Abstract
HE3286, 17α-ethynyl-5-androstene-3β, 7β, 17β-triol, is a novel synthetic compound related to the endogenous sterol 5-androstene-3β, 7β, 17β-triol (β-AET), a metabolite of the abundant adrenal steroid dehydroepiandrosterone (DHEA). HE3286 has shown efficacy in clinical studies in impaired glucose tolerance and type 2 diabetes, and in vivo models of types 1 and 2 diabetes, autoimmunity, and inflammation. Proteomic analysis of solid-phase HE3286-bound bead affinity experiments, using extracts from RAW 264.7 mouse macrophage cells, identified 26 binding partners. Network analysis revealed associations of these HE3286 target proteins with nodes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for type 2 diabetes, insulin, adipokine, and adipocyte signaling. Binding partners included low density lipoprotein receptor-related protein (Lrp1), an endocytic receptor; mitogen activated protein kinases 1 and 3 (Mapk1, Mapk3), protein kinases involved in inflammation signaling pathways; ribosomal protein S6 kinase alpha-3 (Rsp6ka3), an intracellular regulatory protein; sirtuin-2 (Sirt2); and 17β-hydroxysteroid dehydrogenase 1 (Hsd17β4), a sterol metabolizing enzyme.
Introduction
β-AET is a metabolite of dehydroepiandrosterone (DHEA), the most abundant adrenal sterol in the human metabolome [1]. In rodent studies, the anti-inflammatory activities of exogenous β-AET and DHEA [2] are similar. For β-AET, anti-inflammatory activity in rodent models of multiple sclerosis [3], ulcerative colitis [4], lung inflammation and septic shock [2] has been reported. β-AET also opposes certain glucocorticoid (GC) actions in vitro [5] and decreases corticosteroid-induced bone loss in the mouse thermal injury model [6]. Elevated levels of GC are associated with glucose dysregulation and hyperglycemia. Since 11β-HSD1 is involved in the formation of β-AET [7] and GC activity, increased levels of β-AET in obese subjects may relate to the increased levels of this enzyme in obesity [8]. This implies a negative feedback loop, where increased 11β-HSD1 expression results in simultaneous amplification of GC signaling and β-AET formation, which may function to counter undesired aspects of GC signaling and maintain homeostasis. In conditions of GC-associated pathology, this relationship between GC and β-AET may be unbalanced. Furthermore, given that rodents convert DHEA and β-AED into metabolites with significantly more sites of oxidation [9], species differential metabolism may explain why activities observed for these sterols have not translated into human benefit [2], [9].
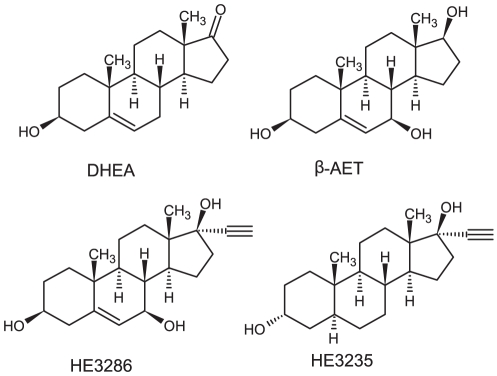
HE3286, an orally bioavailable synthetic compound (Figure 1), is pharmacologically distinct from androgens, estrogens, corticosteroids, and peroxisome proliferators [10]. HE3286 has demonstrated anti-inflammatory activities similar to β-AET in murine models of ulcerative colitis [4], multiple sclerosis [4], type 1 diabetes mellitus [11], chronic obstructive pulmonary disease [12], and rheumatoid arthritis [4], [13], [14], [15]. All of these models involve attenuation of NF-κB activity and decreased levels of pro-inflammatory cytokines, including TNFα. HE3286 exhibits anti-inflammatory and insulin sensitizing activities in preclinical db/db and ob/ob mouse, fa/fa ZDF rat models of diabetes [10], [16], and in a rare example of species translation for sterols, in contrast to DHEA or β-AET, HE3286 is active in impaired glucose tolerant and type 2 diabetes mellitus human subjects [17].
