Post-transcriptional gene regulation by RNA-binding proteins (RBPs) is increasingly recognized to have strong implications for cell physiology. In this paper, the authors took a global approach to elucidate the previously suggested role of the yeast La-related protein (LARP) Slf1p in copper homeostasis. Use of RNA-binding protein immunopurification-microarray (RIP-Chip) allowed the identification of RNA targets for Slf1p, including a set of copper-related transcripts. Interaction of Slf1p with those RNA targets is dependent on the La-motif (LAM), which is the RNA-binding domain for the LARP proteins. Thus, Slf1 stabilizes messages for proteins involved in copper homeostasis and confers increased resistance of cells to copper toxicity. These results highlight the importance of post-transcriptional regulation of factors/pathways implicated in copper homeostasis by a cytoplasmic RBP.
Keywords: RNA binding, La-related protein, copper homeostasis, microarray, yeast
Abstract
The La-motif (LAM) is an ancient and ubiquitous RNA-binding domain defining a superfamily of proteins, which comprises the genuine La proteins and La-related proteins (LARPs). In contrast to La, which binds and stabilizes pre-tRNAs and other RNA polymerase III transcripts, data on function and RNA targets of the LARPs have remained scarce. We have undertaken a global approach to elucidate the previously suggested role of the yeast LARP Slf1p in copper homeostasis. By applying RNA-binding protein immunopurification-microarray (RIP-Chip) analysis, we show that Slf1p and its paralog Sro9p copurify with overlapping sets of hundreds of functionally related mRNAs, including many transcripts coding for ribosomal proteins and histones. Interestingly, among these potential RNA targets were also mRNAs coding for proteins critical for protection of cells against elevated copper concentrations. Mutations introduced in the conserved aromatic patch of the LAM in Slf1p drastically impaired both association with its targets and Slf1-mediated protection of cells against toxic copper concentrations. Furthermore, we show that Slf1p stabilizes copper-related mRNA targets in a LAM-dependent manner. These results provide the first evidence for post-transcriptional regulation of factors/pathways implicated in copper homeostasis by a cytoplasmic RBP.
INTRODUCTION
Slf1 and Sro9 are paralogous RNA-binding proteins (RBPs) that contain the well-conserved La-motif (LAM), a RNA-binding domain found in all eukaryotes except for protists from the Plasmodium genus (Bousquet-Antonelli and Deragon 2009). The superfamily of LAM-containing proteins has been divided into two distinct subfamilies: the genuine, or “authentic,” La proteins and the La-related proteins (LARPs) (Bousquet-Antonelli and Deragon 2009). Human La was first described as an autoantigen in patients suffering from Sjogren's syndrome and systemic lupus erythematosus (Mattioli and Reichlin 1974; Reichlin 1981). Cloning of La revealed that besides the LAM, the genuine La proteins also contain RNA recognition motifs (RRMs), which are juxtapositioned to the LAM (Chambers and Keene 1985). La proteins are known to function in important areas/pathways of noncoding RNA (ncRNA) metabolism. They recognize terminal 3′ oligouridines (3′-UUU-OHs) that are present in newly synthesized RNA polymerase (RNAP) III transcripts such as tRNA precursors and pre-5S ribosomal RNA (rRNA) (for reviews, see Wolin and Cedervall 2002; Bayfield et al. 2010). La assists in the folding of these transcripts and protects them from 3′-exonucleolytic degradation. Recent cocrystal structures of the N-terminal part of human La bound to short RNAs ending in 3′ oligo(U) revealed that the LAM, in concert with the adjacent RRM, forms a binding pocket for terminal Us making specific contacts to the second last U and the 3′-OH group of the last nucleotide (Teplova et al. 2006; Kotik-Kogan et al. 2008). While these data, along with mutational analysis (Alfano et al. 2004; Dong et al. 2004), showed how structural features of the LAM enable La to bind ncRNA targets, they cannot explain implications of La in the translation and metabolism of cellular and viral RNAs not ending in 3′-UUU-OH, such as an internal ribosome entry site (Holcik and Sonenberg 2005) or 5′-terminal oligopyrimidine tract (Schwartz et al. 2004) containing RNAs (Wolin and Cedervall 2002).
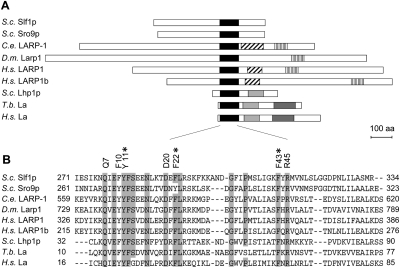
In contrast to the genuine La proteins, studies on LARPs have been rare until recently, even though they fulfill important physiological functions (Bayfield et al. 2010). Based on evolutionary as well as structural characteristics, LARPs have been grouped into four distinct classes: LARPs 1, 4, 6, and 7 (Bousquet-Antonelli and Deragon 2009). Therefore, LARP7 proteins are closest related to genuine La proteins bearing canonical RRMs, while most LARPs contain a RRM-like (RRM-L) domain adjacent to the LAM. In addition, Bousquet-Antonelli et al. identified domains further specifying the different LARPs, most of them with unknown functions like the DM15 motif among LARP1 proteins (Fig. 1A; Nykamp et al. 2008; Bousquet-Antonelli and Deragon 2009). Moreover, except for LARP7, which specifically binds to and stabilizes 7SK small nuclear RNA (snRNA) (He et al. 2008), LARPs appear to function in mRNA rather than ncRNA metabolism. For instance, Caenorhabditis elegans LARP1 regulates the level of mRNA coding for components of the Ras-MAPK–signaling pathway (Nykamp et al. 2008) as well as FEM-3, a message important for sex determination (Zanin et al. 2010). Drosophila melanogaster and human LARP1 proteins affect translation and decay of specific messages and interact with the cytoplasmic poly(A)-binding protein (Blagden et al. 2009; Burrows et al. 2010). Whereas physical interaction of mRNAs with LARP1 has not been directly demonstrated yet, human LARP6 has recently been shown to interact with a conserved stem–loop located in the 5′ UTR of collagen mRNA, presumably promoting localized translation (Cai et al. 2010). Besides the LAM and a RRM-L domain, LARP6 members additionally comprise an S1-like proteins associated (LSA) motif; all of these domains could participate in RNA binding. Human LARP4 and LARP4b proteins (LARP4b is occasionally also referred to as LARP5) interact with the cytoplasmic poly(A)-binding protein (PABPC1) and have a stimulatory role in translation (Schaffler et al. 2010; Yang et al. 2011). LARP4b is thought to stimulate translation by bridging factors at the 3′ end of mRNAs (e.g., PABPC1) with initiating ribosomes (Schaffler et al. 2010); and LARP4 is thought to promote translation, in part, by stabilizing mRNAs (Yang et al. 2011). Interestingly, with the exception of LARP4, every LARP identified to date contains most of the seven conserved amino acid residues in the LAM that are critical for terminal oligo(U)-OH binding by genuine La proteins (Fig. 1B).
FIGURE 1.
Domain structure of selected LARP1 and La proteins and multiple-sequence alignment of LAMs. (A) Protein domain structure as defined by the National Center for Biotechnology Information Conserved domain finder (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cg). Motifs are depicted as boxes: black, LAM; light gray, RRM1; dark gray, RRM2; diagonally striped, RRM-L5; and vertically striped, DM15 motif. (B) Multiple sequence alignment of LAMs. The entire LAM of Slf1p and Sro9p comprised of 63 and 62 amino acids, respectively, is shown. Residues that are >80% conserved are boxed in gray. The identity and position of residues critical for terminal U binding in La proteins are shown on the top (numbering according to the Slf1 LAM). Asterisks highlight three aromatic amino acids that were changed to alanine in the Slf1 La-mutant. Species name abbreviations are as follows: S.c., Saccharomyces cerevisiae; C.e., Caenorhabditis elegans; D.m., Drosophila melanogaster; H.s., Homo sapiens; and T.b., Trypanosoma brucei.
