37 | 4.8 | 497 |
| Irregular segregation pattern | 2 | 196 | 4 | 119 | 1 | 176 | 7 | 0.8 | 490 |
| Redundant, differences only for missing values | 112 | 83 | 42 | 77 | 96 | 80 | 250 | 32.2 | 240 |
| Double recombinant | 0 | 83 | 1 | 76 | 0 | 80 | 1 | 0.1 | 239 |
| Unique mapped | 83 | 76 | 80 | 239 | |||||
| Markers with solved parentages | 3 | 86 | 3 | 79 | 2 | 82 | 8 | 1.0 | 247 |
| Total uniquely mapped | 86 | 79 | 82 | 247 | |||||
Hybridisation to the expanded array
To gain insight into the applicability of DArT in mapping, two segregating populations were hybridised to the expanded array. All genotypes were hybridised with two to four replicate arrays per genotype, using both Cy-3 and Cy-5 fluorescent labelling. Hybridisation, subsequent processing and data analysis were performed according to Wenzl et al. (2004).
Integration of existing genetic markers into genetic linkage maps
The genetic linkage map of Prima × Fiesta was the first for apple covering all 17 chromosomes (Maliepaard et al. 1998). It consisted of 138 dominant markers (mainly RAPDs and some AFLPs) and 152 essentially co-dominant markers (mainly RFLP, some isozymes and SSRs). Since then, 313 new markers have been added through successive European projects, of which 180 are dominant (mainly AFLP) and 133 are co-dominant, mainly consisting of SSR markers from the gDNA-based CH and Hi series (Liebhard et al. 2003b, Silfverberg-Dilworth et al. 2006) as gathered from expressed sequence tag (EST) sequences (Soglio et al. 2009). Some of the new markers were specifically designed for fruit-quality genes (Costa et al. 2005, 2008, 2010) and allergy genes (Gao et al. 2005a, b, c). The quality of the map was thoroughly validated and improved, using JoinMap® (Van Ooijen and Voorrips 2008). This newly enriched, evaluated and extended map of Prima × Fiesta covers approximately 90% of the apple genome, and was used as a starting point for the mapping of DArT markers.
An alternative method for complexity reduction
We evaluated an alternative complexity reduction method that was applied by Wittenberg et al. (2005) to microbial genomes. This approach involved digestion with two six-base cutters, PstI and EcoRI. A standard adapter was ligated to the PstI ends of the restriction fragments and a long, asymmetric adapter with a 3′-amino (NH2) group on the short strand was ligated to the EcoRI ends. The amino group, combined with PCR suppression (Siebert et al. 1995; Broude et al. 2000), was used to prevent amplification of the EcoRI–EcoRI fragments. Only the PstI–PstI and PstI–EcoRI fragments were amplified. To further reduce the complexity of the genomic representations, a third endonuclease, the four-basepair cutter MboI, was used. No adapters were ligated to the MboI sites. Consequently, the fragments cut by MboI were not amplified (Wittenberg et al. 2005).
For this alternative complexity reduction method, the DNA of the apple selection 1980-015-025, a parent of population 2000–2012 (Fig. 1), was used to construct a library of 6,144 fragments, which were printed onto slides (Wittenberg et al. 2005). Target DNA from this selection’s progeny, population 2000–2012, was assayed with this array, using the same alternative complexity reduction method. The adapters ligated to the target DNA of this progeny differed from those of the parental fragments printed on the slides, in order to prevent hybridisation of adapters to one another (Wittenberg et al. 2005).
The alternative array was used for the genotyping of 244 progenies of population 2000–2012, all of which had also been genotyped with the standard method too. The maternal map of the heterozygous parent 1980-015-025 was constructed using DArT markers from both complexity reduction methods. In addition, several SSR markers were used as references on the linkage map; they were generated according to Patocchi et al. (2009).
Results
Mapping of DArT markers in Prima × Fiesta
The Prima × Fiesta progeny were hybridised to the expanded DArT array for the standard complexity reduction method, which provided 776 polymorphic markers. The call rate for the parental genotypes was 99.2%. The call rate is the percentage of targets that could reliably be assigned a score of ‘0’ or ‘1’ for a given candidate marker. The average call rate for the entire Prima × Fiesta mapping population was 96.7%.
Of the aforementioned 776 Prima × Fiesta DArT markers, 247 (32%) were mapped to a unique position (Fig. 2). The other 68% were eliminated during the mapping process for several reasons (Table 1). Only 4.6% of the markers were discarded due to possible scoring problems. Of these, 3.5% were due to incomplete data on the mapping parents, leaving only 1.1% of the markers being possibly discarded for inadequate scoring, remaining ungrouped or showing irregular segregation patterns. The two latter phenomena could also be due to reasons other than scoring difficulties, such as a lack of marker coverage of the genomic regions or the presence of duplicated loci. Online Resource 1b documents the fact that adding DArT markers did not affect the previous high overall map quality, as measured by the average χ2 value.