Figure 1. Chemical structures of DHEA, β-AET, HE3286 and HE3235 (negative control).
In mouse macrophages, HE3286 suppressed lipopolysaccharide (LPS)-induced nuclear factor kappa B (NF-κB) reporter gene expression, nuclear localization, and insulin receptor substrate-1 (IRS-1) serine phosphorylation. In addition, this compound decreased activation of the pro-inflammatory kinases IkappaB kinase (IKK), Jun N-terminal kinase (JNK) and p38 mitogen-activated kinase (p38 Mapk). HE3286 also attenuated tumor necrosis factor alpha (TNFα-stimulated inflammation and TNFα-induced adipocyte-stimulated macrophage chemotaxis [10], [16]. HE3286 treatment suppressed levels of the chemokine monocyte chemoattractant protein-1 (MCP-1), along with its cognate receptor, C-C motif chemokine receptor-2, in white adipose tissue [10]. Similarly, in liver and adipose tissue, HE3286 down-regulated inflammatory cytokine/chemokine protein levels and decreased macrophage migration into adipose tissue in Zucker diabetic fatty (ZDF) rats [16]. In obese humans with inflammation-induced diabetes, HE3286 decreased inflammatory cytokine production, C-reactive protein (CRP) and post-prandial glucose levels, leading to a decrease in HbA1c. HE3286 was safe with no drug-related side effects [18]. These properties suggest a potential role for endogenous β-AET, which is lost during inflammation-driven diseases, and exogenous HE3286 in limiting chronic inflammation. Thus, HE3286 represents the first in a new class of anti-inflammatory agents with the potential to treat chronic inflammatory conditions. The clinical results from administering HE3286 prompted the exploration of target proteins in the macrophage, a cell type that is central to the activity of HE3286, using stable isotope labeling with amino acids in culture (SILAC) technology in RAW 264.7 mouse macrophages.
Results
There were four method variations run in the SILAC experiments. The first and second variations, referred to as forward (A) and reverse (B) conditions, used a solid-phase HE3286-bead compared to non-sterol modified agarose-beads as a control. The third variation, referred to as forward (C) conditions, used solid-phase HE3286-bead in both light and heavy cell preparations where the heavy cell lysate was pre-incubated with 50 µM HE3286 as a soluble competitor. The fourth variation, forward (D) conditions, used solid-phase HE3286-beads with 50 µM HE3235 (NF-κB inactive), pre-incubated in the heavy cells as a soluble competitor and was similarly compared to DMSO buffer using solid-phase HE3286-beads.
The number of proteins found for each experiment and the number identified with a ratio ≥1.5 are delineated in Table 1. A target decoy database of reverse sequences was used to assure a false discovery rate of <1%. This allowed a comparison of different MS database search algorithms for protein identification as there is a known difference in protein identification using different quantification software [19]. We compared search results, found with the Sequest database followed by DTA select processing, with the !Xtandem database, followed by Peptide Prophet and Protein Prophet. In general, the total number of identified proteins was similar. However, while 874 proteins were found in experiment 2A using !Xtandem is extremely close to the 878 discovered initially using Sequest, only 365 coordinate identifications were established. Notably, the alternative !Xtandem methodology, identified 577 proteins with a ratio of >1.5 compared to 372 using the Sequest/DTA analysis. We chose to use the Sequest/DTA algorithms for comparison of separate experiments, as we believed this would provide fewer false positives. Within each experiment we evaluated MS/MS data for each hit to validate the ratios used in our analysis.