The two LARPs Slf1p and Sro9p in the yeast Saccharomyces cerevisiae are exceptional insofar as they do not comprise any other predicted domains besides the LAM. Most likely originating from an ancient gene duplication (Wolfe and Shields 1997), they are most related to the LARP1 family (Fig. 1B; Bousquet-Antonelli and Deragon 2009). Both proteins have been shown to bind poly(U) and poly(G) RNA in vitro and to associate with polysomes (Sobel and Wolin 1999). Sro9p has been implicated in golgi secretion (Tsukada and Gallwitz 1996), budding (Imai et al. 1996), cytoskeleton formation (Kagami et al. 1997), and RNAP II–mediated transcription (Tan et al. 2000). Recently, it has been reported that Sro9p shuttles between the nucleus and the cytoplasm and that it associates with actively transcribed chromatin, supporting a model in which the RBP guides mRNAs through the process of gene expression, from nuclear transcription to cytoplasmic translation (Rother et al. 2010). However, potential Sro9p mRNA targets have not been identified to date. Much less is known about Slf1p. It was originally described in a complementation study screening for factors suppressing the cup14 mutation (Yu et al. 1996). Cup14 mutants showed increased sensitivity to elevated copper concentrations but grew normally when SLF1 was overexpressed. Likewise, slf1Δ cells were sensitive to elevated copper concentrations and lacked the ability to deplete copper from the medium (Yu et al. 1996).
RBP immunopurification (RIP)-microarray (RIP-Chip) experiments have been previously performed to define potential RNA targets for the genuine La protein in yeast (Lhp1) (Inada and Guthrie 2004). Lhp1p was mainly associated with RNAP III transcripts, but unexpectedly, it also interacted with some mRNAs, mainly coding for proteins acting in translation (Inada and Guthrie 2004). Since Slf1 is linked to copper physiology, we hypothesized that this yeast LARP may preferentially interact with mRNAs and possibly affect the expression of messages coding for proteins linked to copper resistance. We have therefore performed a RIP-Chip analysis to define potential RNA targets of Slf1 and Sro9 in yeast; this represents the first systematic approach to define potential RNA targets for LAM containing proteins outside of the genuine La proteins. We found that both Slf1p and Sro9p were reproducibly associated with mRNAs coding for functionally related proteins but only marginally with ncRNAs, as previously seen for the genuine La protein. Among the potential mRNA targets were messages coding for key regulators of the copper-detoxification system that were specifically associated with Slf1p and that may relate to the copper-resistant phenotypes observed with SLF1 overexpressing cells. We further demonstrate that a functional LAM is critical for the association of Slf1p with mRNAs and for the copper phenotype observed upon SLF1 overexpression. Finally, we provide evidence that Slf1p stabilizes specific mRNA targets from RNA decay in a LAM-dependent manner. In conclusion, we propose that Slf1 enhances the stability of mRNAs, which includes messages coding for proteins involved in copper homeostasis, possibly conferring increased resistance to toxicity caused by excess of copper.
RESULTS
A functional LAM is crucial to mediate the Slf1-specific copper-sensitivity phenotype
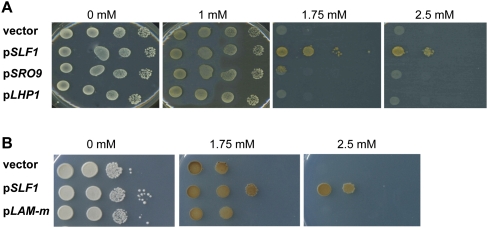
It was previously reported that overexpression of SLF1 can induce “hyper-resistance” of yeast cells to elevated copper salts in the medium (Yu et al. 1996). We could confirm that overexpression of SLF1 on a plasmid confers increased resistance of BY4741 wild-type (WT) cells to CuSO4 (Fig. 2A) but not to cadmium chloride, which is another toxic divalent metal ion (data not shown). Moreover, the copper phenotype was selective for SLF1 as overexpression of neither the paralogs SRO9 nor LHP1, encoding the yeast La protein, changed sensitivity of cells to CuSO4 (Fig. 2A).
FIGURE 2.
Copper resistance of SLF1 overexpressing cells depends on a functional LAM. (A) Yeast cells harboring a plasmid for gal-dependent expression of SLF1, SRO9, LHP1, or the empty vector were serially diluted (1:10) and spotted on SCgal-Ura plates containing indicated concentrations of CuSO4. (B) Same assay comparing WT (pSLF1) and Slf1 La-mutant (pLAM-m) expressing cells. Pictures were taken after incubation of plates for 3–5 d at 30°C.
To test whether the LAM of Slf1 might be involved in this copper-resistance, we recorded the phenotype of WT cells that overexpress a LAM-mutant version of SLF1 (LAM-m). In this mutant, three highly conserved codons for the aromatic amino acids predicted to reside within an aromatic patch on the surface of the LAM were mutated to alanine (Fig. 1B). These residues were previously shown to be critical for high-affinity in vitro binding of genuine La proteins to RNA containing terminal Us (Alfano et al. 2004; Dong et al. 2004; Teplova et al. 2006; Kotik-Kogan et al. 2008). Overexpression of LAM-m did not increase resistance to copper salt (Fig. 2B), indicating that a functional LAM is necessary for SLF1-mediated resistance to copper. Noteworthy, immunoblot analysis against the epitope-tagged proteins revealed that both WT and LAM-m Slf1p were similarly expressed in cells, and hence, the lack of copper resistance is not due to the absence or degradation of the mutant protein (see Fig. 4A,B, below; data not shown).
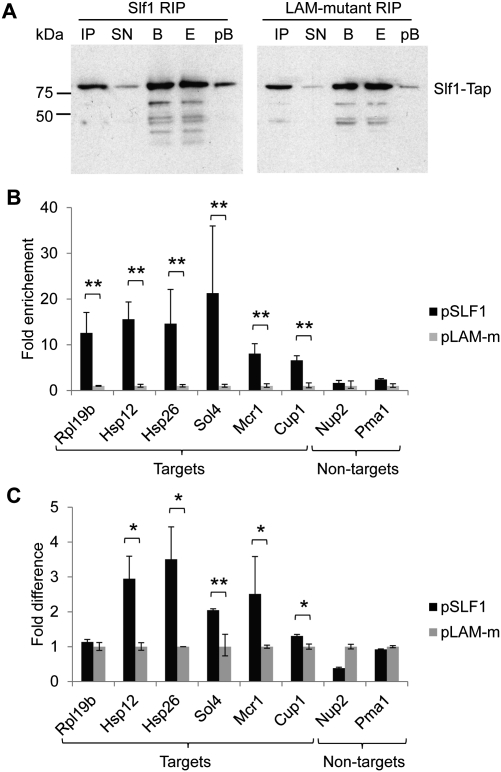
FIGURE 4.
Association of mRNAs with Slf1p depends on a functional LAM. (A) Immunoblot analysis following RIPs with TAP-tagged Slf1 (left) and the Slf1 LAM-mutant (right) expressed in slf1Δ cells using the peroxidase-anti-peroxidase soluble complex (PAP; see Materials and Methods). Lanes are as follows: IP, input yeast extract; SN, supernatant after incubation of extracts with IgG beads; B, captured beads; E, eluate from the beads; and pB, beads after elution of proteins with SDS-EDTA. (B) Four independent RIPs were performed with WT (pSLF1) or LAM-mutant (pLAM-m) SLF1, and the indicated mRNAs were quantified in RIP eluates by real-time qRT-PCR with cycle threshold (Ct) values normalized to mitochondrial 21S rRNA. Depicted is the fold difference in the abundance between WT and LAM-mutant RIP eluates. P-values were calculated based on ΔCt-values (t-test). **P < 0.001. (C) Comparative analysis of mRNA levels in the extracts (=input) used for RIP affinity purifications by qRT-PCR (two biological replicates). Mitochondrial 21S rRNA was used for normalization and averaged data for LAM-m was set to 1 (y-axis). P-values were calculated based on ΔCt-values (t-test). *P < 0.01, **P < 0.001.
