Abstract
Background and Aims
Satellite DNA is a genomic component present in virtually all eukaryotic organisms. The turnover of highly repetitive satellite DNA is an important element in genome organization and evolution in plants. Here we assess the presence and physical distribution of the repetitive DNA E180 family in Medicago and allied genera. Our goals were to gain insight into the karyotype evolution of Medicago using satellite DNA markers, and to evaluate the taxonomic and phylogenetic signal of a satellite DNA family in a genus hypothesized to have a complex evolutionary history.
Methods
Seventy accessions from Medicago, Trigonella, Melilotus and Trifolium were analysed by PCR to assess the presence of the repetitive E180 family, and fluorescence in situ hybridization (FISH) was used for physical mapping in somatic chromosomes.
Key Results
The E180 repeat unit was PCR-amplified in 37 of 40 taxa in Medicago, eight of 12 species of Trigonella, six of seven species of Melilotus and in two of 11 Trifolium species. Examination of the mitotic chromosomes revealed that only 13 Medicago and two Trigonella species showed FISH signals using the E180 probe. Stronger hybridization signals were observed in subtelomeric and interstitial loci than in the pericentromeric loci, suggesting this satellite family has a preferential genomic location. Not all 13 Medicago species that showed FISH localization of the E180 repeat were phylogenetically related. However, nine of these species belong to the phylogenetically derived clade including the M. sativa and M. arborea complexes.
Conclusions
The use of the E180 family as a phylogenetic marker in Medicago should be viewed with caution. Its amplification appears to have been produced through recurrent and independent evolutionary episodes in both annual and perennial Medicago species as well as in basal and derived clades.
Keywords: Medicago, Trigonella, Melilotus, Trifolium, satellite DNA, repetitive E180 family, FISH
INTRODUCTION
With nuclear genome sequencing projects nearing completion, it has become easier to accurately assess the diversity, abundance and chromosomal distribution of repeated DNA sequences in plants. By comparing these data with results from other techniques (e.g. CsCl buoyant density gradient centrifugation, C-banding cytogenetics, Southern blot and dot blot hybridization analysis, molecular cytogenetics), a consolidated picture of plant genomic structure has emerged. Repetitive DNA sequences make up the majority of the nuclear genome, and these sequences consist of many classes of core elements, ranging in size from dinucleotides to more than 10 kb long (Kubis et al., 1998; Heslop-Harrison 2000; Macas et al., 2007).
Satellite DNA (satDNA) is a genomic component present in virtually all eukaryotic organisms. satDNA elements are highly repetitive, non-coding DNA sequences that are organized into long arrays composed of thousands to millions of tandemly arranged units. These arrays represent a large part of the DNA that forms constitutive heterochromatin (Ugarković and Plohl, 2002). Different satDNA sequences can coexist in genomes, forming what has been defined as a library of satDNAs (Fry and Salser, 1977; Mestrovic et al., 1998). satDNA usually forms a higher-order structure, as suggested by its intrinsic ability to induce DNA curvature; this structure might be important in packing the DNA and associated proteins into heterochromatin (Ugarković and Plohl, 2002). Despite the well-recognized roles of some satDNAs (telomeric, centromeric) in the stabilization of chromosome ends and in cellular division mechanisms, their overall biological significance remains unclear (Csink and Henikoff, 1998).
satDNA sequences evolve through evolutionary processes as predicted by the molecular drive model (Dover, 2002). Thus, DNA turnover usually leads to a high intraspecific similarity of arrays belonging to the same satDNA family (homogenization) and low or undetectable interspecific similarity. However, several studies have revealed that there is a balance between satellite homogenization and the persistence of satellite variants. This balance could generate sufficient sequence divergence to cause reproductive isolation between intraspecific lineages, ultimately leading to speciation. On this basis, it has been hypothesized that satDNA families are under selective pressure and evolutionary constraints (Ugarković and Plohl, 2002).
Thus, sequence similarity, genomic distribution and copy number have been claimed to be species-specific, and it is not generally expected that different species will share similar satDNA sequences (Kubis et al., 1998; Suárez-Santiago et al., 2007). However, some satellite sequences have remained unchanged over long evolutionary periods (Abad et al., 1992; Heikkinen et al., 1995). Although the evolution of highly repetitive satDNA is an important element in genome organization and evolution in plants, most research efforts have focused on the isolation, sequence characterization and variation in copy number of satDNA families in model or agronomically important species and closely related species (Contento et al., 2005; Macas et al., 2006; Hemleben et al., 2007; Ambrožová et al., 2011). In contrast, little is known about the genomic distribution of satDNA families and their evolution within a phylogenetic framework.
Medicago is a genus of the legume family (Fabaceae) that includes economically important forage species, e.g. M. sativa (alfalfa), M. scutellata (snail medic) and M. lupulina (black medic), and the model organism for legume biology (M. truncatula). It belongs to the Vicioid clade (Sanderson and Wojciechowski, 1995), a monophyletic group comprising the tribes Cicereae, Trifolieae and Vicieae, and the genus Galega (Galegeae).
The E180 satDNA monomer is an AT-rich repeat sequence of 185–189 bp; this satDNA sequence constitutes about 1 % of the M. sativa genome, roughly about 1·8 × 105 copies (Xia and Erickson, 1993). Calderini et al. (1997) cloned a closely related, tandem repeat sequence (C300) from M. caerulea. The high sequence similarity of the E180 monomer to the C300 monomer (96 %) and the identical estimated copy number present in M. sativa (1·8 × 105 copies) strongly suggest that both E180 and C300 are in fact variants that belong to the same satDNA family (henceforth referred to as the E180 family). Calderini et al. (1997) reported that this repetitive sequence was specific for the members of the M. sativa complex (i.e. M. sativa, M. caerulea and M. falcata) but absent in the woody medic M. arborea, the only other species analysed. The very small sample used by these authors (approx. 85 species included in 12 sections have been recognized in world-wide systematic revisions of the genus, Small and Jomphe, 1989) may not reflect the actual distribution of the repeated DNA family in Medicago. Although contrasting phylogenetic hypotheses have been inferred for Medicago using several nuclear, plastid and mitochondrial DNA sequences, all analyses strongly agree that the M. sativa complex belongs to a derived clade within the genus (Bena et al., 1998a, b; Downie et al., 1998; Bena, 2001; Maureira-Butler et al., 2008; Steele et al., 2010). Therefore, if the E180 satDNA family is specific to the M. sativa complex, then it should have recently appeared during evolution.
