The molecular mechanisms that govern translation initiation to ensure accuracy remain unclear. Here, the authors provide evidence that the subunit-joining step of initiation is controlled in part by a conformational change in the 1408 region of helix h44. Chemical probing of 30S initiation complexes formed with either a cognate (AUG) or near-cognate (AUC) start codon shows that an IF1-dependent enhancement at A1408 is reduced in the presence of AUG. This change in reactivity is due to a conformational change rather than loss of IF1, because other portions of the IF1 footprint are unchanged and high concentrations of IF1 fail to diminish the reactivity difference seen at A1408. These data support a model in which IF1 alters the A1408 region of h44 in a way that makes 50S docking unfavorable, and canonical codon–anticodon pairing in the P site restores h44 to a docking-favorable conformation.
Keywords: ribosome, helix 44, IF1, IF2, IF3, fMet-tRNA, start codon
Abstract
The molecular mechanisms that govern translation initiation to ensure accuracy remain unclear. Here, we provide evidence that the subunit-joining step of initiation is controlled in part by a conformational change in the 1408 region of helix h44. First, chemical probing of 30S initiation complexes formed with either a cognate (AUG) or near-cognate (AUC) start codon shows that an IF1-dependent enhancement at A1408 is reduced in the presence of AUG. This change in reactivity is due to a conformational change rather than loss of IF1, because other portions of the IF1 footprint are unchanged and high concentrations of IF1 fail to diminish the reactivity difference seen at A1408. Second, mutations in h44 such as A1413C stimulate 50S docking and cause reduced reactivity at A1408. Third, streptomycin, which has been shown by Rodnina and coworkers to stimulate 50S docking by reversing the inhibitory effects of IF1, also causes reduced reactivity at A1408. Collectively, these data support a model in which IF1 alters the A1408 region of h44 in a way that makes 50S docking unfavorable, and canonical codon–anticodon pairing in the P site restores h44 to a docking-favorable conformation. We also find that, in the absence of factors, the cognate 30S•AUG•fMet-tRNA ternary complex is >1000-fold more stable than the near-cognate 30S•AUC•fMet-tRNA complex. Hence, the selectivity of ternary complex formation is inherently high, exceeding that of initiation in vivo by more than 10-fold.
INTRODUCTION
Initiation of protein synthesis involves recognition of the start codon by the initiator tRNA in the P site of the ribosome. In bacteria, this process occurs in two major steps and is kinetically controlled by three initiation factors—IF1, IF2, and IF3 (Gualerzi et al. 2001). The first major step is assembly of the 30S initiation complex (30SIC), in which the initiator tRNA (fMet-tRNA) binds the 30S P site and is paired to the start codon, IF2 contacts the 30S shoulder domain and the acceptor end of fMet-tRNA, IF3 interacts with the platform domain near the P site, and IF1 binds the 30S A site (Simonetti et al. 2009). While there is no evidence for an obligate order of ligand binding during 30SIC formation, several studies suggest that, in the predominant pathway, the factors bind before fMet-tRNA and mRNA (Karimi et al. 1999; Pavlov et al. 2008; Milon et al. 2010). The second major step of initiation is docking of the 50S subunit. This involves hydrolysis of GTP by IF2 and dissociation of the factors, yielding a 70S initiation complex (70SIC) ready to enter the elongation phase of translation (Antoun et al. 2003; Grigoriadou et al. 2007a,b; Milon et al. 2008).
Assembly of the 70SIC at the correct start codon is critical because initiation establishes the reading frame for translation. There are two distinct ways in which spurious initiation events can occur. One involves an error in tRNA selection, where the 70SIC is assembled with an elongator tRNA paired to its corresponding codon. The other involves an error in start codon selection, where fMet-tRNA pairs with a codon other than the correct start codon. N-terminal sequencing of products of spurious initiation events suggests that translation in these cases begins with fMet-tRNA (O'Connor et al. 1997, 2001). Furthermore, spurious initiation has been observed to occur from codons termed Class IIA (e.g., AUA, AUC, AUU, ACG, and CUG), which differ from the cognate AUG at only one position (Sacerdot et al. 1996; Sussman et al. 1996). Together, these observations indicate that errors in start codon selection are much more frequent than errors in tRNA selection. The high accuracy of tRNA selection may be largely explained by the fact that IF2 interacts specifically with the acceptor end of fMet-tRNA and discriminates against aminoacyl-tRNAs, while EF-Tu exhibits the opposite binding selectivity (Tanada et al. 1982; Mayer et al. 2001; Antoun et al. 2006a).
Chromosomal mutations that increase spurious initiation have mapped to infC, the gene encoding IF3 (Sacerdot et al. 1996; Sussman et al. 1996; Haggerty and Lovett 1997). Analogous mutations in 16S rRNA have recently been identified, and those with the strongest phenotype cluster to the 790 loop and inhibit IF3 binding, further implicating IF3 in start codon selection (Qin et al. 2007; Qin and Fredrick 2009). In vitro, IF3 negatively regulates both major steps of initiation, strongly inhibiting 70SIC formation in the presence of the near-cognate start codon AUU (Antoun et al. 2006a,b; Grigoriadou et al. 2007b; Milon et al. 2008). While the importance of IF3 in start codon selection is undisputed, the molecular basis for its fidelity function remains unclear. It has been proposed that IF3 “inspects” codon–anticodon pairing in the 30S P site, either directly or indirectly, preferentially destabilizing noncanonical complexes to increase the binding selectivity of the P site (Hartz et al. 1990; La Teana et al. 1993; Meinnel et al. 1999). An alternative view is that IF3 tunes the kinetics of initiation in such a way that the intrinsic difference in stability between a matched and mismatched codon–anticodon helix in the P site is efficiently exploited (Antoun et al. 2006a).
Recent studies indicate that IF1 is also involved in negatively regulating the initiation process to enhance fidelity (Antoun et al. 2006a,b; Milon et al. 2008). It was shown that IF1 slows 50S docking in the presence of AUU and strongly inhibits docking with m002, a model mRNA containing a strong SD closely juxtaposed to the start codon (Milon et al. 2008). These inhibitory effects of IF1 were eliminated with the antibiotic streptomycin. It was hypothesized that IF1 induces a docking-unfavorable conformation of the 30SIC, which can be shifted to a docking-favorable state by cognate codon–anticodon pairing in the P site or by streptomycin. However, the nature of this putative conformational change remained unclear.
In this study, we provide evidence that the subunit-joining step of initiation is controlled in part by a conformational change in h44 of 16S rRNA that occurs in response to start codon recognition. The conformational state of h44 is oppositely influenced by IF1 and streptomycin, providing an explanation for how these ligands exert their effects on 50S docking. The key region of h44 includes A1408, suggesting that the conformational change may directly regulate formation of inter-subunit bridge B2a.