Table 1. Comparison of the number of proteins identified and the number of protein ratios of >1.5 in representative experiments.
| Exp | L/H* | Conditions | #Proteins | #Ratios >1.5 |
| 1A | >1 | (Heavy+Control Bead)+(Light+3286 Bead) | 400 | 141 |
| 1B | <1 | (Light+Control Bead)+(Heavy+3286 Bead) | 580 | 162 |
| 1C | >1 | (Heavy+50 uM HE3286+3286 Bead)+(Light+3286 Bead) | 369 | 8 |
| 1D | = 1 | (Heavy+50 uM HE3235+3286 Bead)+(Light+3286 Bead) | 698 | 131 |
| 2A | >1 | (Heavy+Control Bead)+(Light+3286 Bead) | 878 | 372 |
| 2B | <1 | (Light+Control Bead)+(Heavy+3286 Bead) | 919 | 429 |
| 2C | >1 | (Heavy+50 uM HE3286+3286 Bead)+(Light+3286 Bead) | 942 | 157 |
*L/H: (light isotope label/heavy isotope label) expresses the expected outcome ratio for each set of conditions.
Overall, the methodology initially identified 44 candidate binding partners. These were additionally constrained by censoring against a published list of promiscuous binding partners that have been identified through a retrospective analysis of several SILAC experiments [20]. The final analysis revealed 26 HE3286 binding partners that exceeded a 1.5 binding enrichment ratio threshold, listed in Table 2. A description of protein function for these binding partners is included in supplemental Table S1. In theory, the resulting ratio for the 1D experiment should be ≥1.5 in order for this HE3235 control experiment to uncover proteins that non-specifically bind to sterols. However, as HE3235 is similar in chemical structure to HE3286, there may be shared binding targets between these synthetic sterols that can influence this outcome by incorrectly identifying such proteins as non-specific binding partners to HE3286.
Table 2. Gene product enrichment ratios in mus musculus RAW 264.7 monocyte/macrophage cells using SILAC-based analysis.
| Gene ID | IPI number | Gene | Product | A | B | C | D |
| 66885 | IPI00880948 | Acadsb | acyl-Coenzyme A dehydrogenase | 50.9 | 5.26 | 1.53 | 2.57 |
| 80911 | IPI00857891 | Acox3 | acyl-Coenzyme A oxidase 3, pristanoyl | 46.3 | 4.17 | 1.55 | 3.10 |
| 11564 | IPI00265471 | Adsl | adenylosuccinate lyase | 85.3 | 12.5 | 2.10 | 1.90 |
| 77559 | IPI00662244 | Agl | amylo-1,6-glucosidase, 4-alpha-glucanotransferase | 23.5 | 7.70 | 1.72 | 1.04 |
| 12039 | IPI00331555 | Bckdha | branched chain ketoacid dehydrogenase E1, alpha | 13.7 | 5.26 | 1.52 | 0.98 |
| — | IPI00858053 | Cherp | calcium homeostasis endoplasmic reticulum | 4.10 | 14.3 | 1.91 | 0.81 |
| 234594 | IPI00880243 | Cnot1 | CCR4-NOT transcription complex, subunit 1 | 30.2 | 1.79 | 1.66 | 0.92 |
| 67337 | IPI00116747 | Cstf1 | cleavage stimulation factor, 3′ pre-RNA, subunit 1 | 45.3 | 12.5 | 2.10 | 0.92 |
| 319945 | IPI00226787 | Flad1 | RFad1, flavin adenine dinucleotide synthetase | 16.3 | 12.5 | 1.66 | 0.96 |
| 51886 | IPI00123292 | Fubp1 | far upstream element binding protein 1 | 14.2 | 14.3 | 2.19 | 0.43 |
| 71665 | IPI00387228 | Fuca1 | fucosidase, alpha-L- 1, tissue | 40.5 | 12.5 | 2.28 | * |
| 11991 | IPI00330958 | Hnrnpd | heterogeneous nuclear ribonucleoprotein D | 4.42 | 16.7 | 1.79 | 0.90 |