Here we characterize the presence and genomic distribution of the E180 satDNA family in 70 species of Medicago and its phylogenetically allied genera (Trigonella, Melilotus and the most distantly related Trifolium; all embedded within the vicioid clade; Wojciechowski et al., 2000) using PCR and fluorescence in situ hybridization (FISH) techniques. Our goals were (1) to evaluate the robustness of the two molecular techniques to determine their sensitivity in characterizing repeat DNA families, (2) to gain insight into the karyotype evolution of Medicago using satDNA markers and (3) to evaluate the phylogenetic signal of a satDNA family in a genus where complex hybridization patterns have been hypothesized to play a role in phylogenetic history (Maureira-Butler et al., 2008).
MATERIALS AND METHODS
Plant sampling and DNA extraction
Seed accessions from 40 Medicago taxa (species or subspecies), belonging to eight sections of the genus (Small and Jomphe, 1989, namely sects. Medicago, Dendrotelis, Cartiensae, Spirocarpos, Lupularia, Orbiculares, Platycarpae and Buceras), were available for study. Similarly, seeds of Trigonella (12 taxa), Trifolium (11 taxa) and Melilotus (seven taxa), which are closely related to Medicago, were also included for comparison. Seed material was obtained from a variety of sources (Appendix), and voucher specimens were deposited at the herbarium of the Botanical Garden of Valencia. Seeds were germinated in solid agar in Petri dishes at constant temperature (20 °C) with a 12-h daily regime of white light. Total genomic DNA was isolated from young seedling leaves using the DNAeasy™ Plant Minikit (Qiagen, Hilden, Germany) following the manufacturer's instructions.
Amplification of the E180 satellite family
The published sequence of the M. sativa E180 was retrieved from the PlantSat database (Macas et al., 2002; available at http://w3lamc.umbr.cas.cz/PlantSat/index.html). Specific primers (SAT-180F: 5′TCGATAAGGCTAGGCCACTT3′ and SAT-180R: 5′CCAAAATGGGGGTTAAGTGA3′) were designed to selectively amplify monomers and multiples of the E180 repeat unit. In a pilot study, positive PCR amplification products were obtained from selected species of Medicago that previously reported the presence of this family. PCR was carried out in 20 µL, containing approx. 5 ng of genomic DNA, 0·5 µm of each primer, 2·5 mm MgCl2, 2 mm Tris-HCl (pH 8·0), 5 mm KCl, 0·0001 % bovine serum albumin, 250 µm dNTPs and 1·25 units of DNA polymerase (TaKaRa Ex Taq™, Takara Biotechnology Inc., Valencia, Spain). After an initial denaturation step at 94 °C for 3 min, 40 amplification cycles were performed on the PRIMUS (MWG-Biotech, Ebersberg, Germany) thermal cycler; each cycle consisted of a denaturation step at 94 °C for 30 s, an annealing step at 55 °C for 30 s, and an elongation step at 72 °C for 1 min with a final elongation step of 5 min at 72 °C. Amplified products were visualized on a 1 % (w/v) agarose gel.
In situ hybridization
Root tips from 2–5-d-old seedlings were pre-treated with 2 mm 8-hydroxyquinoline for 2 h at 4 °C, then 2 h at room temperature, fixed in an ethanol/glacial acetic acid (3 : 1) mixture and stored at –20 °C until required. For chromosome observations, the root tips were washed in 10 mm citrate buffer (pH 4·6) and then macerated in a mixture of 2 % (v/v) cellulase (Calbiochem, Darmstadt, Germany) in 10 mm citrate buffer (pH 4·6) and 20 % pectinase (from Aspergillus niger) in 40 % glycerol for 1 h at 37 °C. The in situ hybridization probe was obtained from M. arborea using the primers and PCR conditions described above. The entire range of generated products (from 180-bp monomers to 900-bp E180 pentamers) was labelled with digoxigenin-11-dUTP through a nick translation procedure. FISH was carried out as described by Rosato et al. (2008), except that one stringent wash following the hybridization was at 37 °C in 1× saline sodium citrate for 30 min. These stringency conditions allowed the target sequences of approx. 50 % homology to remain hybridized (Schwarzacher and Heslop-Harrison, 2000).
Karyotype construction
The homologous pairs were arranged based on chromosome length and centromere position, from longest to shortest chromosomes, with the NOR-bearing chromosomes placed following the shortest pair (Bauchan and Hossain, 1997).
RESULTS
PCR amplification of the E180 satellite family in Medicago and related genera
The E180 repeat unit was amplified using PCR in 37 of 40 taxa in Medicago (Appendix). Only the diploids M. blancheana, M. coronata and M. bonarotiana repeatedly failed to produce E180 amplicons (Supplementary Data Fig. S1). Contrary to expectations, PCR products were obtained from M. arborea (and related species from section Dendrotelis), a woody species that was reported to lack the E180 family (Calderini et al., 1997). Our PCR protocols amplified the target satellite in eight out of 12 assayed species of Trigonella, six out of seven analysed species of Melilotus and in only two of the 11 available Trifolium species. As expected, successful PCR amplification usually resulted in a ladder pattern of products (Supplementary Data Fig. S1), suggesting that the E180 family exhibits a tandem repeat organization in the genome, as previously reported (Xia and Erickson, 1993).
Chromosome landmarks defined by FISH using the E180 probe
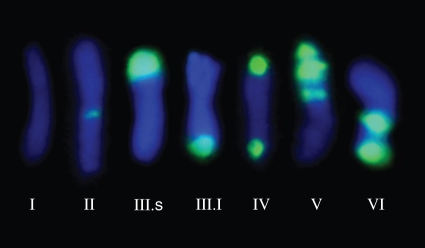
The tandem repeats of the E180 DNA sequence were either completely lacking or predominantly localized at the subtelomeric, intercalary, proximal (close to the centromere) and even pericentromeric (around the centromere) regions of chromosomes. Overall, E180 loci were usually located in only one of the chromosome arms, whereas a few chromosomes showed conspicuous signals in both arms. Up to six chromosome types were detected among the 70 analysed taxa of Medicago, Trigonella, Melilotus and Trifolium in the FISH experiments (Fig. 1, Table 1). Type I chromosomes were defined as those lacking any detectable FISH E180 signal. Type II chromosomes showed fluorescent hybridized signals at the pericentromeric region. Type III chromosomes showed a single and usually strong signal located in the subtelomeric region, either located on the short (III-s) or on the long arm (III-l). Type IV chromosomes presented subtelomeric signals on both chromosomal arms. Type V chromosomes showed three E180 signals located at the subtelomeric, intercalary and proximal regions. Lastly, type VI chromosomes were characterized by two E180 signals located at both the subtelomeric and the intercalary regions.