RESULTS
Mutations in h44 stimulate 70SIC formation
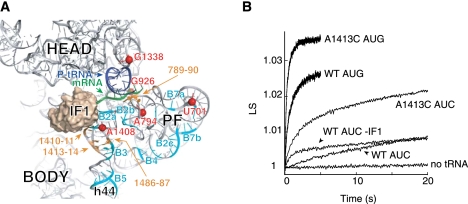
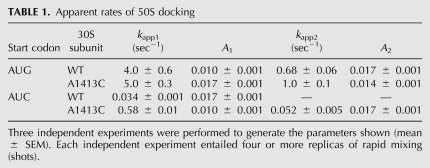
Several mutations that decrease the fidelity of start codon selection were previously identified in h44 (Qin and Fredrick 2009). These mutations lie near the IF1-binding site, between nucleotides that contribute to inter-subunit bridges B2a and B3 (Fig. 1A; Carter et al. 2001; Schuwirth et al. 2005; Selmer et al. 2006). To investigate the effects of these mutations on 70SIC formation in vitro, control (WT) and mutant (A1413C) 30S subunits were purified, and apparent rates of docking of 50S subunits to 30S initiation complexes containing a cognate [30SIC(AUG)] or near-cognate [30SIC(AUC)] start codon were measured, using stopped-flow spectroscopy. Mixing 50S subunits with the WT 30SIC(AUG) resulted in a biphasic increase in light scattering (LS), where the fast (4 sec−1) and slow (0.7 sec−1) phases accounted for ∼40% and 60% of the total signal increase, respectively (Fig. 1B; Table 1). Biphasic increases in LS with similar apparent rates have been reported previously, using 30SICs formed with various mRNAs (Milon et al. 2008). Although the basis of this phenomenon remains unclear, the slow phase may reflect a subpopulation of docking-incompatible complexes that require, for example, an additional component or conformational change before docking can occur (Pavlov et al. 2010). No LS increase was seen in the absence of fMet-tRNA (Fig. 1B), consistent with earlier work demonstrating that 50S docking is completely tRNA-dependent in the presence of IF3 (Antoun et al. 2006b). When the start codon was changed to AUC and the experiment repeated, the rate of 50S docking decreased substantially and the curve became monophasic (kapp = 0.03 sec−1) (Fig. 1B; Table 1). This strong inhibition of 50S docking in the presence of a near-cognate start codon is consistent with previous studies (Milon et al. 2008). Mutation A1413C stimulated 70SIC formation, an effect most pronounced with the near-cognate start codon AUC. For A1413C 30SIC(AUC), the LS increase was biphasic, with kapp1 nearly 20-fold faster than kapp for WT 30SIC(AUC). Values for kapp2 and its amplitude (A2) were similar to the rate and amplitude of the control reaction. In the presence of AUG, mutation A1413C stimulated 70SIC formation somewhat, increasing A1 by 70% and modestly increasing both kapp1 and kapp2. We looked at the effects of two other h44 mutations, A1410G and U1414C. Like A1413C, these mutations stimulated 70SIC formation, particularly in the presence of AUC (data not shown). These data are consistent with the idea that the subunit-joining step of initiation is influenced by the conformation of h44.
FIGURE 1.
Mutation A1413C stimulates 70SIC formation. (A) Tertiary structure of 16S rRNA of the 30S subunit (ribosomal proteins are omitted for clarity), viewed from the subunit interface. The positions of IF1 (tan), P-site tRNA (dark blue), mRNA (green), and nucleotides contributing to intersubunit bridges (cyan, as indicated) are shown. (Orange) Positions of mutations that increase translation from noncanonical start codons (Qin and Fredrick 2009). (Red spheres) Nucleotides identified in this study that exhibit different chemical reactivity in 30SIC(AUG) versus 30SIC(AUC). This image was generated using PDB files 2J02 and 1HR0 (Carter et al. 2001; Selmer et al. 2006). (B) Examples of experiments measuring apparent rates of 50S docking. Control (WT) or mutant (A1413C) 30S subunits (0.075 μM) were pre-incubated for 30 min at 37°C with initiation factors (0.2 μM each), GTP (100 μM), fMet-tRNA (0.2 μM), and mRNA (with start codon AUG or AUC as indicated, 0.5 μM). Then, each 30S complex was rapidly mixed with an equal volume of 50S subunits (0.23 μM) in a stopped-flow apparatus, and scattered light was measured as a function of time. Ordinate values correspond to relative units of light scattering (LS).
TABLE 1.
Apparent rates of 50S docking
It was shown recently that IF1 inhibits 50S docking to 30SICs containing the near-cognate start codon AUU (Milon et al. 2008). This raised the possibility that mutation A1413C stimulated 50S docking indirectly by destabilizing IF1 in the 30SIC. However, the effects of A1413C on docking were not suppressed by adding a 10-fold higher concentration of the factor (2 μM) (data not shown), arguing against the idea that A1413C acts by destabilizing IF1. In our experimental system, omission of IF1 from the WT 30SIC(AUC) increased the apparent rate of 50S docking (Fig. 1B), consistent with the earlier study (Milon et al. 2008). Notably, the stimulatory effect of IF1 omission was less than that of A1413C, suggesting that the mutation does more than reverse the inhibitory effects of IF1.
Structural differences in 30S initiation complexes containing the start codon AUG versus AUC
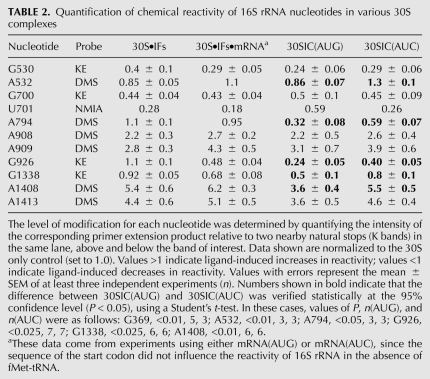
The experiments described above suggested that the rate of 50S docking is influenced by the conformational state of h44. However, whether a conformational change in h44 normally occurs in response to start codon recognition remained unclear. To address this question, we compared the chemical reactivity of 16S rRNA in 30SIC(AUG) versus 30SIC(AUC), using dimethyl sulfate (DMS), kethoxal (KE), and N-methylisatoic anhydride (NMIA) as probes. DMS primarily targets N1 of A, KE targets N1 and N2 of G, and NMIA targets the 2′-OH of ribose (Merryman and Noller 1998; Wilkinson et al. 2006). The former two probes have been used quite extensively to analyze 30S–ligand interactions; however, those studies were largely performed in the context of binary or ternary complexes as opposed to the complete 30SIC. We performed initial experiments in which the 30SIC(AUG) and 30SIC(AUC) were probed in parallel, rRNA was extracted, and modifications in the 16S rRNA molecule were identified and quantified by primer extension. Of ∼110 nucleotides targeted by these probes, six were differentially modified in the 30SIC(AUG) and 30SIC(AUC) reactions (Fig. 1A; Table 2), data that were confirmed in subsequent experiments (see below). One of the nucleotides identified was A1408, which lies in h44 and contributes to inter-subunit bridge B2a.