Identification and characterization of Slf1p and Sro9p RNA-targets
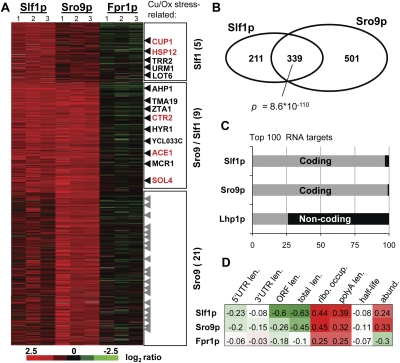
Since overexpression of SLF1 but not SRO9 results in increased copper resistance of cells, we reasoned that a comparative analysis of the putative RNA targets of both proteins could reveal specific Slf1-dependent transcripts, which may contribute to copper resistance. We therefore performed RIP with C-terminal tandem-affinity purification (TAP)–tagged Slf1 and Sro9 proteins expressed under the control of their endogenous promoters, and determined associated RNAs with yeast DNA oligo arrays (see Materials and Methods). To this end, we competitively hybridized Cy5-labeled complementary DNAs (cDNAs) obtained by reverse transcription from copurified RNA and Cy3-labeled cDNAs received from extract RNA (input). In this setup, the ratio of the two cDNA populations at a given array element reflects the enrichment of the corresponding RNA during the RIP procedure (see Materials and Methods; Gerber et al. 2004, 2006). We performed three independent affinity isolations with Slf1-TAP and Sro9-TAP. To control for unspecific binding of RNA to the beads or the affinity-tag, we also performed RIP with Fpr1-TAP, a peptidyl-prolyl cis-trans isomerase not binding to RNA (San Paolo et al. 2009). Based on the high degree of similarity between Slf1p and Sro9p (37% overall amino acid identity, 57% within the LAM), we expected that the two RBPs would be associated with similar sets of target RNAs. Indeed, among the 550 and 840 transcripts that were reproducibly associated with either Slf1 or Sro9, respectively, 339 transcripts were associated with both proteins (P = 8.6 × 10−110, Fisher's exact test) (for procedure to select associated transcripts, see Materials and Methods; Fig. 3A,B; for a list of selected transcripts, see Supplemental Table 1). Most of the respective features (>95%) represented mRNAs, whereas ncRNAs such as rRNAs, tRNAs, and snoRNAs were clearly underrepresented (Fig. 3C). This result is therefore in contrast to data obtained in similar RIP-Chip experiments defining the potential RNA targets for the genuine La protein in yeast (Lhp1), which mainly associated with RNAP III transcripts (Inada and Guthrie 2004). Lhp1 is a nuclear protein, whereas Sro9 and Slf1 are mainly cytoplasmic proteins (see below), and therefore, the protein's localizations are in line with the preferential selection for nuclear and cytoplasmic RNA targets, respectively. However, whether the different RNA associations seen among LARPs depend on their localization, on different RNA-binding specificities, or on additional proteins associated with RNP complexes in vivo needs further investigation.
FIGURE 3.
Slf1p and Sro9p bind to overlapping sets of mRNAs. (A) Heat map depicting the log2 ratios for RNA enrichment in three biological RIP replicates with Slf1p, Sro9p, and the negative control Fpr1p (columns). Rows are features representing transcripts and were grouped into Slf1-specific (211 transcripts), Sro9 and Slf1 associated transcripts (339), and Sro9-specific targets (501 transcripts). Each target group is ranked according to increasing P-values. The color code indicates the degree of enrichment (green–red color scale). mRNAs related to copper-homeostasis (red) and/or response to oxidative stress (black) are indicated to the right (for complete list, see Supplemental Table 2). (B) Venn-diagram depicting the overlap between Slf1p and Sro9p mRNA targets. The P-value was calculated with the Fisher's exact test. (C) Bar chart showing the number of coding and noncoding RNA targets of Slf1p, Sro9p, and Lhp1p (Inada and Guthrie 2004) among the top 100 targets (according to log2 enrichment). (D) Pearson correlation coefficients between RIP-enrichments and selected mRNA characteristics. Abbreviations are as follows: Len., the length of the 5′ UTR, 3′ UTR, open reading frame (ORF) and the poly(A) tail (polyA); ribo., ribosome occupancy; half-life, mRNA half-life; and abund., mRNA abundance.
We next searched for common features determining the association of messages with Slf1p and/or Sro9p. We found that the global enrichment of mRNAs during RIP with either RBP positively correlated with ribosome occupancy (Arava et al. 2003), poly(A) tail length (Beilharz and Preiss 2007), and mRNA abundance (Wang et al. 2002); whereas particularly for Slf1p, a negative correlation was observed to the length of mRNAs/coding sequences (Fig. 3D; Nagalakshmi et al. 2008). Thus, the “typical” mRNA associated with yeast LARPs is short in length, abundant, and well translated and bears a relatively long poly(A) tail. Of note, the observed tendency for the selection of well-expressed mRNAs was mainly driven by ribosome and histone encoding transcripts (see below). However, a substantial fraction of Slf1p (29%) and Sro9p (37%) mRNA targets is composed of low abundant messages (one or fewer mRNA copies per cell (Wang et al. 2002), indicating that interactions do not solely reflect expression levels.
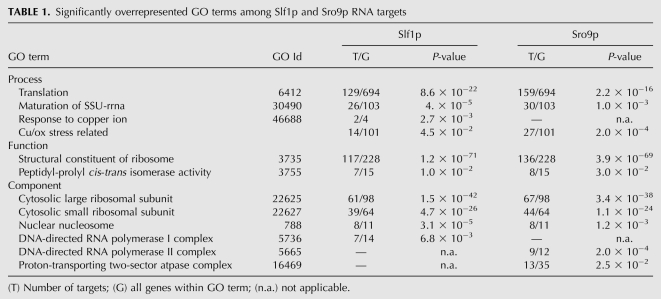
Because many RBPs bind to functionally related sets of messages, which could comprise “post-transcriptional operons” or “RNA regulons” (Keene 2007), we searched for significantly enriched Gene Ontology (GO) terms among proteins encoded by the group of messages that were reproducibly associated with Slf1p and Sro9p (Table 1). Both LARPs were significantly associated with most of the messages coding for components of the small and large subunits of cytosolic ribosomes (Slf1p, 61% of the subunit encoding messages; Sro9p, 68%) (Table 1). Association with transcripts for ribosomal proteins has also been found for Lhp1 (Inada and Guthrie 2004), indicating regulatory functions for all LAM-containing proteins in processes related to protein synthesis. Slf1p as well as Sro9p were also associated with all eight yeast mRNAs coding for histones (GO term “nuclear nucleosome”), which was not observed for yeast Lhp1 (Inada and Guthrie 2004). Slf1p and Sro9p target lists also revealed some distinct GO terms: Among the Sro9-specific ones appeared the “DNA directed RNAP II core complex” (P < 0.0002), whereas Slf1 targets were enriched for mRNAs coding for the DNA directed RNAP I core complex (P < 0.0068). Slf1, but not Sro9-targets, were affiliated with the GO term “response to copper ion” (P = 0.002, Fisher's exact test), which includes two transcripts (CUP1, ACE1) that encode key regulators of the cellular response to excess copper. Thereby, CUP1 was exclusively associated with Slf1p (P-value [Slf1] = 0.002, P-value [Sro9] = 0.33), while ACE1 was also among the Sro9p targets.
TABLE 1.
Significantly overrepresented GO terms among Slf1p and Sro9p RNA targets
In regard of the copper-resistance phenotype mediated by Slf1, we set out to systematically look for copper-stress-relevant targets and compiled a list of 101 genes coding for proteins involved in copper-homeostasis and/or response to oxidative stress, including enzymes engaged in the oxidative branch of the pentose phosphate pathway (PPP) (for a list of the 101 genes, see Supplemental Table 2). We included oxidative stress-response genes because oxidative stress is one of the main toxic effects caused by copper (Cadenas 1989; Jamieson 1998; Avery 2001) and promotes the PPP-mediated production of nicotinamide adenine dinucleotide phosphate (NADPH) (Shanmuganathan et al. 2004). Fourteen of these 101 genes were among Slf1p-associated messages and hence were slightly overrepresented (P = 0.045, Fisher's exact test). Five of those, CUP1, HSP12, TRR2, URM1, and LOT6, were only associated with Slf1p but not with Sro9p (Fig. 3A). CUP1 encodes a copper metallothionein, which confers high resistance to copper and cadmium (Winge et al. 1985; Jeyaprakash et al. 1991). HSP12 codes for a copper-induced heat shock protein increasing membrane stability under a variety of stress conditions (van Bakel et al. 2005; Welker et al. 2010), and knock-out cells show hypersensitivity to elevated copper concentrations (van Bakel et al. 2005). TRR2, URM1, and LOT6 have not been directly linked to copper homeostasis but play roles in the oxidative stress response (Pedrajas et al. 1999; Goehring et al. 2003; Sollner et al. 2007). Among the nine copper and oxidative stress–related targets that were commonly associated with Slf1p and Sro9p were CTR2, involved in mobilization of vacuolar copper stores (Rees et al. 2004); ACE1 coding for a copper binding transcription factor that activates CUP1 expression (Thiele 1988; Welch et al. 1989); and SOL4, which encodes the second enzyme of the PPP (Godon et al. 1998; Grant 2008) and gets down-regulated upon copper deprivation (van Bakel et al. 2005).