Fig. 1.
Types of chromosome landmarks revealed by the absence (type I) or presence (types II to VI) of E180 sites using FISH.
Table 1.
Positive FISH E180 patterns in species of the Vicioid clade
| 2n | Type I | Type II | Type III | Type IV | Type V | Type VI | No. of sites | No. of chromosomes | Sites/genome | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sect. Dendrotelis | ||||||||||
| M. arborea | 32 | – | – | 28 | 4 | – | 2 | 40 | 32 | 10 |
| M. citrina | 48 | 10 | – | 34 | 4 | – | – | 42 | 38 | 7 |
| M. strasseri | 32 | – | – | 26 | 4 | – | 2 | 38 | 32 | 9·5 |
| Sect. Medicago | ||||||||||
| M. caerulea | 16 | – | 2 | – | – | 10 | 4 | 40 | 16 | 20 |
| M. falcata | 16 | 8 | 6 | – | – | – | 2 | 10 | 8 | 5 |
| M. sativa | 32 | 2 | – | 8 | – | 10 | 12 | 74 | 30 | 18·5 |
| M. glutinosa | 32 | 4 | – | 10 | 2 | – | 14 | 42 | 28 | 10·5 |
| M. hemicycla | 32 | 6 | 4 | 16 | – | 6 | – | 38 | 26 | 9·5 |
| M. polychroa | 32 | 2 | – | 12 | – | 14 | – | 54 | 30 | 13,5 |
| Sect. Spirocarpos | ||||||||||
| Subsect. Rotatae | ||||||||||
| M. rugosa | 30 | – | 30 | – | – | – | – | 30 | 30 | 8 |
| M. scutellata | 32 | – | 32 | – | – | – | 32 | 32 | 8 | |
| Subsect. Leptospireae | ||||||||||
| M. disciformis | 16 | – | 16 | – | – | – | – | 16 | 16 | 8 |
| M. laciniata | 16 | 8 | 8 | – | – | – | – | 8 | 8 | 4 |
| Trigonella noeana | 32 | 28 | – | 4 | – | – | – | 4 | 4 | 1 |
| T. geminiflora | 44 | 42 | 2 | – | – | – | – | 2 | 2 | 0·5 |
Sporophytic chromosome number, types of chromosome landmarks, number of FISH E180 sites per diploid genome, chromosomes with FISH sites and number of FISH sites per base genome are indicated. The species are listed according to their ploidy level.
Genomic distribution of the E180 family in the Vicioid clade using FISH
Examination of the mitotic chromosomes of Medicago by FISH using the orthologous E180 probe revealed a different pattern as compared with the results obtained using the PCR approach (Appendix). In fact, only 13 out of 40 analysed taxa (four out of 28 diploid species, all eight analysed tetraploid species and the single hexaploid accession examined) revealed the presence of the highly repeated E180 DNA sequence using conventional FISH techniques (Table 1). Analogous results were obtained with Trigonella, where weak FISH signals on a chromosome type II pair in T. geminiflora, and two type III-s chromosome pairs were observed in T. noeana. Lastly, negative in situ hybridization results were obtained in the karyotypes of all species of Melilotus (seven) and Trifolium (11) analysed (Appendix).
Patterns of FISH localization in the phylogenetic clade including the crop M. sativa
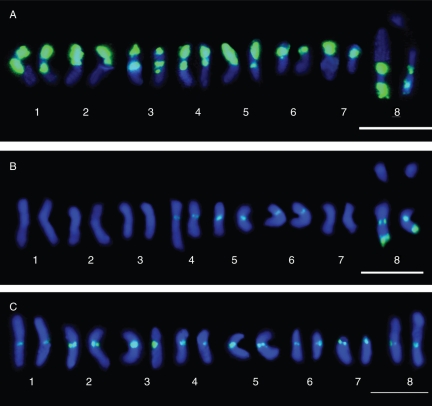
Not all 13 Medicago species analysed showed FISH localization of the E180 satellite repeat were phylogenetically related. However, nine of these species belong to the phylogenetically derived clade including the M. sativa and M. arborea complexes. Two of the diploid taxa of the M. sativa complex (M. caerulea and M. falcata) showed contrasting E180 signals in their karyotypes. M. caerulea showed the highest number of sites (40 sites) and the strongest hybridization signals in all of the type V chromosomes. Interestingly, M. falcata showed the lowest number of hybridization sites (ten sites), located in three type II chromosome pairs and one type VI chromosome pair (Fig. 2). The accession of M. sativa showed 74 E180 sites and the strongest FISH signals in 15 out of its 16 chromosome pairs, including type III (four pairs), type V (five pairs) and type VI (six pairs). The presence of type III chromosomes in M. sativa distinguished this taxon from the related diploid taxa, M. caerulea and M. falcata, which lack them. The three species belonging to section Dendrotelis (M. arborea complex) showed the presence of chromosome types I, III, IV and VI, but each species had a unique karyological pattern. The E180 FISH karyotype of the closely related tetraploids M. arborea and M. strasseri was virtually identical, only differing in the number of type III chromosomes (14 and 13, respectively). Similarly, the hexaploid M. citrina differed from both species by the presence of five pairs of type I chromosomes and the absence of type VI chromosomes (Table 1, Fig. 2).
Fig. 2.
Karyotypes of the FISH pattern of the E180 probe in three diploid Medicago species. (A) M. caerulea; (B) M. falcata; (C) M. disciformis. Scale bars = 10 µm.
DISCUSSION
Contrasting results between PCR and FISH-based methods in detecting satDNA in plant genomes
Obtaining comprehensive and reliable data to assess the structure, sequence variation and genomic distribution of highly repetitive DNA sequences is a challenging task due to technical limitations. On the one hand, given the length of the tandem-repeat arrays and the usually high levels of similarity of these arrays, DNA sequencing (and contig assembling) is usually not possible when performing genomic sequencing (unless whole-genome shotgun sequencing is applied). This usually leads to gaps of uncharted regions of repetitive DNA and, ultimately, to an inability to assess the extent of copy number distribution and sequence homogeneity of the repeat units in a large portion of the nuclear genome (Figueroa and Bass, 2010). On the other hand, the detection of repetitive sequences and their genomic distribution by conventional FISH techniques is hampered by the minimum number of copies of the target sequence that are present, not in the genome as a whole, but at each individual chromosomal locus (Navajas-Pérez et al., 2009). As the average resolution limit of FISH mapping in metaphase chromosomes is approx. 4 Mb, we would not be able to detect E180 arrays composed of less than approx. 500 copies (Hans de Jong et al., 1999).