| 50926 | IPI00755892 | Hnrpdl | heterogeneous nuclear ribonucleoprotein D-like | 24.8 | 1.92 | 5.05 | 0.82 |
| 15488 | IPI00331628 | Hsd17b4 | hydroxysteroid (17-beta) dehydrogenase 4 | 4.97 | 4.76 | 1.76 | 0.95 |
| 16971 | IPI00119063 | Lrp1 | low density lipoprotein receptor-related protein 1 | 9.77 | 9.09 | 2.25 | 0.55 |
| 26394 | IPI00123518 | Lypla2 | lysophospholipase 2 | 2.16 | 2.50 | 2.10 | 0.93 |
| 26413 | IPI00119663 | Mapk1 | mitogen-activated protein kinase 1 | 18.6 | 6.70 | 1.94 | 1.00 |
| 26417 | IPI00230277 | Mapk3 | mitogen-activated protein kinase 3 | 27.8 | 6.20 | 1.80 | 2.13 |
| 76890 | IPI00126892 | Memo1 | mediator of cell motility 1 | 4.63 | 3.85 | 1.74 | 1.08 |
| 23983 | IPI00128904 | Pcbp1 | poly(rC) binding protein 1 | 2.87 | 12.5 | 1.62 | 0.28 |
| 231329 | IPI00320034 | Polr2b | polymerase (RNA) II polypeptide B | 12.4 | 12.5 | 1.99 | 0.27 |
| 110651 | IPI00114333 | Rps6ka3 | ribosomal protein S6 kinase polypeptide 3 | 7.57 | 4.17 | 1.60 | 1.02 |
| 27054 | IPI00317604 | Sec23b | Sec23b | 11.0 | 9.09 | 1.88 | 0.78 |
| 218811 | IPI00229483 | Sec24c | Sec24 related gene family, member C | 6.62 | 11.1 | 1.67 | 0.42 |
| 64383 | IPI00110265 | Sirt2 | Sirtuin 2 | 51.2 | 8.33 | 1.80 | 1.11 |
| 19704 | IPI00420949 | Upf1 | UPF1 regulator of nonsense transcripts | 19.4 | 10.0 | 2.15 | 0.61 |
*This protein was not identified in either sample in this experiment.
Discussion
Specific HE3286 macrophage binding partners were revealed through a combination of solid phase capture and SILAC-based proteomic analysis that resulted in the identification of a set of low abundance proteins including signal transduction-related proteins and transcription factors. The list of specific HE3286 target proteins is more consistent with a biological mechanism operating through a biosystem network paradigm instead of a single target approach. When compared to other strategies for target identification where large lists of proteins are often identified, SILAC affinity experiments produce relatively fewer candidates for inspection. Because the two separate samples are combined before processing and are subjected to an identical experimental protocol, sample background effects are eliminated. Experimental strategies that use soluble competitors significantly reduce the number of potential binding partners; however, multiple binding interactions remain. Functional analyses to establish contributions of potential target proteins to the observed biological phenomena are then required in order to define the most pertinent targets associated with HE3286 biological activities.
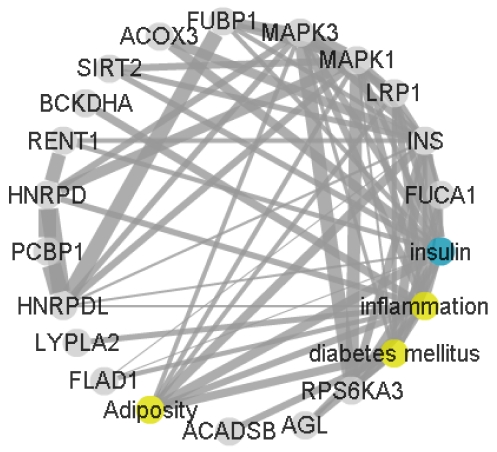
Our hypothesis, built on in vivo and clinical evidence was that pathways with known association to obesity and insulin resistance; namely insulin signaling, TNFα and LPS signaling and adipose tissue signaling resulting in the release of chemokines, should be effected by our list of associated proteins. Data visualized as a network can be easier to interpret than long lists of proteins, interactions and correlations (Figure 2) [21].