TABLE 2.
Quantification of chemical reactivity of 16S rRNA nucleotides in various 30S complexes
A1408 exhibits reduced reactivity in the presence of AUG
Previous DMS and KE footprinting studies showed that binding of IF1 to the 30S subunit protects several nucleotides (e.g., G530, A1492, and A1493) and enhances the reactivity of others (e.g., A908, A909, A1408, A1413, and G1487) (Moazed et al. 1995; Dahlquist and Puglisi 2000). These data can be rationalized in light of the cocrystal structure of the IF1-bound 30S subunit (Carter et al. 2001). IF1 forms specific contacts to G530, A1492, and A1493. These interactions require that the latter two nucleotides flip out from h44, which increases the exposure of A1408, lying on the opposite strand. IF1 also distorts h44 further “down,” which alters backbone contacts to A908–A909 and disrupts the noncanonical pair A1413–G1487, explaining the chemical enhancements of these bases.
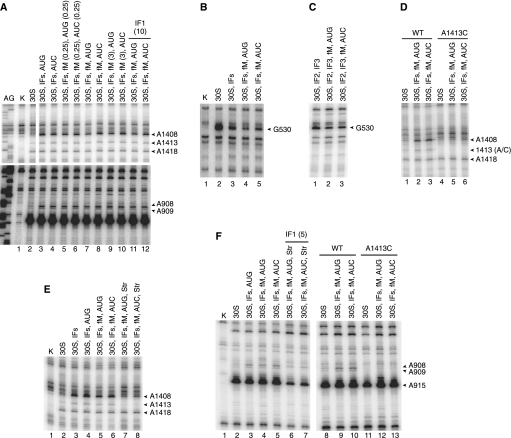
In the 30SIC(AUG), A1408 was found to be hyper-reactive to DMS, but the level of modification was reduced by nearly 50% compared with that in 30SIC(AUC) or in complexes formed in the absence of fMet-tRNA (Fig. 2A; Table 2). Several observations indicate that the reduced reactivity of A1408 is due to a conformational change rather than a decrease in the level of IF1 binding. First, the IF1-dependent enhancements at A908 and A909 were indistinguishable among the complexes (Fig. 2A; Table 2). Second, no loss of protection was seen at G530 in 30SIC(AUG) (Fig. 2B; Table 2). While some degree of protection at G530 in the 30SIC is probably due to P-tRNA (Fig. 2C), in line with previous observations (Moazed and Noller 1990), IF1 is presumably responsible for much of the observed protection in the 30SIC (Fig. 2B). Third, when a 20-fold higher concentration of IF1 (10 μM) was used, the same modification pattern was obtained (Fig. 2A, lanes 11,12). Together, these observations suggest that cognate start codon recognition alters the conformation of A1408 in the 30SIC. Excess fMet-tRNA (3 μM) also failed to change the pattern of reactivity at A1408 (Fig. 2A, lanes 9,10), hence the effects of the mismatch between codon and anticodon cannot be readily suppressed by promoting fMet-tRNA binding.
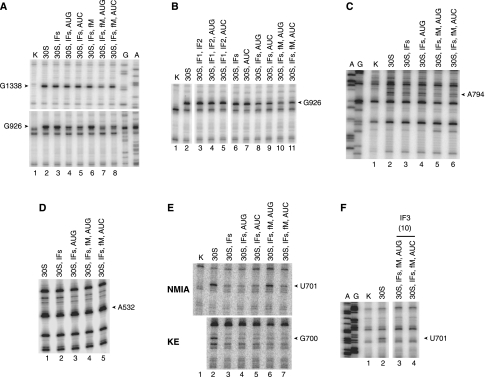
FIGURE 2.
Start codon recognition, mutation A1413C, and streptomycin reduce the reactivity of A1408 in the context of the 30SIC. Complexes were formed by incubating 30S subunits (30S, 0.25 μM) in the absence or presence of various ligands ([IFs] initiation factors, 0.5 μM each; [AUG] mRNA with AUG start codon, 1.0 μM; [AUC] mRNA with AUC start codon, 1.0 μM; [fM] fMet-tRNA, 0.5 μM; [Str] streptomycin, 20 μM) as indicated. GTP (100 μM) was present in all cases. Numbers in parentheses specify micromolar concentrations of ligands when they differ from the standard conditions listed above. Complexes were treated with DMS or KE, and RNA was extracted and analyzed by primer extension. (Lanes A,G) Sequencing reactions; (lanes K) analyses of untreated 30S subunits. (A) DMS probing of the 1408 and 908 regions. (B,C) KE probing of the 530 region. (D) DMS probing of the 1408 region of wild-type (WT) and mutant (A1413C) 30S subunits. (E) DMS probing of the 1408 region in the presence of streptomycin (Str). (F) Effects of streptomycin and mutation A1413C on DMS reactivity in the 915 region.
A1408 exhibits reduced reactivity in the presence of mutation A1413C
Mutation A1413C alters h44 in a way that promotes 70SIC formation (see above). To determine whether A1413C and cognate start codon recognition similarly influence the structure of h44, DMS probing experiments were performed with 30SICs in the presence of the mutation (Fig. 2D). Indeed, in complexes formed with A1413C subunits, the reactivity of A1408 was markedly reduced, regardless of the start codon sequence. Mutation A1410G, which also increases the apparent rate of 50S docking, shows a similar reduction in A1408 reactivity (data not shown). These data suggest that both mutations cause h44 to adopt a conformation that resembles the state normally induced by start codon recognition.
A1408 exhibits reduced reactivity in the presence of streptomycin
Rodnina and coworkers proposed that IF1 negatively regulates initiation by inducing a docking-unfavorable state of the 30SIC, which can be shifted to a docking-favorable state by cognate codon–anticodon pairing in the P site or by streptomycin (Milon et al. 2008). Our finding that start codon recognition reduces the IF1-dependent enhancement of A1408 prompted us to investigate the effects of streptomycin on the structure of h44. We probed 30SIC(AUG) and 30SIC(AUC) with DMS in the presence and absence of streptomycin. Because streptomycin decreases the affinity of IF1 by about twofold (Milon et al. 2008), we used an excess of IF1 (5 μM) in this experiment to ensure high occupancy of its binding site. In the presence of streptomycin, A1408 reactivity was substantially reduced in 30SIC(AUG) and 30SIC(AUC) (Fig. 2E, lanes 7,8), effects strikingly similar to those of A1413C (Fig. 2D). Nucleotides forming direct contacts with the antibiotic (e.g., 913–915) were also protected, as were A908 and A909, which lie near the A1413–G1487 region of h44 (Fig. 2F). Mutation A1413C similarly caused protection of A908 and A909. These data show that streptomycin and A1413C similarly influence the structure of h44 and lend further support to the idea that regulation of 50S docking involves a conformational change in the A1408 region of h44.