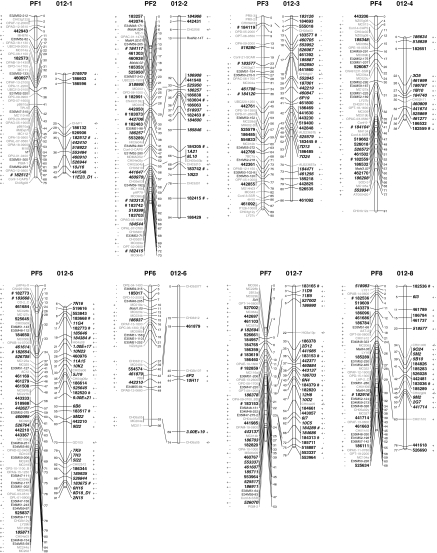
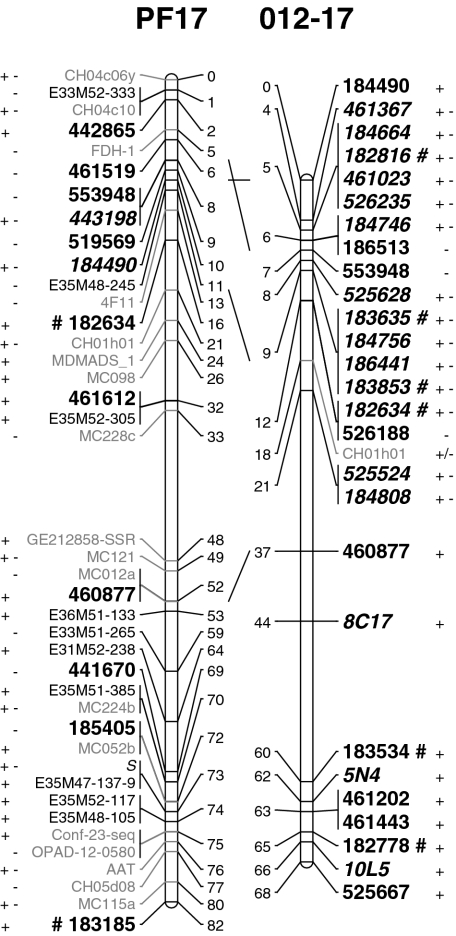
Fig. 2.
Alignment of the 17 linkage groups of the mapping populations Prima × Fiesta (PF) and 2000–2012 (012). DArT markers are displayed in bold, those segregating in both parents are in italics and those generated with the alternative complexity reduction method are underlined. The + and − symbols next to a marker name indicate that the DArT marker was polymorphic in the mother and father, respectively. The # symbols indicate the DArT markers that have been sequenced
A considerable number of clones exhibited identical segregation patterns in Prima × Fiesta compared to other clones, and therefore did not provide a higher genetic resolution in the map. As a result, a total of 364 (54%) of the 677 clones (=776–99; Table 1) were classified as redundant.
Genome coverage
The current DArT array provided moderate genome coverage, as illustrated in Fig. 2. Many markers clustered, producing several short genomic segments containing multiple markers, such as a segment of 4 cM at the top of linkage group (LG) 7 that contained eight unique Prima-specific DArT markers and one marker for both parental cultivars. However, several extended regions had no or very few markers, such as the entire LG1 of Prima, which contained only one Prima-specific and one common marker. Genome coverage was estimated using the integrated map of Fig. 2 as a reference and the following thresholds: (1) only parent-specific markers were considered, as markers common to both parents carry little genetic information, and (2) a single marker covers 10 cM surrounding its position. In these calculations, the current DArT array offered sufficient coverage for performing classical quantitative trait loci (QTL) mapping studies on around 55% of the Prima and 60% of the Fiesta genome. If a single marker were to sufficiently cover a larger window of 30 cM, then the genome coverage for Prima and Fiesta would be 76 and 74%, respectively.
Suitability of DArT markers for map alignment
To examine the power of DArT markers for aligning maps, the second mapping population, 2000–2012, was hybridised to the same DArT array used for Prima × Fiesta. Additionally, several previously mapped SSR markers were included to confirm the DArT marker alignment. A similar number of polymorphic DArT markers was obtained (774 in 2000–2012 vs. 776 in Prima × Fiesta), in addition to a similar call rate (97.3%) and a similar redundancy level, leading to a comparable number of unique single locus markers (260 vs. 247).
The two mapping populations were shown to have 70 common polymorphic DArT markers that consistently aligned homologous linkage groups with regard to their identity and orientation for all 17 linkage groups of apple (Fig. 2). Several cases of minor differences in marker order were observed, depicted as the crossing lines in Fig. 2. The overall consistency in the identity and orientation of the linkage groups and the marker order show that the DArT markers support the alignment of the mapping populations.
DNA sequencing of DArT markers and redundancy estimation
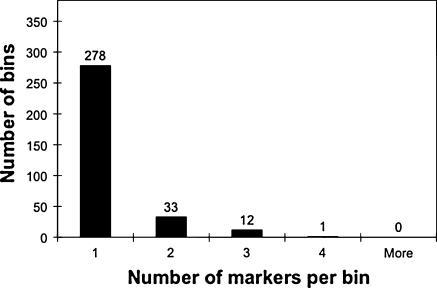
In the mapping process, DArT markers were classified as redundant when they showed identical scores among the progeny, leading to clustering at one genetic position. Such clustering may be caused by high DNA sequence similarity or tight genetic linkage of the markers. To obtain an estimate of sequence-based redundancy, we sequenced 384 clones from the PstI/AluI DArT array. We performed a local pairwise “blast” of all of these sequences and then clustered them into bins of highly similar sequences (e < 1.0E-50), using a Perl script developed in-house (DArT PL unpublished). As Fig. 3 shows, the number of clones per bin varied from one (278 bins) to four (one bin only). There were 324 sequence bins identified; therefore, the percent of redundant clones in the initial sampling of the 384 clones was 15.6% when based on the sequences. Since these 384 sequenced clones represent only 2.6% of the 14,592 clones used for printing on the slides, the real redundancy was higher. Fitting the observed data from Fig. 3 to a Poisson distribution, we deduced that sequence similarity led to approximately 50% redundancy within the total set of clones.
Fig. 3.
Frequency distribution of 384 DNA-sequenced DArT markers from the standard PstI/AluI method. If DNA sequences of one or more clones were highly homologous to each other, then these clones were clustered into a common bin. The 384 markers provided 324 bins
Comparison of these DNA-sequences to sequences in NCBI GenBank databases showed that close to 90% of BlastN and TblastX searches returned highly significant similarities to EST sequences (Online Resource 2). This indicates that the PstI/AluI DArT clones were derived mainly from genes or gene-like sequences.