These caveats apply in particular to the characterization of satDNA in plants due to their overall organization as long tandem repeated arrays, the high number of satDNA families that are usually present within a single plant genome and the dynamic turnover of these families, which allows fast amplifications, deletions and genome transpositions within short evolutionary time frames. The dual experimental approach used here (PCR amplification of the E180 unit, and FISH localization on mitotic chromosomes in cases where positive amplifications were obtained) has shown unexpected contrasting results. Thus, whereas the PCR experiments suggest that the E180 satellite family is widespread in Medicago, found in 92·5 % of the screened accessions, only 32·5 % of them showed E180 signals by FISH localization (type II to VI chromosomes, Fig. 1).
Several lines of evidence suggest that these contrasting results are not technical mistakes. First, there were no cases where positive FISH patterns were obtained from samples with negative PCR amplifications. Second, negative FISH results were recorded on independent days, using different experimental batches. Lastly, all slides showing negative E180 FISH patterns were successfully reprobed with two ribosomal multigene families using homologous 18S–28S and 5S coding regions as probes (our unpubl. res.); these families were also isolated from M. arborea, the same species from which the E180 family was isolated and used in the FISH experiments.
Thus, available data suggest that the results obtained are due to the intrinsic sensitivities of the two techniques. The results of the PCR approach, which is more sensitive in detecting in vitro unique or low-copy DNA sequences, suggest that the E180 family is conserved not only in Medicago but also in other related (Trigonella and Melilotus) and even more phylogenetically distant genera (e.g. Trifolium). The absence of the E180 motif in some species of Medicago is intriguing but we are confident that it is not due to PCR vagaries. It could be argued that modification in the primer region, and homogenization and consequently sequence turnover, may change primer binding sites drastically, therefore giving negative PCR amplifications.
Moreover, the FISH method is considered a roughly semi-quantitative methodology, i.e. it indirectly assesses the relative copy number of the DNA probe in the genome. In our studies, stronger hybridization signals are observed in the subtelomeric and interstitial E180 loci than in the pericentromeric loci, suggesting this satellite family has a preferential genomic location. Furthermore, the positive detection, size and intensity, and chromosomal location of the E180 signals observed in a subset of the analysed species in our study suggest phylogenetically independent processes of amplification and deletion of E180 units in Medicago (see below). Our results, although based on a single DNA repeat family, strongly warn against the use of a single methodological approach when dealing with the presence and location of satDNA along closely related evolutionary lineages.
The lack of the E180 family in M. arborea: natural polymorphism or artefact?
Our FISH analyses of the E180 family in the M. sativa complex agree with the number of sites and chromosomal distribution reported by Calderini et al. (1997); their results were obtained using the C300 repeat family as a probe, corroborating the hypothesis that the E180 and C300 repeats belong to the same satDNA family. However, these authors reported that M. arborea lacked the C300 repeat family, as assessed by Southern blot hybridization and FISH analyses.
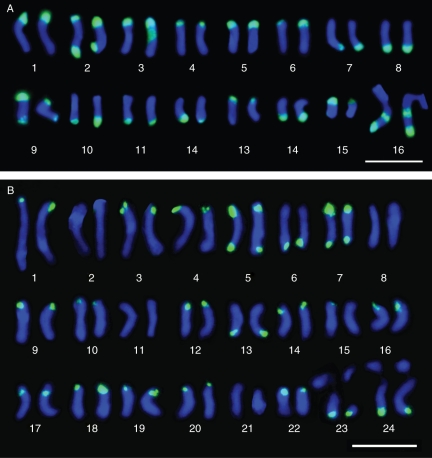
By contrast, our results with the E180 family not only showed its presence in the subtelomeric regions of all chromosomes but also in the related members of section Dendrotelis, the closely tetraploid M. strasseri and the hexaploid M. citrina (Table 1, Fig. 3). It seems unlikely that these contrasting results were due to a misidentification of the accessions used, given the singular morphological features exhibited by M. arborea (a perennial woody medic up to 1·5 m tall). Rather, the origin of the accessions analysed could be the cause of the disagreement between the results obtained by the two teams. Calderini et al. (1997) reported that their M. arborea accession was an experimental strain derived from a mesophyll protoplast culture, whereas our sample originated from the wild. It is possible that, during the establishment of this strain, genomic rearrangements produced a loss, or at least a dramatic reduction in copy number, of the highly repetitive satellite sequence. In fact, Pluhar et al. (2001) reported the reduction in the copy number of the E180 family during callus formation in different genotypes of M. sativa. These authors reasoned that the genomic stress induced by tissue culture might be responsible for the loss of this satDNA. Similar conclusions were anticipated by Lee and Phillips (1988), who indicated that the heterochromatin is particularly vulnerable to aberrations during the cell cycle, and the loss of these repetitive sequences is probably caused by these chromosomal aberrations and genome rearrangements. Thus, failure to detect the E180 family by Calderini et al. (1997) was probably due to the use of a genomically rearranged strain induced by tissue culture, one of the main causes inducing somaclonal variation in plants (Bairu et al., 2011).
Fig. 3.
Karyotypes of the FISH pattern of the E180 probe in polyploid Medicago species. (A) M. arborea, tetraploid; (B) M. citrina, hexaploid. Scale bars = 10 µm.
Preferential amplification of the E180 satDNA family colocalizes with the constitutive heterochromatic regions of the Medicago and Trigonella genomes
The patterns of E180 site distribution in species of Medicago revealed by FISH indicated a non-random distribution. Instead, the FISH patterns revealed that the E180 repeat is preferentially amplified in the prominent C-bands belonging to constitutive heterochromatic regions, as previously reported by various authors (Mariani and Falistocco, 1990, 1991; Falistocco and Falcinelli, 1993; Bauchan and Hossain, 1997, 1999, 2001). Surprisingly, the FISH pattern of the E180 family does not agree with the previously described C-band karyotype and knob distribution in M. citrina (Rosato and Rosselló, 2009) because there are more FISH E180 sites than C-bands. This is probably because some heterochromatin sites in M. citrina may be at the limits of resolution offered by conventional C-banding methods on mitotic chromosomes (1 × 10−2 pg, or 107 bp). To test this hypothesis, we used the less condensed meiotic chromosomes of M. citrina to compare the FISH and C-banding patterns methods outlined in Rosato and Rosselló (2009). The high-quality spread of the pachytene bivalents showed very tiny and weak C-bands at the faint terminal knobs in this species (not detected in somatic mitotic chromosomes) and is consistent with the E180 FISH karyotype (Supplementary Data Fig. S2). The same observation has been reported in different plant species, where the distribution of the satDNA sequence is not always corroborated by the C-banding pattern (Bedbrook et al., 1980; Teoh et al., 1983; Narayan et al., 1985; Sumner, 1998) or the presence of knob-like heterochromatin (Cheng et al., 2001).