Protections attributed to P site tRNA depend on AUG
Several of the nucleotides that showed distinct reactivity in 30SIC(AUG) versus 30SIC(AUC) lie in or near the P site (Fig. 1A). One of these is G1338, which forms a Type II A-minor interaction with nucleotide 41 of the anticodon stem of P site tRNA (Berk et al. 2006). Protection of G1338 by fMet-tRNA was evident in 30SIC(AUG), but no tRNA-dependent protection was seen in the case of 30SIC(AUC) (Fig. 3A; Table 2).
FIGURE 3.
Other 16S rRNA nucleotides exhibiting differential reactivity in complexes with AUG versus AUC. Data are presented as described in the legend to Figure 2. (A,B) KE probing of the 1338 and 926 regions. (C,D) DMS probing of the 794 and 532 regions. (E) NMIA and KE probing of the 700 region. (F) NMIA probing of the 700 region in the presence of 10 μM IF3.
Another differentially modified nucleotide was G926 (Fig. 3A; Table 2). Early footprinting studies suggested that P site tRNA directly protects G926 from KE modification (Moazed and Noller 1990). However, more recent structural studies have revealed that N1 and N2 of G926 donate H-bonds to the phosphate group of the first nucleotide of the P codon of mRNA (Selmer et al. 2006). In our experiments, tRNA-independent protection of G926 was observed in the presence of both mRNA and IF3 but was not seen when either ligand was omitted (Fig. 3B). We infer that mRNA directly protects G926, and IF3 stabilizes the P codon in some way. This interpretation is consistent with evidence that IF3 can reposition mRNA from a “standby” site to a “decoding” site without affecting its overall binding affinity (La Teana et al. 1995). In 30SIC(AUG), G926 was found to be further protected compared with 30S•IFs•mRNA complexes, presumably due to codon–anticodon pairing in the P site. In the case of 30SIC(AUC), no further protection was detected (Fig. 3A,B; Table 2).
Another nucleotide more protected in 30SIC(AUG) than in 30SIC(AUC) was A794 (Fig. 3C; Table 2). This protection depended completely on fMet-tRNA, consistent with its previous assignment as a P-tRNA protection (Moazed and Noller 1990). However, this protection is presumably indirect, since A794 lies >10 Å from P site tRNA (Selmer et al. 2006). In 30SIC(AUG), A794 was strongly protected from DMS. In 30SIC(AUC), protection was still observed but lessened substantially (Table 2).
The reactivity of G532 was also found to differ depending on the start codon. Earlier footprinting studies showed protection of G532 by P site tRNA (Moazed and Noller 1990). This probably corresponds to a tRNA-induced conformational change, since G532 lies away from the P site, on the distal side of the A site (Selmer et al. 2006). We saw protection of G532 in the 30S•IFs complex, which was lost when mRNA was added. Protection was seen again in the context of the 30SIC, but only in the presence of the AUG start codon (Fig. 3D; Table 2).
U701 is less protected from NMIA in the presence of AUG
One target of NMIA, U701, was more reactive in 30SIC(AUG) than in 30SIC(AUC) (Fig. 3E, top panel; Table 2). Previous work has shown that IF3 protects U701 from CMCT modification and from hydroxyl radical cleavage (Muralikrishna and Wickstrom 1989; Dallas and Noller 2001), suggesting that U701 contributes to the IF3-binding site. This raised the possibility that the enhanced reactivity at U701 was due to loss of IF3 in the case of 30SIC(AUG). However, the adjacent G700 was similarly protected from KE in both complexes (Fig. 3E, bottom panel; Table 2), which argued against a difference in the level of bound IF3. To shed light on these seemingly contradictory observations, we monitored NMIA-dependent reactivity of complexes at higher concentrations of IF3 (10 μM) (Fig. 3F). Under these conditions, complete protection of U701 was observed in both complexes, suggesting that the difference observed initially (e.g., in Fig. 3E) reflects a difference in IF3 binding affinity, as opposed to a qualitative difference between the complexes (e.g., conformational change in the platform). This interpretation is consistent with evidence that cognate codon–anticodon pairing destabilizes IF3 in the 30SIC (Antoun et al. 2006b; Milon et al. 2008). The reason why differences in reactivity were observed with NMIA but not KE remains unclear, but we suspect that it has to do with the probes themselves. NMIA is a good general electrophile predicted to modify not only rRNA but also IF3. Hence, NMIA may have decreased the concentration of active IF3 during the course of the reaction and amplified differences in the protection pattern.
Effects of the start codon and mutation A1413C on the extent of fMet-tRNA binding
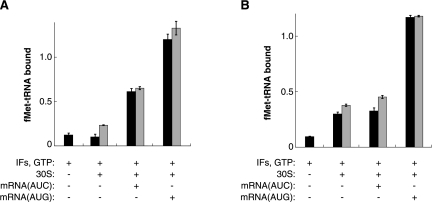
The fact that G1338 and G926 were similarly reactive in the 30SIC(AUC) and 30S•IFs•mRNA complexes suggested that the P site of 30SIC(AUC) is largely unoccupied or only transiently occupied. This raised the question of whether the very slow rate of 70SIC formation in the presence of AUC might be largely explained by the lack of bound fMet-tRNA, since 50S docking is strictly tRNA-dependent. To investigate this, nitrocellulose filter binding was used to compare the level of f-[35S]-Met-tRNA bound in 30SIC(AUG) versus 30SIC(AUC), under conditions identical to those used in the stopped-flow (LS) and chemical probing (CP) experiments (Fig. 4). The effects of A1413C were tested in parallel. Under both LS and CP conditions, the extent of binding was high for 30SIC(AUG), consistent with a fully bound P site. For 30SIC(AUC), the level of binding was reduced by ∼50% under LS conditions and by ∼70% under CP conditions. Under LS conditions, the level of binding was clearly higher than that seen in the absence of mRNA, while under CP conditions, similar binding levels were observed in the absence of mRNA. Little binding was observed in the absence of 30S subunits under either set of conditions. The fact that a substantial proportion of complexes contain fMet-tRNA in the 30SIC(AUC) reactions suggests that the slow observed rate of 70SIC(AUC) formation stems primarily from a kinetic bottleneck during the 50S docking phase of initiation, in line with previous studies (Grigoriadou et al. 2007b; Milon et al. 2008). Consistent with this interpretation, when the stopped-flow experiment measuring 70SIC(AUC) formation was repeated using a 10-fold higher concentration of fMet-tRNA, virtually no increase in docking rate was observed (data not shown). Mutation A1413C had little or no effect on the extent of fMet-tRNA binding in the various 30S complexes (Fig. 4). Importantly, under LS conditions, the level of bound fMet-tRNA was indistinguishable in the control and mutant 30SIC(AUC) complexes, showing that mutation A1413C stimulates 70SIC formation by affecting events subsequent to fMet-tRNA binding (i.e., the subunit-joining phase of the process).