Figure 2. Network of a group of mixed terms using CoPub [21] to mine PubMed literature and generate a force directed weighted network where strongly connected terms have thick edges.
Where once proteins were considered as discrete signals, it is now recognized that many function through complex networks of protein-protein interactions. Using our list of 26 proteins our analysis set increased to 1,569 proteins by including proteins with direct interactions as defined by the BioGRID protein-protein interaction database [22]. KEGG maps for the adipocytokine, toll-like receptor, chemokine and insulin signaling pathways [23] reveal multiple nodes of interaction for these binding partners that are consistent with the observed biological activities of HE3286. The connections of the identified HE3286 binding targets to type 2 diabetes, insulin, adipokine and adipocyte signaling are clear and these KEGG maps are included in a supplemental file Networks S1.
We have subsequently analyzed the data in light of the biological activities and concurrent molecular changes seen with HE3286-treatment protocols. In addition to the anti-inflammatory activities found in preclinical models of autoimmunity and inflammation [4], [11], [12], [13], [14], [15], HE3286 decreases chronic inflammation associated with insulin resistance in rodent models of diabetes [10], [16] and in impaired glucose tolerant and type 2 diabetes mellitus human subjects [17]. HE3286 attenuates NF-κB and TNFα signaling and associated pro-inflammatory cytokines and chemokines, in these settings. Based on these results, we have focused attention on pathways that regulate NF-κB and TNFα stimulation with elaboration of inflammatory cytokines and chemokines. Three particularly important nodes, Lrp1, Mapk1 and Mapk3 were discovered that are consistent with mechanistic explanations for HE3286 anti-inflammatory and insulin-sensitizing activities.
Lrp1 was identified as the only surface receptor that binds to HE3286 on RAW264.7 cells. Lrp1 is regulated by insulin signaling [24], [25] and is important in lipid clearance and glucose tolerance. Additionally, Lrp1 signaling results in anti-inflammatory effects. Mice with an adipocyte-specific inactivation of Lrp1 displayed delayed postprandial lipid clearance, reduced body weight, smaller fat stores, improved glucose tolerance, and resistance to dietary fat-induced obesity and glucose intolerance [26]. Inactivation of the Lrp1 intracellular NpxYxxL motif enhances post prandial dyslipidemia and atherosclerosis, and increases proapoptotic effects via increased secretion of TNFα in macrophages [27].
Lrp1 is one regulator of the tumor necrosis factor receptor-1 (TNFR1) and the IKK-NF-κB pathway. Lrp1 deficient macrophages have decreased NF-κB signaling and MCP-1 expression, due to down-regulation of TNFR1 and inhibition of autocrine TNFR1-initiated cell signaling [28]. Lrp1 binds Apolipoprotien E (ApoE) [29], and ApoE induces the anti-inflammatory M2 phenotype in macrophages [30]. Lrp1 has also been shown to interact with scaffolding and signaling proteins via its intracellular domain in a phosphorylation-dependent manner [31].
In addition to a cell-surface interaction with Lrp1, HE3286 also binds Mapk1 and 3, and this binding likely modulates scaffold interactions, which allows cross-talk between Ras-Erk and Lrp1 signaling. Mapk1 and Mapk3 (Erk2, Erk1 respectively) are integral mediators of inflammatory signal transduction that are central to obesity. Mice deficient in a natural Erk inhibitor, p62, have a high level of Erk activity and develop mature-onset obesity and insulin resistance [32]. Knockout of Erk1 in these mice decreases their adiposity and insulin resistance [33]. Activation of TLR2 and TLR4 with fatty acids [34], [35] contributes to NF-κB activation, increased inflammation, and insulin resistance. Tpl2 kinase is up-regulated in adipose tissue in obese mice and human subjects and is reported to mediate NF-κB and TNFα effects upon Erk activation [36]. Inhibitors of IKKβ or TPL2 eliminated TNFα but not insulin-mediated Erk1 and Erk2 activation and also eliminated IRS-1 Ser636 phosphorylation stimulated by TNFα in 3T3-L1 adipocytes.