Mutations in the aromatic patch of the LAM impair association of copper-related mRNAs with Slf1p
We wondered whether the association of copper-related messages with Slf1p was dependent on a functional LAM, which is critical for the copper resistance of SLF1 overexpressing cells (Fig. 2B). We therefore performed RIP experiments with slf1Δ cells that express either WT or LAM-m SLF1, and measured association of selected mRNA targets by real-time quantitative reverse transcription PCR (qRT-PCR). Both proteins, Slf1, and the La-mutant were expressed at similar levels, and affinity isolations were equally efficient (Fig. 4A). We compared the enrichment of four copper and oxidative stress–related target messages—namely, HSP12, CUP1, SOL4, MCR1—and two unrelated targets RPL19b and HSP26, the latter coding for a low-abundant heat shock protein. For all of them, we consistently measured between 6.6-fold (CUP1) and 21.3-fold (SOL4) enrichment in WT compared with LAM-m Slf1 RIP experiments across four biological replicates (Fig. 4B). Considerably, no significant enrichment was detected for the two nontarget controls NUP2 and PMA1. These results suggests that the LAM—or more precisely conserved residues thought to be critical for 3′-UUU-OH RNA binding in genuine La proteins—is important for the observed associations of Slf1p with specific mRNAs. However, we wish to note that besides the LAM, other yet uncharacterized RNA-binding motifs in Slf1 could also contribute to RNA-binding (O'Connor and Collins 2006). Moreover, it cannot be excluded that the LAM may mediate indirect binding to RNA via a copurifying RBP.
To investigate whether the substantial decrease in mRNA association with the Slf1 La-mutant could also be reflected by altered mRNA expression levels, we further measured the relative changes of steady-state levels of the selected mRNA targets in the extract used for RIP analysis. qRT-PCR analysis revealed that levels of all copper and oxidative stress–related targets were slightly but significantly higher—between 1.3-fold (CUP1; P = 0.042) and 2.9-fold (HSP12; P = 0.01)—in cells expressing WT compared with LAM-m Slf1 (Fig. 4C). The only Slf1p target for which we could not see significant Slf1-mediated up-regulation was RPL19b. Moreover, the mRNA levels of the Slf1p nontargets PMA1 and NUP2 were also not significantly altered. These results suggest that Slf1p positively affects the expression levels of certain target mRNAs in a LAM-dependent manner—possibly due to association with mRNAs. Notably, the association of mRNAs with Slf1 compared with the La-mutant in RIP experiments is still between fivefold and 10-fold compared with changes of mRNA levels in the input. Thus, the differential association in RIP cannot be explained by the slight difference in the expression of copper-related target mRNAs.
Slf1p positively affects steady state levels of mRNA targets
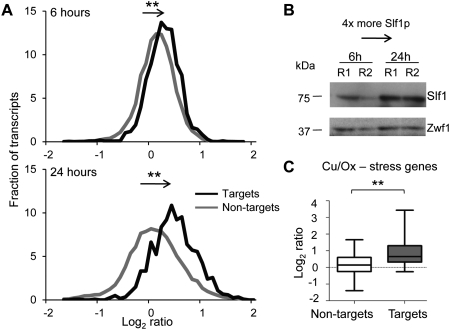
To globally explore the regulatory role of Slf1p on potential target mRNAs, we overexpressed SLF1 on a plasmid in WT cells for 6 and 24 h, and measured the relative changes of mRNA levels compared with control plasmid harboring cells by means of DNA microarrays (Fig. 5A). After 6 h of overexpression, relative expression of Slf1p targets compared to nontargets were on average 1.1-fold increased (P < 0.01, Mann-Whitney test). After 24 h, which further increased expression of Slf1p about four times (Fig. 5B), the Slf1 targets were on average 1.3-fold up-regulated (P < 0.001, Mann-Whitney test). Likewise, 199 Slf1p targets (38% of analyzed targets) were present among the 852 transcripts that were more than 1.5-fold up-regulated after 24 h (P = 5.7 × 10−48, Fisher's exact test), whereas only five Slf1 targets were among the 599 transcripts that were at least 1.5-fold down-regulated (for data for the more than 1.5-fold up- and down-regulated transcripts, see Supplemental Tables 3, 4, respectively). These analyses show that Slf1p preferentially positively affects mRNA expression of selected targets.
FIGURE 5.
Overexpression of SLF1 positively affects Slf1p mRNA target levels. (A) Histogram depicting the fraction (y-axis) of average Cy5/Cy3 fluorescence ratios from duplicate microarray experiments comparing RNA levels in SLF1 overexpressing cells with control cells (empty vector) after 6 h (upper scheme) and 24 h (lower scheme). The bin size for fractions (y-axis) is 0.1 log2 ratios (x-axis). The black line depicts the distribution of Slf1p RNA targets defined from RIP-Chip experiments. The gray line represents nontargets. **P-value < 0.001, Mann-Whitney test. (B) Immunoblot analysis with peroxidase-anti-peroxidase soluble complex (PAP) to detect tagged Slf1p and monitor the expression after 6 and 24 h of induction. Two biological replicates are shown with R1 and R2. Zwf1p is a loading control. (C) Box plots depicting relative differences in mRNA levels of Slf1p nontargets (white) and targets (gray) among copper and oxidative stress–related genes after 24 h of SLF1 overexpression. Averaged relative changes of mRNA expression levels are indicated as log2 ratios. **P-value = 0.0013 (Mann-Whitney test).
Interestingly, 27 of our compiled list of 101 copper and oxidative stress response genes (Supplemental Table 2) were among the 1.5-fold up-regulated genes (P = 1.5 × 10−4, Fisher's exact test). The 14 experimentally defined Slf1 targets of this class were 1.9-fold up-regulated on average and were therefore significantly higher expressed compared with the 87 nontargets of this group (P = 0.0015, Mann-Whitney test) (Fig. 5C). Thus, Slf1p appears to selectively up-regulate target mRNAs coding for proteins involved in copper response. Of note, the greatest up-regulation was observed for the Slf1p-specific copper and oxidative stress–related target HSP12 (11.7-fold).
Slf1p is localized to the cytoplasm and affects the decay of mRNA targets
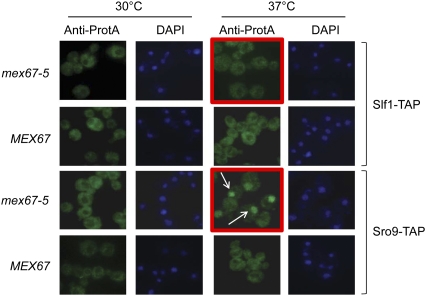
Regulation of mRNA steady-state levels can arise from altered transcription rates and/or mRNA decay. The Slf1p paralog Sro9p is implicated in both transcription and translation (Sobel and Wolin 1999; Tan et al. 2000) and has recently been shown to shuttle between the nucleus and the cytoplasm (Rother et al. 2010). In order to elucidate the mode of Slf1p-mediated mRNA expression regulation, we first investigated the cellular localization of Slf1p under normal growth conditions and after inhibition of mRNA export, a condition in which Sro9p was shown to accumulate in the nucleus (Rother et al. 2010). Slf1 was TAP-tagged in WT and isogenic mex67-5 cells, bearing a temperature-sensitive mutant allele of the mRNA export factor Mex67, which is thought to be the primary mRNA export factor in yeast. In these cells, mRNA export is blocked at nonpermissive temperature (37°C) and poly(A) RNA accumulates in the nucleus (Segref et al. 1997). In contrast to Sro9p, which concentrates in the nucleus of mex67-5 cells at nonpermissive temperature, Slf1p localized to the cytoplasm at both 30°C and 37°C (Fig. 6). In addition, chromatin immunoprecipitation (ChIP) experiments with Slf1-TAP under normal growth conditions or conditions where mRNA export was blocked did not reveal any association with genes coding for specific mRNA targets (data not shown). Thus, in contrast to Sro9p, which is cotranscriptionally recruited to transcribed genes and most likely remains associated with target mRNAs during export to the cytoplasm, our data indicate that Slf1p is a cytoplasmic protein not shuttling to the nucleus and therefore may exert RNA regulatory functions in the cytoplasm.