FIGURE 4.
Effects of the start codon and mutation A1413C on the extent of fMet-tRNA binding. Complexes were formed with f-[35S]-Met-tRNA under LS (A) and CP (B) conditions and filtered through a bilayer of nitrocellulose and nylon membranes to quantify the amount of fMet-tRNA bound. For reactions containing 30S subunits, black bars indicate wild-type subunits and gray bars indicate mutant (A1413C) subunits. Ordinate values correspond to the amount of fMet-tRNA bound per 30S and are corrected for background, which was determined from mock reactions containing f-[35S]-Met-tRNA and buffer only. Data represent the mean ± SEM from three or more independent experiments.
Effects of the start codon and mutation A1413C on the stability of the 30S•mRNA•fMet-tRNA ternary complex
To further characterize mutation A1413C, we used toeprinting to estimate the overall equilibrium constant for 30S•mRNA•fMet-tRNA ternary complex formation (KTC) under several conditions (Fig. 5). An advantage to toeprinting is that it reports specifically on complexes involving the start codon. But one caveat to the method is that detection involves reverse transcriptase (RT), which can disrupt the complex in certain cases, resulting in a decreased toeprint signal. In these experiments, 30S subunits (0.1 μM or 1 μM; for cognate or near-cognate complexes, respectively) were incubated with mRNA (0.01 μM, containing pre-annealed 32P-labeled primer) and fMet-tRNA (various concentrations as indicated) in buffer A (50 mM Tris-HCl at pH 7.5, 30 mM KCl, 70 mM NH4Cl, 7 mM MgCl2, 1 mM DTT; the buffer used in the LS experiments) at 37°C until equilibrium was reached; reverse transcriptase (RT, 2 units) and dNTPs (200 μM each) were added to extend the primer, and the products were resolved by PAGE. The toeprint signal, expressed as a fraction of the total signal [F = toeprint/(toeprint + runoff)], was then plotted as a function of total fMet-tRNA concentration, and the data were fit to the equation F = Fmax[bc/(bc + 1/KTC)], where b and c correspond to the input concentrations of fMet-tRNA and 30S subunits, and Fmax represents the maximal toeprint signal, which presumably reflects the probability that the complex resists disruption by RT (see Materials and Methods).
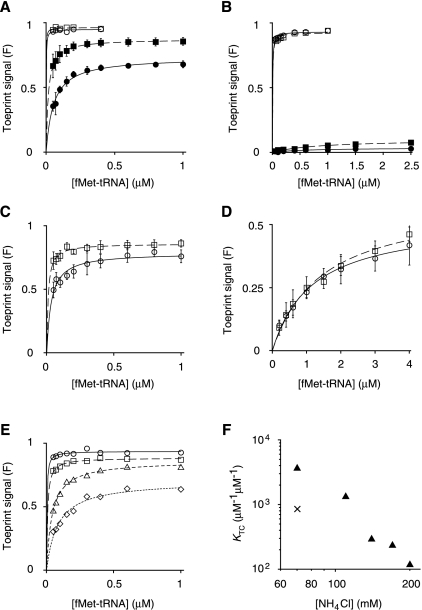
FIGURE 5.
Effect of the start codon on the stability of the 30S•mRNA•fMet-tRNA ternary complex. Toeprinting was used to estimate the equilibrium constant for 30S•mRNA•fMet-tRNA ternary complex formation (KTC) under various conditions. (A–D) Wild-type (circles) or A1413C (squares) 30S subunits (0.1 μM or 1.0 μM) were incubated with fMet-tRNA (various concentrations), mRNA (0.01 μM; with pre-annealed 5′-32P-labeled primer), and GTP (100 μM), in the absence or presence of factors until equilibrium was reached, and then complexes were detected by toeprinting. The toeprint signal (F), expressed as a fraction of the total signal, is plotted as a function of total fMet-tRNA concentration. Data are fit to the equation F = Fmax[bc/(bc + 1/KTC)], where b and c correspond to the input concentrations of fMet-tRNA and 30S subunits, and Fmax represents the maximal toeprint signal. Error bars represent the SEM from three or more independent experiments. (A) Formation of 30S•AUG•fMet-tRNA in the presence of all three factors (filled symbols) or IF1 and IF2 only (open symbols). (B) Formation of 30S•AUC•fMet-tRNA in the presence of all three factors (filled symbols) or IF1 and IF2 only (open symbols). (C) Formation of 30S•AUG•fMet-tRNA in the absence of factors (at 140 mM NH4Cl). (D) Formation of 30S•AUC•fMet-tRNA in the absence of factors. (E) Formation of 30S•AUG•fMet-tRNA in the absence of factors at 70 (○), 110 (□), 170 (△), and 200 (◇) mM NH4Cl. (F) KTC values for 30S•AUG•fMet-tRNA formation (▴), estimated from curves of panels C and E, plotted as a function of NH4Cl concentration. For comparison, the × symbol represents a value 1000-fold greater than the KTC obtained for 30S•AUC•fMet-tRNA formation at 70 mM NH4Cl.
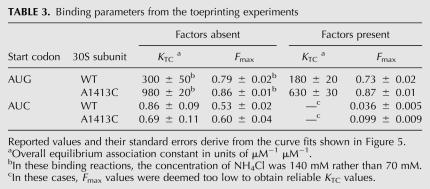
In the presence of factors, the cognate complex was readily detected by toeprinting (Fig. 5A, filled circles). The data yielded an overall equilibrium constant (KTC) of 180 μM−1 μM−1 and an Fmax value of 0.73 (Table 3). This KTC predicts a fully saturated P site under LS conditions, consistent with the nitrocellulose filter binding results (Fig. 4A). Mutation A1413C increased KTC by threefold and increased Fmax to 0.87 (Fig. 5A, filled squares; Table 3), consistent with a modest stabilization of 30SIC(AUG). For the 30SIC(AUC) complexes, the Fmax values were very small (<0.1) (Fig. 5B, closed symbols). Because substantial levels of bound fMet-tRNA were detected by nitrocellulose filter-binding in analogous complexes (Fig. 4A), we suspect that 30SIC(AUC) is too kinetically labile to generate a toeprint. Consistent with this idea, single omission of IF3 increases Fmax for both cognate and near-cognate complexes (Fig. 5A,B, open symbols), and IF3 has been shown to increase the dissociation rate of fMet-tRNA from the P site by 300-fold (Antoun et al. 2006b).
TABLE 3.