We have shown that addition of HE3286 in vitro to Mapk1 in the presence of activated Mek1 does not affect phosphorylation activity of Mapk1 nor does its addition to phosphorylated Mapk1 effect phosphorylation of a peptide substrate (Reading, unpublished observations). This finding suggests that HE3286 binding perturbs scaffolding of Mapk within protein complexes to attenuate Ras-Erk signal transduction rather than by acting through direct inhibition of the kinase's catalytic domain. This may be important to the safety of this molecular class as direct inhibition of kinase catalytic activity by ATP site-dependent inhibitors often leads to undesirable toxicities. The development of “soft drugs” that attenuate signaling has been suggested as an alternative to high affinity molecular entities that neutralize protein action.
While the Lrp1, Mapk1, and Mapk3 are arguably central to the biological phenomena observed for HE3286, other identified binding partners that can also significantly contribute to these effects are enzymes central to glucose metabolism (Agl, glycogen degradation) and fatty acid metabolism (Acadsd, short/branched chain acyl-CoA dehydrogenase: Acox3, acyl-CoA oxidase 3; Hsd17b4, fatty acid β-oxidase; Lypla2, lysophospholipase). Sirt2, a protein of recent interest for its role in intracellular regulation through a mono-ADP-ribosyltransferase activity was also identified as an HE3286 target protein. The steroidogenic enzyme Hsd17b4 oxidation of the C-17 hydroxyl function of C-19 sterols is known to modify the action of 17-hydroxysterols. In a pro-inflammatory environment, oxidation of β-AET deactivates its anti-inflammatory properties thereby permitting the cell to execute its inflammation program. HE3286 a chemically modified version of β-AET, should inhibit the action of this hydroxysteroid dehydrogenase to exert an anti-inflammatory effect.
The SILAC experiments generated a list of 26 target proteins specific to HE3286. This result of multiple target proteins is consistent with our expectations given the diverse biological activities for HE3286 that have been observed in separate in vitro and in vivo models and the involvement of the identified binding partners to these activities. Future additional functional activity assays will serve to detail the contributions of each binding partner to the broad anti-inflammatory, insulin-sensitizing, and lipid regulating activities of HE3286. Experiments with Lrp1 macrophage knockout mice should further establish the role of HE3286 acting upon this binding partner.
Materials and Methods
Materials
All chemicals and solvents, including, l-arginine-13C6 15N4-HCl, l-lysine-13C6 15N2-HCl, 4-iodobenzylamine hydrochloride, 9-fluorenylmethyloxycarbonyl chloride (FMOC-Cl), triethylamine, ethanolamine, palladium diacetate, triphenyl phosphine, cuprous iodide, piperidine, urea, iodoacetamide, ammonium bicarbonate, Tris(2-carboxyethyl)phosphine-HCl (TCEP), sodium chloride, formic acid, THF (anhydrous), DMF (anhydrous), and acetonitrile (ACN, HPLC grade) were purchased from Sigma-Aldrich (St. Louis, MO).
N-hydroxy-succinimide-activated agarose slurry (cat#26200) and SILAC RPMI media (cat#89984) were purchased from Thermo Scientific (Rockfield, IL). HEPES buffer was purchased from Mediatech (Manassas, VA) and the dialyzed fetal bovine serum (FBS) was purchased from Sigma-Aldrich (St. Louis, MO).
Affinity capture beads
The preparation of an amino analog of HE3286 for linkage to NHS-activated agarose beads was completed using a Sonogashira palladium cross-coupling reaction between the C-17 alkynyl group and a FMOC protected 4-iodobenzylamine [37]. The FMOC protecting group was removed following the cross-coupling by heating in piperidine. The HE3286-benzyl amine was coupled to the NHS-activated bead according to standard protocol [38]. For evaluation of non-specific binding of proteins onto the affinity sorbent, we also prepared control agarose beads that were subjected to the same treatments as the HE3286-agarose beads, except the HE3286-amine was replaced with ethanolamine.