Alternative complexity reduction method
The high proportion of redundant markers hampers the statistical and genetic analyses of the data. Therefore, we wondered whether marker redundancy could be decreased by a less stringent complexity reduction method and whether such an alternative approach could also be useful in increasing genome coverage. The standard complexity reduction method was based on PstI/AluI digestion. The alternative method that we examined here was based on PstI/EcoRI/MboI digestion, which resulted in a larger number of fragments, so the chance that a clone was sampled multiple times diminished. The alternative array was used for the genotyping of 244 progenies of population 2000–2012, all of which had also been genotyped with the standard method. The maternal map of the heterozygous parent 1980-015-025 was constructed using DArT markers from both complexity reduction methods. Table 2 shows the number of spotted clones per complexity reduction method, the number of polymorphic maternal markers obtained, the number of maternal markers that could be mapped and the number of ‘unique’ markers. Figure 2 shows the resulting maps. Table 2 and Fig. 2 led to the following conclusions:
The degree of redundancy was lower for the alternative method;
The two methods gave a similar genome coverage;
There are no clear indications that the two methods differ in the genomic regions for which they raise polymorphic markers;
Performance of both methods increased genome coverage compared to application of one method only.
Table 2.
Number of DArT clones and resulting markers from two complexity reduction methods for the maternal map of population 2000–2012
| Complexity reduction method | Number of spotted clones | Number (and percentagea) of segregating markers | Number of mapped markers | Number (and percentageb) of redundant markers | Number (and percentagea) of mapped markers after removal of the redundant ones |
|---|---|---|---|---|---|
| Standard | 14,592 | 547 (3.7%) | 542 | 282 (52%) | 260 (1.8%) |
| Alternative | 6,144 | 105 (1.7%) | 104 | 24 (23%) | 80 (1.3%) |
aas % of the number of spotted clones
bas % of the number of mapped markers
Discussion
Comparison of DArT with other marker technologies
After being initially developed for rice, DArT markers have been developed for many additional plant species (Jaccoud et al. 2001; Xie et al. 2006). These include barley (Wenzl et al. 2004), wheat (Akbari et al. 2006; White et al. 2008), cassava (Xia et al. 2005), Arabidopsis (Wittenberg et al. 2005), pigeon pea (Yang et al. (2006), oat (Tinker et al. 2009), sorghum (Mace et al. 2008) and many others (collated at www.diversityarrays.com/publications.html). The present study demonstrates the performance of DArT technology for low-cost, high-throughput genotyping in apple (Malus).
Several lines of evidence support the utility of DArT for apple genomics studies. First, the call rate and reproducibility appear to be high. Non-DArT marker data required repeated examinations for identification of erroneous data, consuming many months of labour. DArT markers, however, did not require this laborious scrutinising; DArT genotyping was fully automated and more accurate than other marker systems. The non-DArT markers were gel-based systems (i.e. RFLP, RAPD, AFLP, SSR), and were scored manually. Second, the DArT markers integrated smoothly into the existing Prima × Fiesta map. Only one marker could not be placed (0.4%). This percentage is very low compared to that found in other marker systems, such as RFLP, RAPD, AFLP and SSRs. Also, DArT markers were robust among different mapping populations, allowing for map alignment.
The standard DArT array gave similar numbers of non-redundant markers in the two mapping populations, indicating that it is robust over populations. Moreover, the majority of these markers were population-specific, indicating that the extensive pool of clones that are not polymorphic in one population are a vast reservoir of possible new markers in other populations. Thus, the DArT array is applicable over a wide range of mapping populations. The number of unique markers is therefore expected to increase further in more extensive studies.
DArT lends its general applicability for wide germplasm coverage to the hybridisation of PCR fragments that are hundreds of basepairs long. Polymorphisms are based on the presence or absence of restriction sites and are insensitive to SNPs and short indels in the hundreds of basepairs between the restriction sites (Wittenberg et al. 2005). This is reflected by the sequence data. Map-redundant DArT markers were often based on clones that differed in size and sequence but nevertheless showed identical segregation patterns in mapping. For instance, the three markers 183247, 183997 and 184057 showed identical segregation patterns and therefore mapped to the same position (LG10, Prima × Fiesta, LG4, 44 cM; Fig. 2, only 183247 is shown). The three DArT fragments varied in size (479, 469 and 502 bp, respectively) and exhibited some sequence polymorphism, as occurs for different alleles of a single gene. Their map-redundancy was confirmed at the sequence level, as “blasting” matched them to the same gene (Online Resource 2). This insensitivity to SNPs and short indels in the probe makes DArT robust for applications on genetically diverse germplasm, in contrast to the sensitivity of SNP arrays to similar polymorphisms. The proportion of informative markers from SNP arrays drops quickly when applied to germplasm that is genetically distinct from accessions that were used in the design of the SNP array. DArT appears to be less prone to this limitation.
DArT and SNP platforms are both suitable for high-throughput genotyping, benefiting from automated scoring and data quality checks. The first small-scale SNP arrays for Malus were developed recently for Golden Delicious (Micheletti et al. 2011). Recently, initiatives have been undertaken for worldwide collaboration on the development of large-scale, multiple accession-based arrays. We expect that DArT will also play a longer term role because of its own specific advantages, including wide applicability, as discussed above, and low cost even when small numbers of accessions have to be genotyped.