Binding parameters from the toeprinting experiments
In the absence of factors, both the cognate and near-cognate ternary complexes were readily detected (Fig. 5C–E). In buffer A with 1 μM subunits, the binding curves were well defined for the near-cognate complexes (Fig. 5D), giving virtually identical binding parameters for the control and mutant subunits (Table 3). In the same buffer, the cognate complexes were found to be very stable—the fraction of bound mRNA (F) was close to Fmax at all concentrations of fMet-tRNA, even though the 30S subunit concentration was 10-fold lower (0.1 μM) (Fig. 5E, circles). To estimate the stability of the cognate complex relative to the near-cognate complex, we increased the NH4Cl concentration to 110, 140, 170, and 200 mM, destabilizing the complex enough to obtain better-defined KTC values (Fig. 5E,C). From the resulting plot of KTC versus [NH4Cl] (Fig. 5F), we can conservatively estimate that the cognate ternary complex is at least 1000-fold more stable than the near-cognate complex. At 140 mM NH4Cl, mutation A1413C stabilized 30S•AUG•fMet-tRNA modestly (Fig. 5C), increasing KTC by threefold and Fmax to 0.86, effects similar to those observed in the presence of factors (Table 3).
DISCUSSION
In previous work, we isolated several mutations in 16S rRNA that reduce the accuracy of start codon selection in vivo (Qin and Fredrick 2009). These mutations cluster to three regions of the subunit—the neck, the 790 loop, and the “upper” portion of h44 (Fig. 1A). Mutations in the neck region presumably influence the dynamics of the head domain but have yet to be more thoroughly characterized. Mutations in the 790 loop decrease the affinity of IF3 for the 30S subunit by ∼10-fold (Qin et al. 2007; Qin and Fredrick 2009), providing an explanation for their fidelity phenotypes. The mutations in h44 surround the noncanonical A1413–G1487 base pair, which is highly conserved in bacteria. This region of h44 is the center of a local distortion induced by IF1 and lies between nucleotides contributing to inter-subunit bridges B2a and B3. In this study, we further characterize mutation A1413C, which reduces start codon selection in vivo by threefold (Qin and Fredrick 2009). We find that A1413C stimulates 70SIC formation, increasing the apparent rate of 50S docking by 17-fold in the presence of start codon AUC and by less than twofold in the presence of AUG. The large effect on 70SIC(AUC) formation can be attributed to the subunit-joining phase of the reaction because the level of fMet-tRNA bound to the 30S complex prior to mixing is substantial and unaffected by A1413C.
The observation that A1413C strongly stimulates 70SIC formation in the presence of AUC suggested that a conformational change in this vicinity of h44 might normally regulate 50S docking in response to start codon recognition. To investigate this, we compared the chemical reactivity of 16S rRNA in 30SIC(AUG) versus 30SIC(AUC) and identified A1408 as one of a small subset of nucleotides exhibiting differential reactivity in the two complexes. In the presence of IF1, A1408 is highly reactive to DMS but exhibits reduced reactivity upon start codon recognition in the 30SIC. This reduced reactivity is due to a conformational change rather than loss of IF1 because other portions of the IF1 footprint are unchanged and high concentrations of IF1 fail to diminish the reactivity difference seen at A1408. Mutation A1413C and streptomycin also reduce the reactivity of A1408 in the context of the 30SIC, suggesting that each promotes a similar conformational change in h44. Hence, cognate codon–anticodon pairing, A1413C, and streptomycin each stimulates 50S docking and similarly affects h44. These data support a model in which IF1 alters the A1408 region of h44 in a way that makes 50S docking unfavorable, and codon–anticodon pairing in the P site partially reverses the structural changes induced by IF1 to promote 50S docking. This model builds on one proposed by Rodnina and coworkers, based on their studies of IF1 (Milon et al. 2008). Importantly, our findings provide evidence that the key conformational change postulated previously occurs in the A1408 region of h44. A1408 contributes to inter-subunit bridge B2a (Schuwirth et al. 2005), suggesting that conformational changes in h44 may directly regulate formation of B2a and thereby influence 50S docking.
While direct regulation of B2a formation is an attractive model, it is also possible that the structure of h44 influences 50S docking in an indirect manner. For example, changes in h44 caused by A1413C and/or streptomycin may alter the conformation of IF2 and/or fMet-tRNA on the 30S subunit and thereby promote docking. IF2 and fMet-tRNA are known to play critical roles in the control of 50S docking (Grigoriadou et al. 2007a,b; Pavlov et al. 2010), so even small effects on these ligands might substantially influence the rate of 70SIC formation. Clearly, further experiments will be necessary to elucidate the degree to which B2a formation governs the docking reaction.
Earlier chemical probing experiments predicted conformational signaling between the P site and h44. Moazed and Noller (1987) identified six nucleotides in the 30S subunit (A790, G791, A909, A1394, A1413, and A1487) protected from chemical probes by multiple ligands (e.g., P-tRNA, various antibiotics, 50S subunit) (Moazed and Noller 1987). These nucleotides were termed Class III sites and the protections were attributed to ligand-induced conformational changes. Interestingly, three of these nucleotides (A909, A1413, and A1487) localize in or near h44, are protected by either tRNA or streptomycin, and become hyper-reactive in the presence of IF1 (Moazed and Noller 1987; Dahlquist and Puglisi 2000). These data suggest that the conformational change induced by start codon recognition encompasses the 1413–1487 portion of h44 as well as the 1408 region. In the presence of all components of the 30SIC, A1413 appears slightly more protected in the presence of the cognate AUG start codon (Table 2). While this difference was not verified statistically at the 95% confidence level, each independent experiment showed the same trend in which A1413 is less reactive in 30SIC(AUG) compared with 30SIC(AUC), as would be predicted from the earlier probing studies (Moazed and Noller 1987). Hence, we suspect that codon–anticodon pairing in the P site perturbs h44 in a similar way in the absence and presence of initiation factors but that the observed decrease in A1413 reactivity is smaller in the latter case.
In this study, we have also adapted the toeprinting technique to measure the overall equilibrium constant (KTC) for 30S•mRNA•fMet-tRNA ternary complex formation under several conditions. In the presence of all three factors, KTC for 30S•AUG•fMet-tRNA formation is 180 μM−1 μM−1. This value is in good agreement with the equilibrium constant deduced by measuring the kinetics of fMet-tRNA binding in the context of 30SIC(AUG) formation (Antoun et al. 2006b). Unfortunately, 30SIC(AUC) cannot be detected by toeprinting (Fig. 5B), and hence the difference in stability between 30SIC(AUG) and 30SIC(AUC) remains to be determined. We suspect that 30SIC(AUC) is too kinetically labile to inhibit primer extension by RT, since the complex can be detected by nitrocellulose filter binding (Fig. 4A). In the absence of factors, both the cognate and near-cognate ternary complex can be detected by toeprinting. We find that the stability of 30S•AUG•fMet-tRNA is at least 1000-fold higher than 30S•AUC•fMet-tRNA. This intrinsic selectivity of ternary complex formation is more than enough to account for the accuracy of start codon selection in vivo (AUG/AUC ≈ 100) (Sacerdot et al. 1996; Sussman et al. 1996), in contrast to earlier suppositions (La Teana et al. 1993). These data are consistent with the idea that the factors (and IF3 in particular) influence the kinetics of the process in a way that allows the energetic cost of a codon–anticodon mismatch to be effectively exploited, without necessarily enhancing the selectivity of mRNA binding (Antoun et al. 2006a).