These results agree with the overall chromosomal pattern of heterochromatin in angiosperm species (Guerra, 2000). The emerging pattern is that the heterochromatin is preferentially located in similar chromosomal regions regardless of the distance from the centromere. Heitz (1957) hypothesized that the heterochromatin is preferentially distributed in the terminal, interstitial and proximal regions of the chromosomes (termed equilocal distribution). Schweizer and Loidl (1987) postulated that the heterochromatin was preferentially observed on shorter chromosomes (or chromosome arms) as heterochromatic blocks in the telomeric regions, whereas the longer chromosomes show heterochromatin in the intercalary regions. This distribution pattern (termed equidistant localization) was hypothesized to be the consequence of the spatial disposition of the telomeres in the mitotic interphase nucleus (Rabl orientation).
In the analysed Medicago species that had positive FISH E180 signals, they showed both a preferential equilocalization, i.e. localized at pericentromeric (type II) and subtelomeric regions (type III), and a non-equidistant location in the non-homologous chromosomes. The data reviewed by Guerra (2000) suggested that the equidistribution of heterochromatin was facilitated by the Rabl polarization and the heterochromatin–centromere distance. Thus, DNA sequences in different chromosome regions may have the same possibilities of amplification and dispersion due to sharing structural and functional similarity. Ultimately, this pattern should be confirmed when other satDNA families from Medicago species are isolated and physically mapped by FISH. Unfortunately, study of the DNA sequence composition of heterochromatin in Medicago is still in its infancy, and, to date, only three other satDNA families (centromeric, MtR3; pericentromeric, MtR1 and MtR2 isolated from the model plant M. truncatula; Kulikova et al., 2004) are known to be present in the genus.
Can the distribution of the E180 family shed light on the phylogeny and evolutionary patterns in Medicago and related genera?
The DNA library model proposed by Fry and Salser (1977) hypothesized that closely related species share a set of DNA satellite families (satDNA library) differing in copy number and sequence divergence (Ugarković and Plohl, 2002). The highly dynamic nature of satDNA turnover results in considerable fluctuation in satellite copy number and sequence variations, even in closely related species. As a consequence of the amplification of a particular satellite family in a species, that particular family of satDNA becomes highly abundant, while the other families are present only as minor repeats; this leads to species-specific satDNA profiles (Ugarković and Plohl, 2002).
When the above conceptual framework, the experimental constraints imposed by the PCR (lack of information concerning the genomic situation of the repeats) and FISH (detection of satDNA sites above a 4-Mb threshold) techniques used, and the available phylogenetic and taxonomic knowledge of the medicagoid core are considered, the following statements can be made.
Amplification of the E180 family, as inferred by the presence of a positive FISH pattern, appears to have been produced through recurrent and independent evolutionary episodes in both annual and perennial Medicago species as well as in basal and in derived clades, as evidenced by all phylogenetic studies using low-copy and multigene nuclear, and organellar DNA sequences that are so far available (Bena et al., 1998a, b; Downie et al., 1998; Bena, 2001; Maureira-Butler et al., 2008; Steele et al., 2010).
Nevertheless, two phylogenetic points need to be noted. The first implies that the sister polyploid species M. rugosa and M. scutellata (Bena et al., 1998a, b) show the same FISH pattern involving only centromeric bands (type II chromosomes). The second includes the M. sativa and M. arborea complexes. Although no obvious taxonomic relationships between both groups have been postulated and they are traditionally grouped into separate sections (sect. Medicago and sect. Dendrotelis, respectively), their phylogenetic closeness is greater than previously expected and has been repeatedly inferred by a suite of independent DNA sequences (Bena et al., 1998a, b; Bena, 2001; Maureira-Butler et al., 2008; Steele et al., 2010). The M. arborea complex is composed exclusively of polyploid (tetraploid and hexaploid) species but no clues about their origin have been postulated (Rosato et al., 2008). Interestingly, type III, IV and VI chromosomes are present in members of both complexes (Table 1) and have not been found elsewhere in our Medicago sampling, supporting their phylogenetic relationships. Other molecular markers should be used to determine the diploid ancestors from which the polyploid section Dendrotelis within the M. sativa complex originated.
The FISH pattern of the E180 family is species-specific in both the M. arborea and M. sativa complexes
The chromosomes of Medicago species are relatively small and only differ slightly in size, hampering the identification of unknown samples using conventional karyological techniques. Our results have shown that all analysed taxa from both M. arborea and M. sativa (only M. postrata has not been included in our analysis) complexes can be identified using the E180 probe (Table 1), even those showing the same chromosome types, and are therefore taxon-specific.
Nevertheless, we strongly discourage their use as molecular markers in tracing the species contribution in breeding programmes containing somatic hybrids, or plants derived from tissue culture. The chromosomal and molecular rearrangements resulting from these techniques are well known (Bairu et al., 2011), but it has been specifically reported in Medicago that the nuclear ribosomal regions (Crea et al., 1997) and the highly repetitive E180 DNA sequence are affected (Pluhar et al., 2001). The contrasting results obtained for M. arborea using wild accessions (this study) or tissue-cultured plants (Calderini et al., 1997) highlight the need to use natural, non-modified accessions whenever possible.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank the Institut fur Pflanzengenetik and Kulturpflanzenforschung (Gatersleben, Germany) and the Botanischer Garten Berlin-Dahlem (Berlin, Germany) for kindly providing seeds, and an anonymous referee for insightful comments that improved the manuscript. This work was supported by funds from the Spanish Ministry of Education and Science [Project CGL2010-22347-C02-01], the Catalan Government [Consolidated Research Group 2009SGR608] and by a PhD grant from the Spanish Ministry of Education and Science to J.A.G.