Impact of pedigree structure
Although the standard DArT array gave similar numbers of non-redundant markers for both mapping populations (247 for Prima × Fiesta vs. 260 for 2000–2012), differences were observed in the distribution of these markers among the parents. Whereas the proportion of maternal, paternal and bi-parental markers were similar in Prima × Fiesta (35, 32 and 33%, respectively), unbalanced proportions were observed in 2000–2012, with relative under-representation of paternal markers and over-representation of bi-parental markers (35, 20 and 45%). These differences in representation reflect differences in the pedigree structures of these populations (Fig. 1). The father of population 2000–2012 arose from a cross between two sibs, resulting in an expected level of homozygosity of approximately 25%. This parent would yield no segregating markers in these homozygous regions. In addition, the parents of population 2000–2012 have common ancestors, Golden Delicious and Ingrid Marie, reducing levels of allelic diversity. This increases the number of bi-parental markers that segregate for both the father and the mother. Prima and Fiesta lack such recent common ancestors. The distribution of segregating DArT markers among the parents thus reflects the pedigree structures, further supporting the suitability of DArT for genetic studies.
Redundancy
Markers that perfectly co-segregate do not provide additional genetic information, but slow down the mapping process and lead to the statistical overweighting of that specific position.Redundant markers were therefore removed. This reduced the number of polymorphic markers used in the mapping process by about half. Markers that segregated in both populations needed to be preferentially retained to facilitate linkage map alignment.
Clustering of DArT markers can be caused by: (1) an absence of recombination between the markers due to tight genetic linkage; (2) sequence identity; or (3) high sequence similarity derived from different alleles from different apple accessions. The effective population size of Prima × Fiesta was 121, which can only partially explain the current high degree of redundancy in the DArT markers as a result of a lack of recombination between markers. Consequently, sequence identity and high sequence similarity were likely to be significant sources of redundancy. This is consistent with our estimation from Fig. 3 that sequence identity indeed led to approximately 50% redundancy. Based on the sequence information, we conclude that the clustering of PstI–AluI markers is mainly due to sequence similarity among the markers, in addition to genetic linkage.
In classifying markers as “redundant” using a mapping process, more than just similar mapping positions were taken into account. For a proper classification, the parental origin and linkage phase have to be identical too. For example, JoinMap mapped the bi-parental DArT markers 183313 and 184063 within 0.1 cM from each other. Considering the size of the Prima × Fiesta population (n = 121 for the DArT markers) and the type of marker (bi-parental), both markers belonged to the same genetic bin; there was no evidence that they mapped to different positions. Their scores did not indicate any recombination. Although this suggests that they are identical, their linkage phase was different: they were in repulsion phase, indicating that these markers come from different genomic positions. Indeed, DNA sequencing confirmed a substantial sequence difference of the two clones (Online Resource 2).
The positive side of redundancy is that the underlying markers can be regarded as repeats when present on the same slide and thus confirm validity of marker scores. The co-segregation of markers with highly similar sequences highlights the high reliability of the DArT markers.
DArT markers mainly derived from gene rich regions
Our analysis of the sequences shows that 90% of the sequenced markers are highly homologous to mRNA sequences that are deposited in EST databases (Online Resource 2). This indicates that the set of DArT markers is highly enriched for genic regions. This phenomenon has been observed previously in other species like barley (Wenzl et al. 2006), wheat (Akbari et al. 2006) and sorghum (Mace et al. 2008). DArT markers are predominantly located in gene-rich islands in the subtelomeric regions of chromosomes. This is not surprising, as the PstI enzyme is methylation-sensitive (Gruenbaum et al. 1981), and targets hypomethylated, low-copy sequences, which occur primarily in the gene-rich regions (Feng et al. 2010).
Comparison of complexity reduction methods
The standard DArT array developed here provided moderate coverage of the apple genome (Fig. 2), allowing for a quick start in QTL mapping experiments. Coverage, however, was not uniform across the entire genome. Enhancing the array with more clones could increase genome coverage, but using the standard complexity reduction method to do so would not be efficient, due to sequence redundancy. Therefore, we tested an alternative complexity reduction method in order to develop more DArT markers and improve the genome coverage.
Whereas the standard method only amplified PstI–PstI fragments, the alternative method amplified PstI–EcoRI fragments as well as PstI–PstI fragments, yielding higher complexity with the alternative method than with the standard method. We simulated the number of fragments that would be amplified using the published whole genome sequences of Arabidopsis thaliana, and found that the alternative would result in approximately 4.5 times more amplicons in Arabidopsis than the standard method (Mark Fiers, unpublished). The higher complexity of the alternative method and the printing of fewer of these amplicons resulted in less redundancy for population 2000–2012 (23%), compared to the standard method (52%).
Despite the lower redundancy, the percentage of clones leading to unique single locus markers was low for the alternative method (1.3%) compared to the standard method (1.8%; Table 2). The most likely explanation is an unfavourable signal-to-noise ratio due to the higher degree of complexity. The analysis of raw image data, looking at the noise level, supports this explanation (data not presented). Another possible explanation is a reduction in polymorphism due to variation in methylation. Whereas the standard procedure only amplifies PstI–PstI segments, the alternative method also amplifies PstI–EcoRI segments. The EcoRI enzyme is methylation-insensitive, and thus possible polymorphism in methylation is not used. Wittenberg et al. (2005) ascribed up to 8% of the polymorphisms of the alternative method to differences in methylation in Arabidopsis thaliana.
Importantly, a single standard DArT assay was clearly capable of providing reasonable genome coverage, as exemplified by the 17 linkage groups of the Prima × Fiesta map, with non-redundant markers distributed across all of the chromosomes, albeit unevenly (Fig. 2).