This and previous chemical probing work provide evidence for conformational signaling between the P site and h44. A question that remains open is whether this signaling depends specifically on cognate codon–anticodon pairing or more generally on anticodon binding to the 30S P site, which occurs only transiently in the case of 30SIC(AUC). Potentially relevant to this question, we find that, in the absence of factors, mutation A1413C stabilizes the 30S•AUG•fMet-tRNA ternary complex, as indicated by an increase in both KTC and Fmax (Fig. 5C). In contrast, A1413C has no effect on the stability of 30S•AUC•fMet-tRNA (Fig. 5D). A plausible explanation for these results is that A1413C and the matched codon–anticodon helix in the P site each stabilizes a particular conformation of h44, whereas the mismatched codon–anticodon helix fails to do so. This scenario predicts that A1413C would stabilize fMet-tRNA only in the presence of a cognate codon, as we observe. In the future, it will be of interest to determine whether the same trend is seen in the presence of factors.
In summary, this study supports a model in which formation of the 30SIC is strongly influenced by the sequence of the start codon, with the equilibrium shifted strongly leftward (toward disassembly) in the near-cognate case. When a cognate start codon is recognized, the complex is altered in at least two ways: (1) the 1408 region of h44 adopts a docking-favorable conformation, and (2) IF3 becomes destabilized (Antoun et al. 2006b; Milon et al. 2008). Together, these changes promote rapid docking of the 50S subunit and formation of the 70SIC.
MATERIALS AND METHODS
Small ribosomal subunits were purified from Escherichia coli Δ7 prrn strains as described (Qin et al. 2007). Large subunits were purified from E. coli strain CSH142 [F− ara Δ(gpt-lac)5] using the same method, except that the 50S peak was collected. For the toeprinting experiments, 30S subunits were purified from gradient-isolated 70S ribosomes as opposed to crude ribosomes. Purified tRNAfMet (Chemical Block) was aminoacylated and formylated as described (Walker and Fredrick 2008) and stored in 2 mM NaOAc (pH 5.2). The mRNAs, derived from phage T4 gene 32, were made by in vitro transcription and purified as described (Fredrick and Noller 2002). The initiation regions of these mRNAs are as follows: mRNA(AUG), 5′-(N)41-AAAGGAAAUAAAAAUGGUAUACUUUAAAUCU-(N)67-3′; mRNA(AUC), 5′-(N)41-AAAGGAAAUAAAAAUCGUAUACUUUAAAUCU-(N)67-3′; where the Shine-Dalgarno is indicated in bold and the start codon is underlined. Each initiation factor was overexpressed in strain BL21/DE3 from pET24b-based constructs. IF1 was purified using a Resource S column (GE Healthcare) as described (Qin and Fredrick 2009). His-tagged IF2 (α form) and IF3 were purified using Ni-NTA agarose (QIAGEN) as described (Dallas and Noller 2001).
Light scattering experiments were performed essentially as described (Milon et al. 2008). 30SICs were formed by incubating activated 30S subunits (0.075 μM) with fMet-tRNAfMet (0.2 μM), mRNA (0.5 μM), initiation factors (0.2 μM each), and GTP (100 μM) in 1 mL of buffer A (50 mM Tris-HCl at pH 7.5, 30 mM KCl, 70 mM NH4Cl, 7 mM MgCl2, 1 mM DTT) for 30 min at 37°C. For each mixing experiments, 60 μL of 30SIC was rapidly mixed with an equal volume of 50S subunits (0.23 μM; in buffer A), using an SX20 stopped-flow apparatus (Applied Photophysics). Exciting light at 430 nm was applied to the mixing chamber, and scattered light was monitored without a cutoff filter at an angle perpendicular to the excitation beam as a function of time. The data were fit to either a single or double exponential function depending on the nature of the curve.
Chemical modification experiments were performed as described (Merryman and Noller 1998). Complexes were formed by incubating 30S subunits (0.25 μM), fMet-tRNAfMet (0.5 μM), mRNA (1 μM), initiation factors (0.5 μM each), and GTP (100 μM) in buffer B (80 mM K-cacodylate at pH 7.5, 30 mM KCl, 70 mM NH4Cl, 7 mM MgCl2, 1 mM DTT) for 10 min at 37°C and then for 30 min at room temperature. DMS (35 mM; Sigma-Aldrich) or KE (10 mM; USB) was added, the reaction was incubated for 8 min at 37°C, ribosomes were precipitated, and the rRNA was analyzed by primer extension as described (Merryman and Noller 1998). For the KE reactions, K-borate (25 mM, pH 7.0) was added after the probing step to stabilize the conjugate. In NMIA experiments, 80 mM K-HEPES (pH 7.9) replaced K-cacodylate, the probe concentration was 13 mM, and the reaction time was 45 min.
Nitrocelllulose filter binding assays were used to compare the extent of fMet-tRNA binding in various complexes. 30SICs were formed with f-[35S]-Met-tRNA exactly as described above for the light-scattering (LS) or chemical probing (CP) experiments. Binding reactions were filtered through a bilayer of nitrocellulose and nylon membranes (NitroBind and Hybond-N+; GE Healthcare), and the amount of radiolabel on each membrane was quantified to determine the amount of fMet-tRNA bound (Fahlman and Uhlenbeck 2004).