FIGURE 6.
Slf1p is localized to the cytoplasm. Immunostaining of Slf1-TAP and Sro9-TAP in WT (MEX67) and Mex67 mutant cells (mex67-5) at the permissive (30°C) and nonpermissive (37°C) temperatures. White arrows point to accumulated nuclear Sro9p. DNA was visualized by DAPI staining.
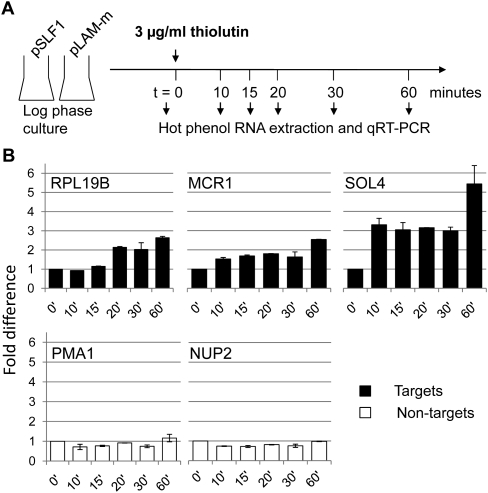
Such a cytoplasmic function may involve implications in mRNA stability control, which can greatly affect mRNA steady-state levels of transcripts (Houseley and Tollervey 2009). To test this hypothesis, we performed RNA decay experiments with slf1Δ cells expressing either WT or LAM-m Slf1 (as a control). We added 3 μg/mL thiolutin (a transcriptional inhibitor for all three RNAPs) (Tipper 1973) to cultures grown to mid-log phase and harvested the cells after 0, 10, 15, 20, 30, and 60 min (Fig. 7A). RNA was isolated from samples, and mRNA levels were determined by qRT-PCR. The two nontargets PMA1 and NUP2 decayed with similar rates in SLF1 and in LAM-m expressing cells (NUP2: t1/2[LAM-m)] = 28 min, t1/2[Slf1] = 32 min; PMA1: t1/2[LAM] = 35 min, t1/2[WT] = 38 min) and were comparable to previously reported half-life rates (Wang et al. 2002). We then monitored the decay of the target RPL19b as well as for the two oxidative stress–related targets SOL4 and MCR1. However, the measurement of RNA decay for the latter transcripts was hampered by an initial up-regulation of their expression after thiolutin addition. Thiolutin and other transcriptional inhibitors are known to impose stress on cells, which prompts residual transcriptional activity to specifically activate genes related to oxidative stress (Grigull et al. 2004). We therefore determined the relative decay, namely, comparing mRNA abundance over time in WT SLF1 and LAM-m expressing cells treated with thiolutin. For all three Slf1 targets, we observed considerable slower degradation in cells expressing WT Slf1 compared with cells expressing the LAM-m (Fig. 7B). These results suggest that the Slf1p-mediated positive regulation of target mRNA levels arises from interference with RNA decay.
FIGURE 7.
Slf1p stabilizes specific mRNA targets. (A) Scheme of RNA decay measurements. Transcription was halted by the addition of thiolutin to slf1Δ cells expressing either WT (pSLF1) or mutant (pLAM-m) Slf1 as a control. (B) Difference of remaining mRNA levels (pSLF1/pLAM-m expressing cells) of Slf1p targets (black) and nontargets (white) at various time points in minutes after thiolutin addition. Representative experiments are shown with standard deviations from three replicate measurements.
DISCUSSION
Copper is an essential trace element for literally all organisms. Its redox properties are critical for cells and make copper an indispensable cofactor for a variety of enzymes acting in electron transfer and antioxidant systems (for reviews, see Pena et al. 1999; Turski and Thiele 2009). However, these beneficial features can easily become threatening to cells as copper concentrations rise: Reactive oxygen species (ROS) and oxidative stress are direct consequences of elevated copper levels (Avery 2001). Cells are therefore equipped with a variety of mechanisms to tightly control intracellular copper levels. This is reflected by the fact that free copper is restricted to less than one atom per cell in S. cerevisiae (Rae et al. 1999). Altered levels of intracellular copper have been associated with a variety of human diseases such as cancer, Alzheimer's, Parkinson's, Creutzfeldt–Jakob disease, and Huntington's (for review, see Tisato et al. 2010). Here we present first evidence for the implication of post-transcriptional gene regulation mediated by a cytoplasmic RBP in the control of copper homeostasis. We have undertaken a global approach to study the roles of the yeast LARPs Slf1p and Sro9p in copper physiology and demonstrate that copper-homeostasis mediated by Slf1p is dependent on a functional LAM. Slf1p stabilizes mRNAs involved in copper response, and we speculate that Slf1 may control a “post-transcriptional operon” that allows cells to deal with elevated copper concentrations.
RNA profiling experiments with SLF1 overexpressing cells showed an increase in the steady-state levels of a significant fraction of our experimentally defined potential Slf1p targets (Fig. 5A). This included messages coding for translation-related proteins as well as factors important for responses to elevated copper concentrations and oxidative stress. Moreover, within the functional group of mRNAs coding for copper and oxidative stress proteins, Slf1p targets were significantly more affected upon SLF1 overexpression compared with nontargets (Fig. 5C). The overlap between up-regulation and Slf1p-association suggests that Slf1p directly stabilizes the affected messages. This interpretation is further supported by the cytoplasmic localization of Slf1p (Fig. 6) and a series of RNA decay experiments (Fig. 7). Importantly, all of the Slf1p-associated transcripts that encode proteins related/connected to copper homeostasis act in processes important for copper detoxification (CUP1, ACE1, HSP12), with the possible exception of CTR2, which is believed to mobilize vacuolar copper stores (Rees et al. 2004). We did not find the association of Slf1p with any messages coding for proteins that are critical in conditions of copper deprivation, such as transcripts of the copper importers Ctr1p, Ctr3p, and Mac1p, the latter controlling transcription of several proteins engaged in copper uptake. Along with the finding that Slf1p stabilizes target transcripts and confers hyper-resistance to toxic copper concentrations upon overexpression, these observations support a model in which Slf1p specifically stabilizes mRNAs coding for proteins important in response to copper stress, facilitating the synthesis of proteins engaged in copper detoxification. Indeed, elevated mRNA levels of the Slf1p targets CUP1 is well known to confer increased resistance against high copper concentrations (Jeyaprakash et al. 1991), and deletion of either ACE1 or HSP12 leads to copper-hypersensitivity (Thiele 1988; van Bakel et al. 2005).
A Slf1p controlled post-transcriptional copper-detoxification operon would be in analogy to a previously identified yeast RBP network triggering coordinated responses to iron deficiency (Puig et al. 2005). Puig et al. (2005) showed that under conditions of low iron, the RBP Cth2 down-regulates mRNAs encoding proteins that participate in iron-dependent processes, ensuring that the availability of limited iron is optimized. Cth2p interacts with AU-rich elements of target mRNAs in the nucleus and guides them to the cytoplasm promoting degradation (Vergara et al. 2010). Thus, it appears that post-transcriptional control of cellular programs dealing with changing concentrations of transition metals could become a common theme in physiology. It will be interesting to determine whether similar LARP-regulated networks for the protection against elevated copper concentrations are present in higher eukaryotes, especially since not only LARPs but also many of the factors/pathways controlling copper homeostasis have been conserved during evolution.