Linked research
The genetic maps shown here are the basis for several QTL studies that are currently underway, examining metabolomics (Khan et al., submitted), disease resistance, fruit quality traits and low allergenicity (Van de Weg et al., in prep.). Additionally, the standard DArT assay is being applied worldwide to a number of mapping populations and to the construction of consensus genetic maps (Van Dyk et al., in prep.). All the markers in this study are being sequenced and will be aligned with the apple genome sequence (Velasco et al. 2010). This will allow for the integration of the genetic positions of phenotypic traits with the whole-genome sequence of apple, thereby aiding in the search for underlying genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was carried out with financial support from the Ministry of Agriculture, Nature and Food Quality, The Netherlands. Unpublished data from the European projects DARE and ISAFRUIT were used, which were supported financially by the Commission of the European Communities under the Thematic Priority FAIR5 of the 4th Framework Programme of RTD (Contract no. FP4-FAIR-CT-1997-3898), and 5–Food Quality and Safety of the 6th Framework Programme of RTD (Contract no. FP6-FOOD–CT-2006-016279), respectively. Also, unpublished data from a Voluntary Contribution project between the Australian National Apple Breeding Program, and Horticulture Australia Ltd, Australia (AP06009) were used. We are grateful to Mark Fiers (Plant and Food Research, New Zealand), for simulating the number of DArT restriction fragments in Arabidopsis, for different combinations of restriction enzymes. We acknowledge also James J. Luby and James M. Bradeen (University of Minnesota) for their contribution in selection of Malus genotypes for construction of libraries for the diversity arrays.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Henk J. Schouten, W. Eric van de Weg these authors contributed equally to this work.
Contributor Information
Henk J. Schouten, Phone: +31-317-481074, FAX: +31-317-418094, Email: henk.schouten@wur.nl
D. Jasper G. Rees, Email: reesj@arc.agric.za
Maria M. Van Dyk, Email: daleen.vandyk@up.ac.za
References
- Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet. 2006;113:1409–1420. doi: 10.1007/s00122-006-0365-4. [DOI] [PubMed] [Google Scholar]
- Aljanabi SN, Forget L, Dookun A. An improved and rapid protocol for the isolation of polysaccharide- and polyphenol-free sugarcane DNA. Plant Mol Biol Rep. 1999;17:1–8. doi: 10.1023/A:1007692929505. [DOI] [Google Scholar]
- Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red colouration in apple skin. Plant Cell Physiol. 2007;48:958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- Broothaerts W. New findings in apple S-genotype analysis resolve previous confusion and request the re-numbering of some S-alleles. Theor Appl Genet. 2003;106:703–714. doi: 10.1007/s00122-002-1120-0. [DOI] [PubMed] [Google Scholar]
- Broude NE, Zhang L, Woodward K, Englert D, Cantor CR. Multiplex allele-specific target amplification based on PCR suppression. Proc Natl Acad Sci USA. 2000;98:206–211. doi: 10.1073/pnas.98.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bus VGM, Laurens FND, Van de Weg WE, Rusholme RL, Rikkerink EHA, Gardiner SE, Bassett HCM, Plummer KM. The Vh8 locus of a new gene-for-gene interaction between Venturia inaequalis and the wild apple Malus sieversii is closely linked to the Vh2 locus in Malus pumila R12740–7A. New Phytol. 2005;166:1035–1049. doi: 10.1111/j.1469-8137.2005.01395.x. [DOI] [PubMed] [Google Scholar]
- Bus VGM, Rikkerink EHA, van de Weg WE, Rusholme RL, Gardiner SE, Bassett HCM, Kodde LP, Parisi L, Laurens FND, Meulenbroek EJ, Plummer KM. The Vh2 and Vh4 scab resistance genes in two differential hosts derived from Russian apple R12740–7A map to the same linkage group of apple. Mol Breed. 2005;15:103–116. doi: 10.1007/s11032-004-3609-5. [DOI] [Google Scholar]
- Cabe PR, Baumgarten A, Onan K, Luby JJ, Bedford DS. Using microsatellite analysis to verify breeding records: a study of ‘Honeycrisp’ and other cold-hardy apple cultivars. Hortic Sci. 2005;40:15–17. [Google Scholar]
- Calenge F, Faure A, Goerre M, Gebhardt C, Van de Weg WE, Parisi L, Durel CE. Quantitative trait loci (QTL) analysis reveals both broad-spectrum and isolate-specific QTL for scab resistance in an apple progeny challenged with eight isolates of Venturia inaequalis. Phytopathology. 2004;94:370–379. doi: 10.1094/PHYTO.2004.94.4.370. [DOI] [PubMed] [Google Scholar]
- Calenge F, Van der Linden CG, van de Weg E, Schouten HJ, Van Arkel G, Denancé C, Durel CE. Resistance gene analogues identified through the NBS-profiling method map close to major genes and QTL for disease resistance in apple. Theor Appl Genet. 2005;110:660–668. doi: 10.1007/s00122-004-1891-6. [DOI] [PubMed] [Google Scholar]