For the toeprinting experiments, 30S subunits (0.1 μM or 1 μM, for cognate or near-cognate complexes, respectively) were incubated at 37°C in buffer A with fMet-tRNA (various concentrations), mRNA (0.01 μM, with pre-annealed 5′-32P-labeled primer), and GTP (100 μM), in the absence or presence of factors (0.5 μM each or 1.5 μM each, for cognate or near-cognate complexes, respectively) until equilibrium was reached. Then AMV reverse transcriptase (Northstar BioProducts, 2 units) and dNTPs (200 μM each) were added, and the resulting extension products were resolved by denaturing PAGE. The incubation times used (120 min in the absence of factors, 4 min in the presence of factors) were chosen after quantifying the toeprint intensity as a function time for each complex in binding reactions with the lowest concentration of fMet-tRNA (0.05 μM). The overall equilibrium constant for 30S•mRNA•fMet-tRNA ternary complex formation (KTC) can be expressed as KTC = [ABC]/[A][B][C], where [ABC] is the concentration of ternary complex, [A] is the concentration of free mRNA, [B] is the concentration of free fMet-tRNA, and [C] is the concentration of free 30S subunits. Under the conditions used, the total (input) concentration of fMet-tRNA and 30S subunits always exceeded that of mRNA by more than fivefold. Hence, [B] ≈ b and [C] ≈ c, where b and c represent the total (input) concentrations of fMet-tRNA and 30S subunits, respectively, and KTC = [ABC]/[A]bc (Eq. 1). The fraction of signal corresponding to the toeprint (F) can be defined as F =[ABC]/([A] + [ABC]) (Eq. 2). Combining Equations 1 and 2 and simplifying gives the hyperbolic function F = bc/(bc + 1/KTC) (Eq. 3). A caveat to the toeprinting technique is that reverse transcriptase (RT) can disrupt certain complexes, resulting in a reduced toeprint signal. This is indicated by a lower plateau in the binding curve from experiments in which b, for example, is varied. To account for this RT effect, Equation 3 is multiplied by the parameter Fmax, which presumably reflects the probability that the complex resists disruption by RT, giving F =Fmax[bc/(bc + 1/KTC)] (Eq. 4). Plots of F versus b were fit to this equation using the program Kaleidograph, to estimate the overall equilibrium constant for ternary complex formation (KTC). Similar values for KTC were obtained when either the concentration of fMet-tRNA (b) or 30S subunits (c) was varied, supporting the general validity of the experimental approach.
ACKNOWLEDGMENTS
We thank C. Squires and S. Quan for the Δ7 prrn strain SQZ10, and M. Ibba, R. Gonzalez, and anonymous reviewers of this manuscript for their helpful feedback. This work was supported by NSF grant MCB 0840996.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.031203.111.
REFERENCES
- Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M 2003. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J 22: 5593–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M 2006a. How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol Cell 23: 183–193 [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M 2006b. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J 25: 2539–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V, Zhang W, Pai RD, Cate JHD 2006. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci 103: 15830–15834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501 [DOI] [PubMed] [Google Scholar]
- Dahlquist KD, Puglisi JD 2000. Interaction of translation initiation factor IF1 with the E. coli ribosomal A site. J Mol Biol 299: 1–15 [DOI] [PubMed] [Google Scholar]
- Dallas A, Noller HF 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8: 855–864 [DOI] [PubMed] [Google Scholar]
- Fahlman RP, Uhlenbeck OC 2004. Contribution of the esterified amino acid to the binding of aminoacylated tRNAs to the ribosomal P- and A-sites. Biochemistry 43: 7575–7583 [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF 2002. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell 9: 1125–1131 [DOI] [PubMed] [Google Scholar]
- Grigoriadou C, Marzi S, Kirillov SV, Gualerzi CO, Cooperman BS 2007a. A quantitative kinetic scheme for 70S translation initiation complex formation. J Mol Biol 373: 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadou C, Marzi S, Pan D, Gualerzi CO, Cooperman BS 2007b. The translational fidelity function of IF3 during transition from the 30S initiation complex to the 70S initiation complex. J Mol Biol 373: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi CO, Brandi L, Caserta E, Garofalo C, Lammi M, La Teana A, Petrelli D, Spurio R, Tomsic J, Pon CL 2001. Initiation factors in the early events of mRNA translation in bacteria. In The ribosome (ed. B Stillman), pp. 363–376. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [DOI] [PubMed] [Google Scholar]
- Haggerty TJ, Lovett ST 1997. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J Bacteriol 179: 6705–6713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, Binkley J, Hollingsworth T, Gold L 1990. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev 4: 1790–1800 [DOI] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M 1999. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell 3: 601–609 [DOI] [PubMed] [Google Scholar]
- La Teana A, Pon CL, Gualerzi CO 1993. Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc Natl Acad Sci 90: 4161–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A, Gualerzi CO, Brimacombe R 1995. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA 1: 772–782 [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Stortchevoi A, Kohrer C, Varshney U, RajBhandary UL 2001. Initiator tRNA and its role in initiation of protein synthesis. In The ribosome (Cold Spring Harbor symposia on quantitative biology LXVI), pp. 195–206. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [DOI] [PubMed] [Google Scholar]
- Meinnel T, Sacerdot C, Graffe M, Blanquet S, Springer M 1999. Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J Mol Biol 290: 825–837 [DOI] [PubMed] [Google Scholar]
- Merryman C, Noller HF 1998. Footprinting and modification-interference analysis of binding sites on RNA. In RNA:protein interactions (ed. CWJ Smith), Vol. 192, pp. 237–253. Oxford University Press, Oxford [Google Scholar]
- Milon P, Konevega AL, Gualerzi CO, Rodnina MV 2008. Kinetic check point at a late step in translation initiation. Mol Cell 30: 712–720 [DOI] [PubMed] [Google Scholar]
- Milon P, Garotti M, Konevega AL, Wintermeyer W, Rodnina MV, Gualerzi CO 2010. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep 11: 312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327: 389–394 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF 1990. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J Mol Biol 211: 135–145 [DOI] [PubMed] [Google Scholar]
- Moazed D, Samaha RR, Gualerzi C, Noller HF 1995. Specific protection of 16 S rRNA by translational initiation factors. J Mol Biol 248: 207–210 [DOI] [PubMed] [Google Scholar]
- Muralikrishna P, Wickstrom E 1989. Escherichia coli initiation factor 3 protein binding to 30S ribosomal subunits alters the accessibility of nucleotides within the conserved central region of 16S rRNA. Biochemistry 28: 7505–7510 [DOI] [PubMed] [Google Scholar]
- O'Connor M, Thomas CL, Zimmermann RA, Dahlberg AE 1997. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res 25: 1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M, Gregory ST, Rajbhandary UL, Dahlberg AE 2001. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA 7: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Antoun A, Lovmar M, Ehrenberg M 2008. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J 27: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov MY, Zorzet A, Andersson DI, Ehrenberg M 2010. Activation of initiation factor 2 by ligands and mutations for rapid docking of ribosomal subunits. EMBO J 30: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Fredrick K 2009. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S ribosomal RNA. Mol Microbiol 71: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Abdi NM, Fredrick K 2007. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA 13: 2348–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M 1996. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol Micro 21: 331–346 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, Yusupova G, Klaholz BP, Yusupov M 2009. A structural view of translation initiation in bacteria. Cell Mol Life Sci 66: 423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman JK, Simons EL, Simons RW 1996. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol Micro 21: 347–360 [DOI] [PubMed] [Google Scholar]
- Tanada S, Kawakami M, Nishio K, Takemura S 1982. Interaction of aminoacyl-tRNA with bacterial elongation factor Tu : GTP complex: effects of the amino group of amino acid esterified to tRNA, the amino acid side chain, and tRNA structure. J Biochem 91: 291–299 [DOI] [PubMed] [Google Scholar]
- Walker SE, Fredrick K 2008. Preparation and evaluation of acylated tRNAs. Methods 44: 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616 [DOI] [PubMed] [Google Scholar]