Protein capture
The mouse macrophage cell line RAW 264.7 (ATCC CRL-2278) cells were grown for at least 6 cell divisions in media supplemented with 5% dialyzed FBS, in a humidified air atmosphere with 5% CO2. All standard SILAC media preparation and labeling steps were followed as previously described [39]. Separate RAW 264.7 cell cultures, SILAC labeled with either light or heavy amino acids, were lysed in ice-chilled Tris buffer (50 mM, pH 8) that included protease inhibitors. Light and heavy lysate protein concentrations were estimated using the DC Protein Assay (Bio-Rad, Hercules CA). Affinity enrichment mixtures were incubated overnight (4°C, 16 h) on an end-over-end rotator, and beads were combined for work-up following necessary individual washing steps.
Proteins enriched in the SILAC affinity pull-down experiments were reduced and alkylated on bead, in 5 mM TCEP and 10 mM iodoacetamide, respectively. Trypsin was then added and the vials were incubated at 37°C overnight on a thermomixer.
Protein analysis by LC-MS
The LC/MS analysis followed a standard protocol for the MuDPIT technique described in the referenced protocol [40]. There were 5 salt gradients flushed through the capillary column prior to a gradient HPLC method that eluted peptides fractions into the Mass Spec for MS and MS/MS spectra collection. Tandem mass spectra were searched using the Sequest algorithm (v 3.0) against the mouse database (ipi.MOUSEv368.fasta) from the European Bioinformatics Institute (EBI).
SILAC study design
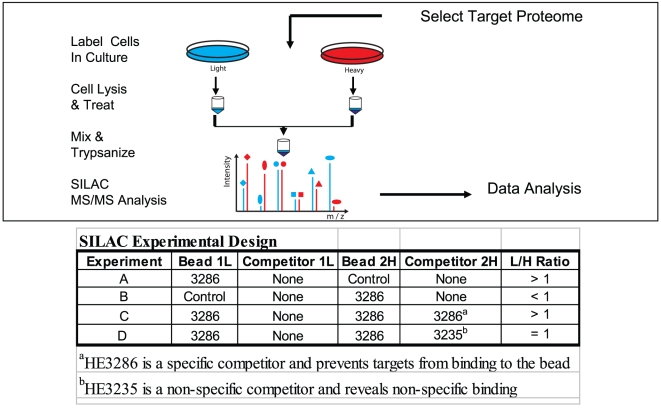
The experimental design used to identify HE2386 binding partners in the RAW 264.7 cell proteome (Figure 3) was adapted to this application from a previously described method [41]. For this purpose, HE3286 was coupled by an amine substituted linker to a solid phase agarose bead to retain proteins that could be classified as candidate binding partners (i.e., HE3286 target proteins). Most proteins non-specific to HE3286 were washed off the beads and candidate binding partners that remained adsorbed to the solid phase were subsequently released and captured for identification. This resulting mixture will contain direct target proteins to HE3286 or alternatively secondary accessory binders (due to protein-protein interactions) such as those involved in scaffold assembly of protein complexes. Control beads were used in each experiment to identify non-specific absorption by the agarose bead, and these non-specific absorbed proteins were removed from the candidate list by subtracting the two data sets. Thorough mixing of the solid phase with microliter quantities of the reagents and cell extracts was an important element to reduce the variability in each experiment. In another experiment, soluble HE3286 was used as a competitor to inhibit bead binding of candidate target proteins. Subtracting the soluble competitor data set from a buffer control produced a decreased ratio (>1.5) for HE2386-specific binding partners in comparison to the previous list of candidate binding partners and also identified non-specific binding proteins. The soluble competitor experiment is sensitive to the presence of residual competitor during workup.
Figure 3. The experimental strategy for identification of proteins that bind HE3286 in the RAW 264.7 mouse macrophage proteome.