- Chagné D, Carlisle CM, Blond C, Volz RK, Whitworth CJ, Oraguzie NC, Crowhurst RN, Allan AC, Espley RV, Hellens RP, Gardiner SE. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics. 2007;8:212. doi: 10.1186/1471-2164-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagné D, Gasic K, Crowhurst RN, Han Y, Bassett HC, Bowatte DA, Lawrence TJ, Rikkerink EHA, Gardiner SE, Korban SS. Development of a set of SNP markers present in expressed genes of the apple. Genomics. 2008;92:353–358. doi: 10.1016/j.ygeno.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Costa F, Stella S, Van de Weg WE, Guerra W, Cecchinel M, Dallavia J, Koller B, Sansavini S. Role of the genes Md-ACO1 and Md-ACS1 in ethylene production and shelf life of apple (Malus domestica Borkh) Euphytica. 2005;141:181–190. doi: 10.1007/s10681-005-6805-4. [DOI] [Google Scholar]
- Costa F, Stella S, Van de Weg WE, Dondini L, Pratesi D, Musacchi S, Sansavini S. Map position and functional allelic diversity of Md-Exp7, a new putative expansin gene associated with fruit softening in apple (Malus × domestica Borkh.) and pear (Pyrus communis) Tree Genet Genome. 2008;4:575–586. doi: 10.1007/s11295-008-0133-5. [DOI] [Google Scholar]
- Costa F, Peace P, Stella S, Serra S, Musacchi S, Bazzani M, Sansavini S, Van de Weg WE (2010) QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in an apple mapping population. J Exp Bot 61:3029–3039 [DOI] [PMC free article] [PubMed]
- Dondini L, Pierantoni L, Gaiotti F, Chiodini R, Tartarini S, Bazzi C, Sansavini S. Identifying QTLs for fire-blight resistance via a European pear (Pyrus communis L.) genetic linkage map. Mol Breed. 2004;14:407–418. doi: 10.1007/s11032-004-0505-y. [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:1–15. [Google Scholar]
- Espley RV, Brendolise C, Chagné D, Green S, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KM, Patocchi A, Rezzonico F, Mathis F, Durel CE, Fernández-Fernández F, Boudichevskaia A, Dunemann F, Stankiewicz-Kosyl M, Gianfranceschi L, Komjanc M, Lateur M, Madduri M, Noordijk Y, Van de Weg WE (2010) Genotyping of pedigreed apple breeding material with a genome covering set of SSRs: Trueness to type of cultivars and their parentages. Mol Breed. doi:10.1007/s11032-010-9502-5
- Feng S, Jacobsen S, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZS, Van de Weg WE, Schaart JG, Schouten HJ, Tran DH, Kodde L, Van der Meer IM, Van der Geest AHM, Kodde J, Breiteneder H, Hoffmann-Sommergruber H, Bosch D, Gilissen LJWJ. Genomic cloning and linkage mapping of the Mal d 1 (PR-10) gene family in apple (Malus domestica) Theor Appl Genet. 2005;111:171–183. doi: 10.1007/s00122-005-2018-4. [DOI] [PubMed] [Google Scholar]
- Gao ZS, Van den Weg WE, Schaart JG, Van der Meer IM, Kodde L, Laimer M, Breiteneder H, Hoffmann-Sommergruber K, Gilissen LJWJ. Linkage map positions and allelic diversity of two Mal d 3 (non-specific lipid transfer protein) genes in the cultivated apple (Malus domestica) Theor Appl Genet. 2005;110:479–491. doi: 10.1007/s00122-004-1856-9. [DOI] [PubMed] [Google Scholar]
- Gao ZS, Van de Weg WE, Schaart JG, Van Arkel G, Breiteneder H, Hoffmann-Sommergruber K, Gilissen LJWJ. Genomic characterization and mapping of the apple allergen genes Mal d 2 (thaumatin-like protein) and Mal d 4 (profilin) Theor Appl Genet. 2005;111:1087–1097. doi: 10.1007/s00122-005-0034-z. [DOI] [PubMed] [Google Scholar]
- Gardiner SE, Bus VGM, Rusholme RL, Chagné D, Rikkerink EHA. Apple. In: Kole C, editor. Genome mapping and molecular breeding in plants, Vol 4. Fruits and nuts. Berlin: Springer; 2007. pp. 1–62. [Google Scholar]
- Gianfranceschi L, Koller B, Seglias N, Kellerhals M, Gessler C. Molecular selection in apple for resistance to scab caused by Venturia inaequalis. Theor Appl Genet. 1996;93:199–204. doi: 10.1007/BF00225746. [DOI] [PubMed] [Google Scholar]
- Gisbert AD, Martínez-Calvo J, Llácer G, Badenes ML, Romero C. Development of two loquat [Eriobotrya japonica (Thunb.) Lindl.] linkage maps based on AFLPs and SSR markers from different Rosaceae species. Mol Breed. 2009;23:523–538. doi: 10.1007/s11032-008-9253-8. [DOI] [Google Scholar]
- Gruenbaum Y, Cedar H, Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981;11:2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R. Microsatellite in Malus × domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet. 1997;94:249–254. doi: 10.1007/s001220050407. [DOI] [Google Scholar]
- He Q, Li XW, Liang GL, Ji K, Guo QG, Yuan WM, Zhou GZ, Chen KS, Van de Weg WE, Gao ZS (2010) Genetic diversity and identity of Chinese loquat cultivars/accessions (Eriobotrya japonica) using mapped SSR markers from apple. Plant Mol Biol Rep. doi:10.1007/s11105-010-0218-9
- Heller-Uszynska K, Uszynski G, Huttner E, Evers M, Carling J, Caig V, Aitken K, Jackson P, Piperidis G, Cox M, Gilmour R, D’Hont A, Butterfield M, Glaszmann J, Kilian A (2010) Diversity Arrays Technology effectively reveals DNA polymorphism in a large and complex genome of sugarcane. Mol Breed. doi:10.1007/s11032-010-9460-y
- Hemmat M, Weeden NF, Brown SK. Mapping and evaluation of Malus × domestica microsatellites in apple and pear. J Am Soc Hort Sci. 2003;128:515–520. [Google Scholar]
- Hokanson SC, Szewc-McFadden AK, Lamboy WF, McFerson JR. Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus × domestica Borkh. core subset collection. Theor Appl Genet. 1998;97:671–683. doi: 10.1007/s001220050943. [DOI] [Google Scholar]