In genuine La proteins or LARPs, RNA-binding activity is mediated by the LAM and an adjacent RRM (Goodier et al. 1997; Ohndorf et al. 2001; Alfano et al. 2004; Dong et al. 2004; Horke et al. 2004). Therefore, seven residues within the LAM mediate high-affinity binding to terminal Us, but only one residue within the juxtapositioned RRM (Fig. 1B; Dong et al. 2004; Teplova et al. 2006; Kotik-Kogan et al. 2008). Although all of these seven residues are fully conserved in Slf1p and Sro9p (Fig. 1B), these proteins were mainly associated with mRNAs that are supposed to bear a poly(A) tail at the 3′ end. Moreover, given that neither Slf1p nor Sro9p associates with poly(A) RNA in vitro (Sobel and Wolin 1999), it seems also unlikely that these proteins recognize the 3′ hydroxyl groups of poly(A) tails in mRNAs, and thus, Slf1/Sro9 may interact with nonterminal sequences in the mRNA.
In contrast to genuine La proteins, neither Slf1p nor Sro9p contains a RRM or any other bioinformatically detectable domain besides the LAM domain (Bousquet-Antonelli and Deragon 2009). Although the LAM itself appears to contribute to selective mRNA binding (Fig. 4), Slf1p and Sro9p may also comprise a cyptic and as-yet-uncharacterized RNA-binding moiety. In this regard, we were not able to see direct and specific interactions of recombinant Slf1p with mRNA targets applying in vitro RNA-binding assays (e.g., RNA pull-down assays) but rather found unspecific binding to RNA, which could be in part mediated by residues outside of the LAM (L Schenk and AP Gerber, unpubl.). Furthermore, the lack of reconstitution of selective RNA–protein interactions in vitro could indicate that the integrity of endogenously formed RNPs is crucial for the identification of relevant in vivo mRNA targets or that additional factors may contribute to selectivity. In the future, it will certainly be of great interest to further characterize the RNA elements and critical amino acid residues that define RNA-protein interactions by LARPs.
MATERIALS AND METHODS
Yeast strains, plasmids, and media
SLF1-TAP (YDR515w), SRO9-TAP (YCL037c), and FPR1-TAP (YNL135c) strains used for RIP (Ghaemmaghami et al. 2003); the isogenic WT strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0); and the SLF1 deletion strain (slf1Δ) were obtained from Open Biosystems (Winzeler et al. 1999). The integrity of the TAP-tagged and slf1 deletion strain was verified by PCR. The strains SRO9-TAP MEX67 and SRO9-TAP mex67-5 have been described earlier (Rother et al. 2010). Strains SLF1-TAP MEX67 (MATα; ura3-52; ade2-1; his3-11,15; leu2-3,112; trp1-1; SLF1-TAP::TRP1-KL, mex67::HIS3; pUN100-MEX67) and SLF1-TAP mex67-5 (MATα; ura3-52; ade2-1; his3-11,15; leu2-3,112; trp1-1; SLF1-TAP::TRP1-KL, mex67::HIS3; pUN100-mex67-5) were obtained by integration of the TAP-tag at the C-terminal end of the SLF1 ORF into the genome of the MEX67 shuffle strain by homologous recombination (Puig et al. 2001). The correct insertion of the TAP tag was also verified by PCR. The strain was transformed with plasmid pUN100-MEX67 or pUN100-mex67-5 (Segref et al. 1997), and pRS316-MEX67 was shuffled out on 5-fluoroorotic acid–containing plates.
Plasmids pBG1805-Slf1 (pSLF1), pBG1805-Sro9 (pSRO9), and pBG1850-Lhp1 (pLHP1) were used to express C-terminally hexa-histidine (6His), hemagglutinin- (HA), and ZZ-domain (ZZ) triple-tagged proteins under the control of a galactose (gal) inducible promoter (Gelperin et al. 2005). The plasmids were obtained from Open Biosystems. A LAM-mutant plasmid of Slf1 (pLAM-m) was constructed by replacing nucleotides 816–1351 of the SLF1 ORF with a fragment, where codons for tyrosine 282 (TAT to GCA), phenylalanine 293 (TTT to GCA), and phenylalanine 314 (TTT to GCA) were mutated to alanine. The DNA fragment was synthesized by Mr. Gene (http://mrgene.com) and cloned into pSLF1 via EagI and EcoRI restriction sites.
Yeast cells were grown in synthetic complete (SC) medium or in SC medium lacking uracil (SC-Ura) supplemented with 2% glucose (Sherman 2002). Gal-dependent expression of an ORF inserted into the pBG1805 plasmid was induced with SC-Ura containing 3% gal and 1% glucose (SCgal-Ura).
Copper-phenotype studies
WT cells (BY4741) were transformed with pBG1805 (empty vector), pSLF1, pLAM-m, pSRO9, or pLHP1 and were grown overnight in SC-Ura at 30°C. Cells equivalent to an optical density at 600 nm (OD600) of 1 were serially diluted (1:10) and spotted onto SCgal-Ura plates containing different concentrations of CuSO4. Plates were further incubated for 3–5 d at 30°C.
RBP affinity isolations
RIP of tagged proteins was carried-out as described (Scherrer et al. 2011). In brief, 1 L cultures of cells expressing endogenously tagged SLF1-, SRO9-, or FPR1-TAP (negative control) were grown in SC to an OD600 of 0.6–0.9 at 30°C, while cells overexpressing WT or LAM-m Slf1-6His-HA-ZZ on a plasmid were grown in SCgal-Ura. Cells were collected by centrifugation, and tagged proteins were captured from cell-free extract with immunoglobulin G coupled Agarose beads (Sigma) for 2 h at 4°C. The TAP-tagged proteins were cleaved-off from the beads by incubation with 80 units of acTEV protease (Invitrogen) for 2 h at 16°C. Slf1-6His-HA-ZZ fusion proteins expressed from plasmids were released from the beads with 500 μL of SDS-EDTA solution (50 mM Tris–HCl at pH 8.0, 140 mM KCl, 10 mM EDTA, 1% SDS) for 10 min at 65°C under strong agitation (1,400 rpm; Eppendorf Thermomixer Compact). The suspension was transferred into a 1.2-mL micro-spin column and centrifuged for 4 min at 2,000g, collecting the eluate. RNA was purified from the extract (input) and from the eluate with the RNeasy Mini/Micro Kit (Qiagen). At least three independent affinity purifications were performed with each strain.
SLF1 overexpression
BY4741 cells bearing plasmid pBG1805-Slf1 or control plasmid pBG1805 were grown in SC-Ura overnight. Cultures were diluted to an OD600 of 0.1 and were further grown in 60 mL of SCgal-Ura for 6 or 24 h at 30°C. Cells were pelleted and washed twice with bi-distilled water, and RNA was immediately isolated from cells by hot phenol extraction and DNAse I digested (Kohrer and Domdey 1991). Slf1Δ cells overexpressing WT or LAM-m Slf1 were grown and harvested after 24 h as described above.
DNA microarrays and data analysis
Microarray data are available at the Stanford Microarray Database (SMD) (Hubble et al. 2009) or at the Gene Expression Omnibus with accession no. GSE27312. Procession of yeast oligo microarrays and hybridization with fluorescently labeled cDNAs was essentially performed as described (Halbeisen et al. 2009). Five micrograms of total RNA isolated from the extract (input) and 50% (∼500 ng) of the affinity purified RNA was reverse transcribed with a mixture of random nonamer and dT20V primers in the presence of 5-(3-aminoallyl)-dUTP and natural dNTPs; cDNAs were covalently linked to Cy3 and Cy5 NHS-monoesters (GE HealthSciences, catalog no. RPN5661), respectively, and competitively hybridized to yeast oligo arrays. Likewise, 7.5 μg of total RNA was labeled to measure relative changes of transcript levels in SLF1 overexpressing cells (pSLF1) compared with cells expressing a control plasmid pBG1805 (see below). Microarrays were scanned with an Axon Instruments Scanner 4200A and analyzed with GenePix Pro 5.1 (Molecular Devices), and data were deposited and computer normalized at the SMD. After filtering for signal over background (>2.5) in the channel measuring total RNA (Cy3), the log2 median ratios from three independent affinity isolations of Slf1, Sro9, and Fpr1 were retrieved from the SMD and exported into Microsoft Excel. To select transcripts that were specifically enriched with either Slf1p or Sro9p, we extracted all features that met the following criteria: average log2 ratio >0.6 in Slf1 or Sro9 affinity isolates, P-value <0.05 (Student t-test) to Fpr1 control isolates, and average log2 ratio in Fpr1 control isolates of <0.2 to remove unspecifically enriched features. Of the 8091 analyzed features, 606 features met the criteria in Slf1 affinity isolates and 923 in Sro9 affinity isolates, representing 550 and 840 different transcripts, respectively. For SLF1 overexpression analysis, log2 median ratios from biological replicates were filtered for regression correlation <0.5 and signal over background >2.5 in both channels with Acuity 4.0 software (Molecular Devices), leaving 7760 features.