APPENDIX
Species and accessions used in this study.
| 2n | x | PCR | Accession | Origin | |
|---|---|---|---|---|---|
| Medicago | |||||
| Sect. Dendrotelis | |||||
| M. arborea L. | 32 | 4x | + | BGV-VAL196335 | n.a. |
| M. citrina (Font Quer) Greuter | 48 | 6x | + | BGV-VAL196337 | Spain |
| M. strasseri Greuter, Matthäs & Risse | 32 | 4x | + | BGB-1579 | Crete |
| Sect. Medicago | |||||
| M. caerulea Less. ex Ledeb. | 16 | 2x | + | IPK-MED2640 | n.a. |
| M. falcata L. | 16 | 2x | + | IPK-MED172 | n.a. |
| M. sativa L. | 32 | 4x | + | BGB-2047 | Germany |
| M. glutinosa M. Bieb. | 32 | 4x | + | IPK-MED148 | n.a. |
| M. hemicycla Grossh. | 32 | 4x | + | IPK-MED149 | n.a. |
| M. marina L. | 16 | 2x | + | BGV-VAL196733 | France |
| M. suffruticosa Ram. ex DC. | 16 | 2x | + | IPK-MED174 | n.a. |
| M. polychroa Grossh. | 32 | 4x | + | IPK-MED143 | Russia |
| Sect. Carstiensae | |||||
| M. carstiensis Wulf. | 16 | 2x | + | IPK-MED152 | n.a. |
| Sect. Spirocarpos | |||||
| Subsect. Pachyspireae | |||||
| M. aculeata Wield. | 16 | 2x | + | IPK-MED50 | Morocco |
| M. littoralis Rohde | 14 | 2x | + | IPK-MED192 | Portugal |
| M. murex Willd. | 14 | 2x | + | IPK-MED15 | Greece |
| M. rigidula (L.) All. | 16 | 2x | + | IPK-MED102 | Bulgaria |
| M. tornata (L.) Mill. | 16 | 2x | + | IPK-MED211 | n.a. |
| M. truncatula Gaertn. | 16 | 2x | + | IPK-MED28 | Greece |
| M. turbinata (L.) All. | 16 | 2x | + | IPK-MED211 | n.a. |
| Subsect. Rotatae | |||||
| M. blancheana Boiss. | 16 | 2x | – | BGV-VAL40861 | Spain |
| M. bonarotiana Arcang. | 16 | 2x | – | IPK-MED108 | n.a. |
| M. rotata Boiss. | 16 | 2x | + | IPK-MED105 | Israel |
| M. rugosa Desr. | 30 | 4x | + | IPK-MED54 | n.a. |
| M. scutellata (L.) Mill. | 32 | 4x | + | IPK-MED176 | Italy |
| Subsect. Intertextae | |||||
| M. ciliaris (L.) Krocker | 16 | 2x | + | IPK-MED182 | Egypt |
| M. intertexta (L.) Mill. | 16 | 2x | + | IPK-MED60 | Portugal |
| M. granadensis Willd. | 16 | 2x | + | IPK-MED107 | n.a. |
| M. muricoleptis Tin. | 16 | 2x | + | IPK-MED187 | n.a. |
| Subsect. Leptospireae | |||||
| M. arabica (L.) Huds. | 16 | 2x | + | IPK-MED40 | n.a. |
| M. coronata (L.) Bart. | 16 | 2x | – | IPK-MED110 | Turkey |
| M. disciformis DC | 16 | 2x | + | IPK-MED104 | Bulgaria |
| M. laciniata (L.) Mill. | 16 | 2x | + | IPK-MED62 | Morocco |
| M. minima (L.) Bart. | 16 | 2x | + | IPK-MED680 | Morocco |
| M. polymorpha L. | 14 | 2x | + | IPK-MED42 | Italy |
| M. praecox DC. | 14 | 2x | + | BGB-2045 | Greece |
| Sect. Lupularia | |||||
| M. lupulina L. | 16 | 2x | + | IPK-MED4 | Italy |
| Sect. Orbiculares | |||||
| M. orbicularis (L.) Bart. | 16 | 2x | + | IPK-MED7 | Greece |
| Sect. Platycarpae | |||||
| M. cretacea M. Bieb. | 16 | 2x | + | IPK-MED151 | n.a. |
| Sect. Buceras | |||||
| Subsect. Erectae | |||||
| M. polyceratia (L.) Trautv. | 28 | 4x | + | IPK-TRIG24 | Portugal |
| Subsect. Reflexae | |||||
| M. monspeliaca (L.) Trautv. | 16 | 2x | + | IPK-TRIG30 | n.a. |
| Trigonella | |||||
| T. balansae Boiss. & Reut. | 16 | 2x | + | IPK-TRIG76 | Greece |
| T. caerulea (L.) Ser. | 16 | 2x | – | IPK-TRIG96 | Georgia |
| T. caerulescens (M. Bieb.) Halács. | 16 | 2x | + | IPK-TRIG83 | Turkey |
| T. calliceras Fish. ex M. Bieb | 16 | 2x | – | IPK-TRIG15 | n.a. |
| T. corniculata L. | 16 | 2x | + | IPK-TRIG18 | India |
| T. cretica (L.) Boiss. | 16 | 2x | – | IPK-TRIG8 | n.a. |
| T. geminiflora Bunge | 44 | 4x | + | IPK-TRIG31 | Uzbekistan |
| T. graeca Boiss. | 16 | 2x | + | IPK-TRIG67 | Greece |
| T. grandiflora Bunge | 16 | 2x | – | IPK-TRIG111 | Kazakhstan |
| T. noeana Boiss. | 32 | 4x | + | IPK-TRIG28 | n.a. |
| T. procumbens (Besser) Rchb. | 16 | 2x | + | IPK-TRIG77 | Hungary |
| T. rechingeri Sirj. | 16 | 2x | + | IPK-TRIG68 | Greece |
| Melilotus | |||||
| M. alba Medik. | 16 | 2x | + | IPK-MEL45 | Albania |
| M. indica (L.) All. | 16 | 2x | + | IPK-MEL44 | Tunisia |
| M. infesta Guss. | 16 | 2x | + | IPK-MEL37 | Italy |
| M. officinalis (L.) Pall. | 16 | 2x | + | IPK-MEL38 | Uzbekistan |
| M. segetalis (Brot.) Sér. in DC. | 16 | 2x | + | IPK-MEL41 | Italy |
| M. sulcata Desf. | 16 | 2x | – | IPK-MEL16 | n.a. |
| M. wolgica Poir. | 16 | 2x | + | IPK-MEL18 | n.a. |
| Trifolium | |||||
| T. alexandrinum L. | 16 | 2x | + | IPK-TRIF312 | Italy |
| T. angustifolium L. | 16 | 2x | + | IPK-TRIF263 | Italy |
| T. arvense L. | 14 | 2x | + | IPK-TRIF258 | Hungary |
| T. campestre Schreb. | 14 | 2x | + | IPK-TRIF41 | n.a. |
| T. cherleri Jusl. | 10 | 2x | + | IPK-TRIF256 | France |
| T. incarnatum L. | 14 | 2x | + | IPK-TRIF17 | n.a. |
| T. lupinaster L. | 28 | 4x | + | IPK-TRIF262 | Russia |
| T. physodes Steven ex M. Bieb. | 16 | 2x | + | IPK-TRIG261 | Portugal |
| T. scabrum L. | 16 | 2x | – | IPK-TRIF272 | Italy |
| T. striatum L. | 14 | 2x | + | IPK-TRIF23 | n.a. |
| T. subterraneum L. | 16 | 2x | – | IPK-TRIF259 | USA |
Chromosome number, ploidy level (x), and positive (+) and negative (–) PCR amplifications of the E180 satDNA family of the analysed accessions are also reported. IPK: Institut fur Pflanzengenetik and Kulturpflanzenforschung, Gatersleben; BGB: Botanischer Garten Berlin-Dahlem; BGV: Botanical Garden of Valencia University. n.a.= not available.