- Igarashi M, Abe Y, Hatsuyama Y, Ueda T, Fukasawa-Akada T, Kon T, Kudo T, Sato T, Suzuki M. Linkage maps of the apple (Malus × domestica Borkh.) cvs ‘Ralls Janet’ and ‘Delicious’ include newly developed EST markers. Mol Breed. 2008;22:95–118. doi: 10.1007/s11032-008-9159-5. [DOI] [Google Scholar]
- Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 2001;29:e25. doi: 10.1093/nar/29.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenis K, Keulemans J, Davey M. Identification and stability of QTLs for fruit quality traits in apple. Tree Genet Genome. 2008;4:647–661. doi: 10.1007/s11295-008-0140-6. [DOI] [Google Scholar]
- Kilian A, Huttner E, Wenzl P, Jaccoud D, Carling J, Caig V, Evers M, Heller-Uszynska K, Cayla C, Patarapuwadol S, Xia L, Yang S, Thomson B (2005) The fast and the cheap: SNP and DArT-based whole genome profiling for crop improvement. In: Tuberosa R, Phillips RL, Gale M (eds) Proceedings of the international congress in the wake of the double helix: from the green revolution to the gene revolution, May 27–31, 2003, Bologna, Italy. Avenue Media, pp 443–461
- Kim CS, Lee CH, Shin JS, Chung YS, Hyung NI. A simple and rapid method for isolation of high quality genomic DNA from fruit trees and conifers using PVP. Nucleic Acids Res. 1997;25:1085–1086. doi: 10.1093/nar/25.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GJ, Lynn JR, Dover CJ, Evans KM, Seymour GB. Resolution of quantitative trait loci for mechanical measures accounting for genetic variation in fruit texture of apple (Malus pumila Mill.) Theor Appl Genet. 2001;102:1227–1235. doi: 10.1007/s001220000530. [DOI] [Google Scholar]
- Koller B, Lehmann A, McDermott JM, Gessler C. Identification of apple cultivars using RAPD markers. Theor Appl Genet. 1993;85:901–904. doi: 10.1007/BF00225036. [DOI] [PubMed] [Google Scholar]
- Koller B, Gianfranceschi L, Seglias N, McDermott J, Gessler C. DNA markers linked to Malus floribunda 821 scab resistance. Plant Mol Biol. 1994;26:597–602. doi: 10.1007/BF00013746. [DOI] [PubMed] [Google Scholar]
- Liebhard R, Kellerhals M, Pfammatter W, Jertmini M, Gessler C (2003a) Mapping quantitative physiological traits in apple (Malus × domestica Borkh.) Plant Mol Biol 52:511–526 [DOI] [PubMed]
- Liebhard R, Koller B, Gianfranceschi L, Gessler C. Creating a saturated reference map for the apple (Malus × domestica Borkh.) genome. Theor Appl Genet. 2003;106:1497–1508. doi: 10.1007/s00122-003-1209-0. [DOI] [PubMed] [Google Scholar]
- Mace ES, Xia L, Jordan DR, Halloran K, Parh DK, Huttner E, Wenzl P, Kilian A. DArT markers: diversity analyses and mapping in Sorghum bicolor. BMC Genomics. 2008;9:26. doi: 10.1186/1471-2164-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliepaard C, Alston FH, Van Arkel C, Brown LM, Chevreau E, Dunemann F, Evans S, Gardiner KM, Guilford P, Van Heusden AW, Janse J, Laurens F, Lynn JR, Manganaris AG, Den Nijs APM, Periam N, Rikkerink E, Roche P, Ryder C, Sansavini S, Schmidt H, Tartarini S, Verhaegh JJ, Vrielink-van Ginkel M, King GJ. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor Appl Genet. 1998;97:60–73. doi: 10.1007/s001220050867. [DOI] [Google Scholar]
- Micheletti D, Troggio M, Zharkikh A, Costa F, Malnoy M, Velasco R, Salvi S (2011) Genetic diversity of the genus Malus and implications for linkage mapping with SNPs. Tree Genet Genome. doi:10.1007/s11295-011-0380-8
- Moriya S, Iwanami H, Kotoda N, Takahashi S, Yamamoto T, Abe K. Development of a marker-assisted selection system for columnar growth habit in apple breeding. J Jpn Soc Hort Sci. 2009;78:279–287. doi: 10.2503/jjshs1.78.279. [DOI] [Google Scholar]
- N’Diaye A, Van de Weg WE, Kodde LP, Koller B, Dunemann F, Thiermann M, Tartarini S, Gennari F, Durel CE. Construction of an integrated consensus map of the apple genome based on four mapping populations. Tree Genet Genome. 2008;4:727–743. doi: 10.1007/s11295-008-0146-0. [DOI] [Google Scholar]
- Norelli JL, Farrell RE, Bassett JRCL, Baldo AM, Lalli DA, Aldwinckle HS, Wisniewski ME. Rapid transcriptional response of apple to fire blight disease revealed by cDNA suppression subtractive hybridization analysis. Tree Genet Genome. 2009;5:27–40. doi: 10.1007/s11295-008-0164-y. [DOI] [Google Scholar]
- Patocchi A, Fernández-Fernández F, Evans K, Gobbin D, Rezzonico F, Boudichevskaia A, Dunemann F, Stankiewicz-Kosyl M, Mathis-Jeanneteau F, Durel CE, Gianfrancesci L, Costa F, Toller C, Cova V, Mott D, Komjanc M, Barbaro E, Kodde L, Rikkerink E, Gessler C, van de Weg WE. Development and test of 21 multiplex PCRs composed of SSRs spanning most of the apple genome. Tree Genet Genome. 2009;5:211–223. doi: 10.1007/s11295-008-0176-7. [DOI] [Google Scholar]
- Peil A, Garcia-Libreros T, Richter K, Trognitz FC, Trognitz B, Hanke MV, Flachowsky H. Strong evidence for a fire blight resistance gene of Malus robusta located on linkage group 3. Plant Breed. 2007;126:470–475. doi: 10.1111/j.1439-0523.2007.01408.x. [DOI] [Google Scholar]
- Richards CM, Volk GM, Reilley AE, Henk AD, Lockwood DR, Reeves PA, Forsline PL. Genetic diversity and population structure in Malus sieversii, a wild progenitor species of domesticated apple. Tree Genet Genome. 2009;5:1614–2942. doi: 10.1007/s11295-008-0190-9. [DOI] [Google Scholar]