Western blots and immunofluorescence
Proteins were separated by SDS–polyacrylamide gel electrophoresis and subsequently transferred to nitrocellulose membranes (BioRad) with a semi-dry electrophoretic transfer cell (Trans Blot SD, BioRad). The membranes were blocked in phosphate-buffered saline–0.05% Tween-20 (PBST) containing 5% low-fat milk and probed over night at 4°C with indicated antibodies and developed with horse radish peroxidase (HRP)-coupled secondary antibodies and the ECL Plus Western Blotting Detection System (Amersham). The peroxidase anti-peroxidase soluble complex (PAP) reagent (Sigma; 1:5.000) was used for detection of the protein A moiety in the ZZ/TAP-tag; rabbit anti-Zwf1p (Sigma, 1:5000) was a loading control.
The subcellular localization of Slf1p and Sro9p under conditions where mRNA export is inhibited was analyzed using strains SLF1-TAP MEX67, SLF1-TAP mex67-5, SRO9-TAP MEX67, and SRO9-TAP mex67-5 by indirect immunofluorescence according to the method described by Rother et al. (2010). 4′,6-diamidino-2-phenylindole (DAPI) and fluorescent staining were analyzed with a Leica DMI6000B fluorescence microscope.
mRNA decay experiments
Slf1Δ cells bearing plasmids pSLF1 or pLAM-m were grown in 100 mL of SCgal–Ura to OD600 = 0.5–0.7, and transcription was stopped by addition of 3 μg/mL thiolutin to the media. Total RNA was isolated with hot-phenol from samples of cells taken after 0, 10, 15, 20, 30, and 60 min of thiolutin addition. Relative mRNAs expression changes were analyzed by qRT-PCR (see below). Therefore, exponential decay curves were fit to points representing the means of two biological replicates, and half-lives were calculated based on the equations described by best-fit curves in Excel. Relative decay was determined comparing mRNA steady-state levels after indicated time points comparing WT and LAM-m overexpressing cells.
SYBR green qRT-PCR analysis
RT was performed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem) with 3–10 ng of affinity purified RNA (RIP experiments) or 50 ng of total RNA isolated from cells. Quantitative PCR was performed with the SYBR Green PCR Master Mix (Applied Biosystems) in an Applied Biosystems 7900HT fast real-time PCR system. Primers sequences are as follows: Rpl19B, 5′-AGACTTGCCGCTTCTGTTGT-3′ and 5′-ACGATGGTTCCGTTCTTGAC-3′; Hsp12, 5′-AAGGTCGCTGGTAAGGTTCA-3′ and 5′-GCTTGGTCTGCCAAAGATTC-3′; Hsp26, 5′-GACTTGTCCCTGTTCCCATC-3′ and 5′-GACACCAGGAACCACGACTT-3′; Sol4, 5′-ATCTTACTCGGATGCGGAGA-3′ and 5′-GCCTTGGGCGAATTATTACA-3′; Mcr1, 5′-CAGCTGGTCGTCAAGCATTA-3′ and 5′-GGTTGCCACTTCCATTTCAT-3′; Cup1, 5′-GAAGGTCATGAGTGCCAAT-3′ and 5′-ATTTCCCAGAGCAGCATGAC-3′; Nup2, 5′-ACATTTGGCTCCTCTGCACT-3′ and 5′-TTTCCGAATGAGAAGGATGG-3′; Pma1, 5′-GGTTACTGCCGTTGTCGAAT-3′ and 5′-TTCCCAGTGACCTTCACCTC-3′; and 21s rRNA, 5′-ATAATTGAGGTCCCGCATGA-3′ and 5′-CTTTCCGTCTTGCTGAAGGT-3′. The following temperature program was used: 50°C for 2 min; 95°C for 15 min, followed by 35 cycles of the sequence 95°C for 15 sec and 58°C for 1 min. Ct-values were normalized to 21S mitochondrial rRNA, whose Ct values were invariant between bait and control in all qRT-PCR experiments performed. Fold changes and standard deviations were calculated according to the ΔΔCt method (Pfaffl 2001). T-tests were performed on ΔCt-values with Excel.
Bioinformatic tools and data processing
Multiple alignments of Slf1p and Sro9p with LARP1- and genuine La representatives were performed using a constraint-based alignment tool (COBALT, www.ncbi.nlm.nih.gov/tools/cobalt) (Papadopoulos and Agarwala 2007). Amino acid sequences were downloaded from the Saccharomyces Genome Database (SGD, www.yeastgenome.org; Cherry et al. 1998) and the NCBI protein database (www.ncbi.nlm.nih.gov/protein). The NCBI Conserved domain finder (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cg) was used to identify domains within the La proteins and LARPs.
Significantly enriched GO terms were identified with the GO Term Finder at SGD (version 0.83) using the default background set (featuring all transcripts in the database that have GO annotations, including ncRNAs, pseudogenes, and transposable elements). A list of genes implicated in copper and oxidative stress response was compiled by downloading all genes affiliated with the GO terms “copper ion binding, response to copper ion, cellular copper ion homeostasis, copper ion import, copper ion transport, cellular response to oxidative stress” and “pentose-phosphate shunt, oxidative branch” (for a compilation of these genes, see Supplemental Table 2).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. K. Harshman and his team at the Center for Integrative Genomics (University of Lausanne, Switzerland) for production of the yeast oligo arrays. We further thank Dr. Bernhard Dichtl and members of the Gerber and Detmar laboratories for fruitful discussions, Dr. Jochen Imig for technical help, and Dr. Marko Jovanovic for critical reading of the manuscript. This work was supported by grants from the Human Frontier Science Program Organization (CDA0048-2005-C) and the Bonizzi-Theler Foundation to A.P.G. and from the European Research Council (ERC) and the Sonderforschungsbereich SFB646 to K.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.028506.111.