LITERATURE CITED
- Abad JP, Carmena M, Baars S, et al. Dodeca satellite: a conserved G+C rich satellite from the centromeric heterochromatin of Drosophila melanogaster. Proceedings of the National Academy of Sciences, USA. 1992;89:4663–4667. doi: 10.1073/pnas.89.10.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrožová K, Mandáková T, Bureš P, et al. Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Annals of Botany. 2011;107:255–268. doi: 10.1093/aob/mcq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairu MW, Aremu AO, Van Staden J. Somaclonal variation in plants: causes and detection methods. Plant Growth Regulation. 2011;63:147–173. [Google Scholar]
- Bauchan GR, Hossain MA. Karyotypic analysis of C-banded chromosomes of diploid alfalfa (Medicago sativa ssp. coerulea and ssp. falcata) Journal of Heredity. 1997;88:533–537. doi: 10.1093/oxfordjournals.jhered.a023152. [DOI] [PubMed] [Google Scholar]
- Bauchan GR, Hossain MA. Constitutive heterochromatin DNA polymorphisms in diploid Medicago sativa ssp. falcata. Genome. 1999;42:930–935. doi: 10.1139/g99-038. [DOI] [PubMed] [Google Scholar]
- Bauchan GR, Hossain MA. Distribution and characterization of heterochromatic DNA in the tetraploid African population alfalfa genome. Crop Science. 2001;41:1921–1926. [Google Scholar]
- Bedbrook JR, Jones J, O'Dell M, Thompson RD, Flavell RB. A molecular description of telomeric heterochromatin in Secale species. Cell. 1980;19:545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- Bena G. Molecular phylogeny supports the morphologically based taxonomic transfer of the “medicoid” Trigonella species to the genus Medicago L. Plant Systematics and Evolution. 2001;229:217–236. [Google Scholar]
- Bena G, Jubier M-F, Olivieri I, Lejeune B. Ribosomal external and internal transcribed spacers: combined use in the phylogenetic analysis of Medicago (Leguminosae) Journal of Molecular Evolution. 1998a;46:299–306. doi: 10.1007/pl00006306. [DOI] [PubMed] [Google Scholar]
- Bena G, Lejeune B, Prosperi J-M, Olivieri I. Molecular phylogenetic approach for studying life-history evolution: the ambiguous example of the genus Medicago L. Proceedings of Royal Society of London B. 1998b;265:1141–1151. doi: 10.1098/rspb.1998.0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderini O, Pupilli F, Paolocci F, Arcioni S. A repetitive and species-specific sequence as a tool for detecting the genome contribution in somatic hybrids of the genus Medicago. Theoretical and Applied Genetics. 1997;95:734–740. [Google Scholar]
- Cheng Z, Stupar RM, Gu M, Jiang J. A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma. 2001;110:24–31. doi: 10.1007/s004120000126. [DOI] [PubMed] [Google Scholar]
- Contento A, Heslop-Harrison JS, Schwarzacher T. Diversity of a major repetitive DNA sequence in diploid and polyploid Triticeae. Cytogenetics and Genome Research. 2005;109:34–42. doi: 10.1159/000082379. [DOI] [PubMed] [Google Scholar]
- Crea F, Calderini O, Nenz E, Cluster , Damiani F, Arcioni S. Chromosomal and molecular rearrangements in somatic hybrids between tetraploid Medicago sativa and diploid Medicago falcata. Theoretical and Applied Genetics. 1997;95:1112–1118. [Google Scholar]
- Csink AK, Henikoff S. Something from nothing: the evolution and utility of satellite repeats. Trends in Genetics. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive. Trends in Genetics. 2002;18:587–589. doi: 10.1016/s0168-9525(02)02789-0. [DOI] [PubMed] [Google Scholar]
- Downie SR, Downie DSK, Rogers EJ, Zujewski HL, Small E. Multiple independent losses of the plastid rpoC1 intron in Medicago (Fabaceae) as inferred from phylogenetic analyses of nuclear ribosomal DNA internal transcribed spacer sequences. Canadian Journal of Botany. 1998;76:791–803. [Google Scholar]
- Falistocco E, Falcinelli M. Karyotype and C-banding in Medicago noëana Boiss., Leguminosae. Cytologia. 1993;58:151–154. [Google Scholar]
- Figueroa DM, Bass HW. A historical and modern perspective on plant cytogenetics. Briefings in Functional Genomics. 2010;9:95–102. doi: 10.1093/bfgp/elp058. [DOI] [PubMed] [Google Scholar]
- Fry K, Salser W. Nucleotide sequences of HS-alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell. 1977;12:1069–1084. doi: 10.1016/0092-8674(77)90170-2. [DOI] [PubMed] [Google Scholar]
- Guerra M. Patterns of heterochromatin distribution in plant chromosomes. Genetics and Molecular Biology. 2000;23:1029–1041. [Google Scholar]
- Hans de Jong J, Fransz P, Zabel P. High resolution FISH in plants – techniques and applications. Trends in Plant Science. 1999;4:258–263. doi: 10.1016/s1360-1385(99)01436-3. [DOI] [PubMed] [Google Scholar]
- Heikkinen E, Launonen V, Muller E, Bachmann L. The pvB370 BamHI satellite DNA family of the Drosophila virilis group and its evolutionary relation to mobile dispersed genetic pDv elements. Journal of Molecular Evolution. 1995;41:604–614. doi: 10.1007/BF00175819. [DOI] [PubMed] [Google Scholar]
- Heitz E. Tendeloo HJC, editor. Die Chromosomenstruktur im kern während der kernteilung und der entwicklung des organismus. Conference on chromosomes. 1957 Willink: Zwolle, 5–26. [Google Scholar]
- Hemleben V, Kovarik A, Torres-Ruiz RA, Volkov RA, Beridze T. Plant highly repeated satellite DNA: molecular evolution, distribution and use for identification of hybrids. Systematics and Biodiversity. 2007;5:277–289. [Google Scholar]