- Shulaev V, Korban SS, Sosinski B, Abbott AG, Aldwinkle HS, Folta KM, Iezzoni A, Main D, Arús P, Dandekar M, Lewers K, Brown SK, Davis TM, Gardiner SE, Potter D, Veilleux RE. Multiple models for Rosaceae genomics. Plant Physiol. 2008;147:985–1003. doi: 10.1104/pp.107.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silfverberg-Dilworth E, Matasci CL, Van de Weg WE, Van Kaauwen MPW, Walser M, Kodde LP, Soglio V, Gianfranceschi L, Durel CE, Costa F, Yamamoto T, Koller B, Gessler C, Patocchi A. Microsatellite markers spanning the apple (Malus × domestica Borkh.) genome. Tree Genet Genome. 2006;2:202–224. doi: 10.1007/s11295-006-0045-1. [DOI] [Google Scholar]
- Soglio V, Costa F, Molthoff JW, Weemen-Hendriks WMJ, Schouten HJ, Gianfranceschi L. Transcription analysis of apple fruit development using cDNA microarrays. Tree Genet Genome. 2009;5:685–698. doi: 10.1007/s11295-009-0219-8. [DOI] [Google Scholar]
- Soriano JM, Joshi SG, Van Kaauwen M, Noordijk Y, Groenwold R, Henken B, Van de Weg WE, Schouten HJ. Identification and mapping of the novel apple scab resistance gene Vd3. Tree Genet Genome. 2009;5:475–482. doi: 10.1007/s11295-009-0201-5. [DOI] [Google Scholar]
- Tinker NA, Kilian A, Wight CP, Heller-Uszynska K, Wenzl P, Rines HW, Bjørnstad, Howarth CJ, Jannink JL, Anderson JM, Rossnagel BG, Stuthman DD, Sorrells ME, Jackson EW, Tuvesson S, Kolb FL, Olsson O, Federizzi LC, Carson ML, Ohm HW, Molnar SJ, Scoles GJ, Eckstein PE, Bonman JM, Ceplitis A, Langdon T (2009) New DArT markers for oat provide enhanced map coverage and global germplasm characterization. BMC Genomics 21 10:39 [DOI] [PMC free article] [PubMed]
- Van Dyk MM, Soeker MK, Labuschagne IF, Rees DJG (2010) Identification of a major QTL for time of initial vegetative budbreak in apple (Malus × domestica Borkh.). Tree Genet Genome 6: 489–502
- Van Ooijen JW, Voorrips RE (2008) JoinMap ® 4. Software for the calculation of genetic linkage maps in experimental populations of diploid species. Kyazma, Wageningen, The Netherlands
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagné D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouzé P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–841 [DOI] [PubMed]
- Weeden NF, Lamb RC. Identification of apple cultivars by isozyme phenotypes. J Am Soc Hort Sci. 1985;110:509–515. [Google Scholar]
- Wenzl P, Carling J, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A. Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA. 2004;101:9915–9920. doi: 10.1073/pnas.0401076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesná J, Cakir M, Poulsen D, Wang J, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A. A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics. 2006;7:206. doi: 10.1186/1471-2164-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Law JR, MacKay I, Chalmers KJ, Smith JSC, Kilian A, Powell W. The genetic diversity of UK, US and Australian cultivars of Triticum aestivum measured by DArT markers and considered by genome. Theor Appl Genet. 2008;116:439–453. doi: 10.1007/s00122-007-0681-3. [DOI] [PubMed] [Google Scholar]
- Wittenberg AHJ, Van der Lee T, Cayla C, Kilian A, Visser RGF, Schouten HJ. Validation of the high throughput marker technology DArT using the model plant Arabidopsis thaliana. Mol Genet Genomics. 2005;274:30–39. doi: 10.1007/s00438-005-1145-6. [DOI] [PubMed] [Google Scholar]
- Wittenberg AHJ, Van der Lee TAJ, Ben M’Barek S, Ware SB, Goodwin SB, Kilian A, Visser RGF, Kema GHJ, Schouten HJ (2009) Meiosis drives extraordinary genome plasticity in the haploid fungal plant pathogen Mycosphaerella graminicola. PLoS ONE 4(6): e5863. doi:10.1371/journal.pone.0005863 [DOI] [PMC free article] [PubMed]
- Xia L, Peng K, Yang S, Wenzl P, de Vicente MC, Fregene M, Kilian A. DArT for high-throughput genotyping of cassava (Manihot esculenta) and its wild relatives. Theor Appl Genet. 2005;110:1092–1098. doi: 10.1007/s00122-005-1937-4. [DOI] [PubMed] [Google Scholar]
- Xie Y, McNally K, Li C-Y, Leung H, Zhu Y-Y. A high-throughput genomic tool: diversity array technology complementary for rice genotyping. J Int Plant Biol. 2006;48:1069–1076. doi: 10.1111/j.1744-7909.2006.00298.x. [DOI] [Google Scholar]
- Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, Matsuta N. SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet. 2001;102:865–870. doi: 10.1007/s001220000524. [DOI] [Google Scholar]
- Yamamoto T, Kimura T, Soejima J, Sanada T, Ban Y, Hayashi T. Identification of quince varieties using SSR markers developed from pear and apple. Breed Sci. 2004;54:239–244. doi: 10.1270/jsbbs.54.239. [DOI] [Google Scholar]
- Yang S, Pang W, Ash G, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A. Low level of genetic diversity in cultivated pigeon pea compared to its wild relatives is revealed by diversity arrays technology (DArT) Theor Appl Genet. 2006;113:585–595. doi: 10.1007/s00122-006-0317-z. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Barrett BH. Md-ACS1 and Md-ACO1 genotyping of apple (Malus × domestica Borkh.) breeding parents and suitability for marker-assisted selection. Tree Genet Genome. 2008;4:555–562. doi: 10.1007/s11295-007-0131-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.