REFERENCES
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci 100: 3889–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery SV 2001. Metal toxicity in yeasts and the role of oxidative stress. Adv Appl Microbiol 49: 111–142 [DOI] [PubMed] [Google Scholar]
- Bayfield MA, Yang R, Maraia RJ 2010. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta 1799: 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Gatt MK, Archambault V, Lada K, Ichihara K, Lilley KS, Inoue YH, Glover DM 2009. Drosophila Larp associates with poly(A)-binding protein and is required for male fertility and syncytial embryo development. Dev Biol 334: 186–197 [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Deragon JM 2009. A comprehensive analysis of the La-motif protein superfamily. RNA 15: 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows C, Abd Latip N, Lam SJ, Carpenter L, Sawicka K, Tzolovsky G, Gabra H, Bushell M, Glover DM, Willis AE, et al. 2010. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res 38: 5542–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas E 1989. Biochemistry of oxygen toxicity. Annu Rev Biochem 58: 79–110 [DOI] [PubMed] [Google Scholar]
- Cai L, Fritz D, Stefanovic L, Stefanovic B 2010. Binding of LARP6 to the conserved 5′ stem-loop regulates translation of mRNAs encoding type I collagen. J Mol Biol 395: 309–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Keene JD 1985. Isolation and analysis of cDNA clones expressing human lupus La antigen. Proc Natl Acad Sci 82: 2115–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. 1998. SGD: Saccharomyces Genome Database. Nucleic Acids Res 26: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Chakshusmathi G, Wolin SL, Reinisch KM 2004. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J 23: 1000–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79 doi: 10.1371/journal.pbio.0020079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci 103: 4487–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS 2003. Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Godon C, Lagniel G, Lee J, Buhler JM, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J 1998. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem 273: 22480–22489 [DOI] [PubMed] [Google Scholar]
- Goehring AS, Rivers DM, Sprague GF Jr 2003. Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot Cell 2: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Fan H, Maraia RJ 1997. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol Cell Biol 17: 5823–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM 2008. Metabolic reconfiguration is a regulated response to oxidative stress. J Biol 7: 1 doi: 10.1186/jbiol63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol Cell Biol 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen RE, Scherrer T, Gerber AP 2009. Affinity purification of ribosomes to access the translatome. Methods 48: 306–310 [DOI] [PubMed] [Google Scholar]
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q 2008. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 29: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N 2005. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Horke S, Reumann K, Schulze C, Grosse F, Heise T 2004. The La motif and the RNA recognition motifs of human La autoantigen contribute individually to RNA recognition and subcellular localization. J Biol Chem 279: 50302–50309 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Hubble J, Demeter J, Jin H, Mao M, Nitzberg M, Reddy TB, Wymore F, Zachariah ZK, Sherlock G, Ball CA 2009. Implementation of GenePattern within the Stanford Microarray Database. Nucleic Acids Res 37: D898–D901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai J, Toh-e A, Matsui Y 1996. Genetic analysis of the Saccharomyces cerevisiae RHO3 gene, encoding a rho-type small GTPase, provides evidence for a role in bud formation. Genetics 142: 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada M, Guthrie C 2004. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc Natl Acad Sci 101: 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson DJ 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14: 1511–1527 [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Welch JW, Fogel S 1991. Multicopy CUP1 plasmids enhance cadmium and copper resistance levels in yeast. Mol Gen Genet 225: 363–368 [DOI] [PubMed] [Google Scholar]
- Kagami M, Toh-e A, Matsui Y 1997. SRO9, a multicopy suppressor of the bud growth defect in the Saccharomyces cerevisiae rho3-deficient cells, shows strong genetic interactions with tropomyosin genes, suggesting its role in organization of the actin cytoskeleton. Genetics 147: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Kohrer K, Domdey H 1991. Preparation of high molecular weight RNA. Methods Enzymol 194: 398–405 [DOI] [PubMed] [Google Scholar]
- Kotik-Kogan O, Valentine ER, Sanfelice D, Conte MR, Curry S 2008. Structural analysis reveals conformational plasticity in the recognition of RNA 3′ ends by the human La protein. Structure 16: 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli M, Reichlin M 1974. Heterogeneity of RNA protein antigens reactive with sera of patients with systemic lupus erythematosus. Description of a cytoplasmic nonribosomal antigen. Arthritis Rheum 17: 421–429 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykamp K, Lee MH, Kimble J 2008. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA 14: 1378–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM, Collins K 2006. A novel RNA binding domain in tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol 26: 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohndorf UM, Steegborn C, Knijff R, Sondermann P 2001. Contributions of the individual domains in human La protein to its RNA 3′-end binding activity. J Biol Chem 276: 27188–27196 [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079 [DOI] [PubMed] [Google Scholar]
- Pedrajas JR, Kosmidou E, Miranda-Vizuete A, Gustafsson JA, Wright AP, Spyrou G 1999. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J Biol Chem 274: 6366–6373 [DOI] [PubMed] [Google Scholar]
- Pena MM, Lee J, Thiele DJ 1999. A delicate balance: homeostatic control of copper uptake and distribution. J Nutr 129: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45 doi: 10.1371/journal.pbio.0020079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110 [DOI] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV 1999. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284: 805–808 [DOI] [PubMed] [Google Scholar]
- Rees EM, Lee J, Thiele DJ 2004. Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J Biol Chem 279: 54221–54229 [DOI] [PubMed] [Google Scholar]
- Reichlin M 1981. Current perspectives on serological reactions in SLE patients. Clin Exp Immunol 44: 1–10 [PMC free article] [PubMed] [Google Scholar]
- Rother S, Burkert C, Brunger KM, Mayer A, Kieser A, Strasser K 2010. Nucleocytoplasmic shuttling of the La motif-containing protein Sro9 might link its nuclear and cytoplasmic functions. RNA 16: 1393–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet 5: e1000555 doi: 10.1371/journal.pgen.1000555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffler K, Schulz K, Hirmer A, Wiesner J, Grimm M, Sickmann A, Fischer U 2010. A stimulatory role for the La-related protein 4B in translation. RNA 16: 1488–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer T, Femmer C, Schiess R, Aebersold R, Gerber AP 2011. Defining potentially conserved RNA regulons of homologous zinc-finger RNA-binding proteins. Genome Biol 12: R3 doi: 10.1186/gb-2011-12-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EI, Intine RV, Maraia RJ 2004. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5′-terminal oligopyrimidine mRNA metabolism. Mol Cell Biol 24: 9580–9591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 16: 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmuganathan A, Avery SV, Willetts SA, Houghton JE 2004. Copper-induced oxidative stress in Saccharomyces cerevisiae targets enzymes of the glycolytic pathway. FEBS Lett 556: 253–259 [DOI] [PubMed] [Google Scholar]
- Sherman F 2002. Getting started with yeast. Methods Enzymol 350: 3–41 [DOI] [PubMed] [Google Scholar]
- Sobel SG, Wolin SL 1999. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell 10: 3849–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner S, Nebauer R, Ehammer H, Prem A, Deller S, Palfey BA, Daum G, Macheroux P 2007. Lot6p from Saccharomyces cerevisiae is a FMN-dependent reductase with a potential role in quinone detoxification. FEBS J 274: 1328–1339 [DOI] [PubMed] [Google Scholar]
- Tan Q, Li X, Sadhale PP, Miyao T, Woychik NA 2000. Multiple mechanisms of suppression circumvent transcription defects in an RNA polymerase mutant. Mol Cell Biol 20: 8124–813311027282 [Google Scholar]
- Teplova M, Yuan YR, Phan AT, Malinina L, Ilin S, Teplov A, Patel DJ 2006. Structural basis for recognition and sequestration of UUU(OH) 3′ termini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell 21: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele DJ 1988. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol Cell Biol 8: 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipper DJ 1973. Inhibition of yeast ribonucleic acid polymerases by thiolutin. J Bacteriol 116: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisato F, Marzano C, Porchia M, Pellei M, Santini C 2010. Copper in diseases and treatments, and copper-based anticancer strategies. Med Res Rev 30: 708–749 [DOI] [PubMed] [Google Scholar]
- Tsukada M, Gallwitz D 1996. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J Cell Sci 109: 2471–2481 [DOI] [PubMed] [Google Scholar]
- Turski ML, Thiele DJ 2009. New roles for copper metabolism in cell proliferation, signaling, and disease. J Biol Chem 284: 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Strengman E, Wijmenga C, Holstege FC 2005. Gene expression profiling and phenotype analyses of S. cerevisiae in response to changing copper reveals six genes with new roles in copper and iron metabolism. Physiol Genomics 22: 356–367 [DOI] [PubMed] [Google Scholar]
- Vergara SV, Puig S, Thiele DJ 2010. Early recruitment of AU-rich element (ARE)-containing mRNAs determines their cytosolic fate during iron deficiency. Mol Cell Biol 31: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO 2002. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci 99: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J, Fogel S, Buchman C, Karin M 1989. The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. EMBO J 8: 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker S, Rudolph B, Frenzel E, Hagn F, Liebisch G, Schmitz G, Scheuring J, Kerth A, Blume A, Weinkauf S, et al. 2010. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol Cell 39: 507–520 [DOI] [PubMed] [Google Scholar]
- Winge DR, Nielson KB, Gray WR, Hamer DH 1985. Yeast metallothionein. Sequence and metal-binding properties. J Biol Chem 260: 14464–14470 [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–7139192896 [Google Scholar]
- Wolin SL, Cedervall T 2002. The La protein. Annu Rev Biochem 71: 375–403 [DOI] [PubMed] [Google Scholar]
- Yang R, Gaidamakov SA, Xie J, Lee J, Martino L, Kozlov G, Crawford AK, Russo AN, Conte MR, Gehring K, et al. 2011. La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol Cell Biol 31: 542–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Farrell RA, Stillman DJ, Winge DR 1996. Identification of SLF1 as a new copper homeostasis gene involved in copper sulfide mineralization in Saccharomyces cerevisiae. Mol Cell Biol 16: 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanin E, Pacquelet A, Scheckel C, Ciosk R, Gotta M 2010. LARP-1 promotes oogenesis by repressing fem-3 in the C. elegans germline. J Cell Sci 123: 2717–2724 [DOI] [PubMed] [Google Scholar]