- Heslop-Harrison JS. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. The Plant Cell. 2000;12:617–635. doi: 10.1105/tpc.12.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikova O, Geurts R, Lamine M, et al. Satellite repeats in the functional centromere and pericentromeric heterochromatin of Medicago truncatula. Chromosoma. 2004;113:276–283. doi: 10.1007/s00412-004-0315-3. [DOI] [PubMed] [Google Scholar]
- Lee M, Phillips RL. The chromosomal basis of somaclonal variation. Annual Review of Plant Physiology and Plant Molecular Biology. 1988;39:413–437. [Google Scholar]
- Macas J, Meszaros T, Nouzova M. PlantSat: a specialized database for plant satellite repeats. Bioinformatics. 2002;18:28–35. doi: 10.1093/bioinformatics/18.1.28. [DOI] [PubMed] [Google Scholar]
- Macas J, Navrátilová A, Koblížková A. Sequence homogenization and chromosomal localization of VicTR-B satellites differ between closely related Vicia species. Chromosoma. 2006;115:437–447. doi: 10.1007/s00412-006-0070-8. [DOI] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navrátilová A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. http://dx.doi.org/10.1186/1471-2164-8-427 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani A, Falistocco E. Chromosome studies in 2n=14 and 2n=16 type of Medicago murex. Genome. 1990;33:159–163. [Google Scholar]
- Mariani A, Falistocco E. Cytogenetic analysis of Medicago rugosa and Medicago scutellata. Journal of Genetic and Breeding. 1991;45:111–116. [Google Scholar]
- Maureira-Butler IJ, Pfeil BE, Muangprom A, Osborn TC, Doyle JJ. The reticulate history of Medicago (Fabaceae) Systematic Biology. 2008;57:466–482. doi: 10.1080/10635150802172168. [DOI] [PubMed] [Google Scholar]
- Mestrovic N, Plohl M, Mravinac B, Ugarković D. Evolution of satellite DNAs from the genus Palorus experimental evidence for the ‘library’ hypothesis. Molecular Biology and Evolution. 1998;15:1062–1068. doi: 10.1093/oxfordjournals.molbev.a026005. [DOI] [PubMed] [Google Scholar]
- Narayan RKJ, Ramachandran C, Raina SN. The distribution of satellite DNA in the chromosome complements of Vicia species (Leguminosae) Genetica. 1985;66:115–121. [Google Scholar]
- Navajas-Pérez R, Quesada del Bosque ME, Garrido-Ramos MA. Effect of location, organization, and repeat-copy number in satellite-DNA evolution. Molecular and Genetic Genomics. 2009;282:395–406. doi: 10.1007/s00438-009-0472-4. [DOI] [PubMed] [Google Scholar]
- Pluhar SA, Erickson L, Pauls KP. Effects of tissue culture on a highly repetitive DNA sequence (E180 satellite) in Medicago sativa. Plant Cell, Tissue and Organ Culture. 2001;67:195–199. [Google Scholar]
- Rosato M, Rosselló JA. Karyological observations in Medicago section Dendrotelis (Fabaceae) Folia Geobotanica. 2009;44:423–433. [Google Scholar]
- Rosato M, Castro M, Rosselló JA. Relationships of the woody Medicago species (section Dendrotelis) assessed by molecular cytogenetic analyses. Annals of Botany. 2008;102:15–22. doi: 10.1093/aob/mcn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ, Wojciechowski MF. Molecular phylogenetic analysis of a temperate legume clade (Fabaceae) American Journal of Botany. 1995;82:159. (Supplement) [Google Scholar]
- Schmidt T, Heslop-Harrison JS. Genomes, genes and junk: the large scale organization of plant chromosomes. Trends in Plant Science. 1998;3:195–199. [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. Practical in situ hybridization. Oxford: Bios; 2000. [Google Scholar]
- Schweizer D, Loidl J. A model for heterochromatin dispersion and the evolution of C-band patterns. Chromosomes Today. 1987;9:61–74. [Google Scholar]
- Small E, Jomphe M. A synopsis of the genus Medicago (Leguminosae) Canadian Journal of Botany. 1989;67:3260–3294. [Google Scholar]
- Steele KP, Ickert-Bond SM, Zarre S, Wojciechowski MF. Phylogeny and character evolution in Medicago (Leguminosae): evidence from analyses of plastid TRNK/MATK and nuclear GA3OX1 sequences. American Journal of Botany. 2010;97:1142–1155. doi: 10.3732/ajb.1000009. [DOI] [PubMed] [Google Scholar]
- Suárez-Santiago VN, Blanca G, Ruiz-Rejón M, Garrido-Ramos MA. Satellite-DNA evolutionary patterns under a complete evolutionary scenario: the case of Acrolophus subgroup (Centaurea L., Compositae) from the western Mediterranean. Gene. 2007;404:80–92. doi: 10.1016/j.gene.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Sumner AT. The mitotic chromosome. Advances in Genome Biology. 1998;5:211–261. [Google Scholar]
- Teoh B, Hutchinson J, Miller TE. A comparison of the chromosomal distribution of cloned repetitive DNA sequences in different Aegilops species. Heredity. 1983;51:635–641. [Google Scholar]
- Ugarković D, Plohl M. Variation in satellite DNA profiles – causes and effects. EMBO Journal. 2002;21:5955–5959. doi: 10.1093/emboj/cdf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski MF, Sanderson MJ, Steele KP, Liston A. Molecular phylogeny of the “Temperate Herbaceous Tribes” of Papilionoid legumes: a supertree approach. In: Herendeen PS, Bruneau A, editors. Advances in legume systematics. Vol. 9. Kew, UK: Royal Botanic Gardens; 2000. pp. 277–298. [Google Scholar]
- Xia X, Erickson L. An AT-rich satellite DNA sequence, E180, in alfalfa (Medicago sativa) Genome. 1993;36:427–432. doi: 10.1139/g93-058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.