Cells are grown in light (natural isotope) growth medium or heavy medium, containing amino acids enriched in stable isotopes (13C and 15N). Both the heavy and light lysates were incubated with small molecule loaded beads or beads without drug (control) to enrich proteins using the drug as an affinity probe. Alternatively, the addition of a soluble form of the drug is added to compete with the affinity matrix for binding to target proteins. Proteins bound to the affinity matrix are digested on bead to peptides and quantified by LC-MS/MS. Target proteins are identified by analyzing the resulting abundance ratios XIC light/XIC heavy.
In a separate approach for identifying non-specific binding proteins, the structurally similar sterol 17 α -ethynyl -5α-androstane-3α, 17β-diol (HE3235), which was inactive in our in vitro biochemical assay for NF-κB inhibition, was used as a non-specific soluble competitor. This eliminated proteins that bound to the solid phase with low affinity, which are presumed to be non-specific in nature and unrelated to the observed biological activity for HE3286. Importantly, HE3235 in the soluble competitor experiment may compete with specific HE3286-bound target proteins if binding sites for their respective binding partners are structurally similar. This complication may eliminate a participating target protein and produce a false negative result. Variability inherent in the SILAC method is resolved through experimental repetition.
Supporting Information
Brief description of the 26 gene products bound by HE3286.
(XLS)
Interactions from the BioGRID protein-protein interaction database (v. 3.1.74) suggest 1,569 possible proteins under the influence of HE3286. Using this expanded set of proteins, multiple nodes within pathways found in the KEGG database suggest that HE3286 may be involved in various pathways linked to inflammation. In the included three example pathways, Adipocytokine Signaling, Insulin Signaling, and Type II Diabetes Mellitus, nodes are highlighted in green for proteins found in this SILAC study and nodes are highlighted in red if the targets are involved in a protein-protein interaction. The two column tables below each pathway list the HE3286 bound proteins in the first column (black) while the partnered protein (listed in BioGRID) is shown in the second column, highlighted in the same color as in the pathway above.
(PDF)
Acknowledgments
The experimental procedures and mass spectrometry were conducted in the laboratories of Professor Ben Cravatt in The Scripps Research Institute with the technical guidance of Dr. Sara Tulley.
Footnotes
Competing Interests: The authors have read the journal's policy and have the following conflicts: All authors declare that they are employees of Harbor Biosciences who funded this study. All authors own stock in Harbor Biosciences. Harbor Biosciences owns US patent No. 7,776,845, which is related to HE3286. In addition, Dr. Frincke sits on the Board of Harbor Biosciences. HE3286 is in clinical development, but there is no marketed product to declare. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials, as detailed online in the guide for authors.
Funding: No current external funding was received for this study.
References
- 1.Celec P, Starka L. Dehydroepiandrosterone - is the fountain of youth drying out? Physiol Res. 2003;52:397–407. [PubMed] [Google Scholar]
- 2.Auci DL, Reading CL, Frincke JM. 7-Hydroxy androstene steroids and a novel synthetic analogue with reduced side effects as a potential agent to treat autoimmune diseases. Autoimmun Rev. 2009;8:369–372. doi: 10.1016/j.autrev.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Offner H, Zamora A, Drought H, Matejuk A, Auci D, et al. A synthetic androstene derivative and a natural androstene metabolite inhibit relapsing-remitting EAE. J Neuroimmunol. 2002;130:128. doi: 10.1016/s0165-5728(02)00214-x. [DOI] [PubMed] [Google Scholar]
- 4.Ahlem C, Auci D, Mangano K, Reading C, Frincke J, et al. HE3286: a novel synthetic steroid as an oral treatment for autoimmune disease. Ann N Y Acad Sci. 2009;1173:781–790. doi: 10.1111/j.1749-6632.2009.04798.x. [DOI] [PubMed] [Google Scholar]
- 5.Loria RM. Antiglucocorticoid function of androstenetriol. Psychoneuroendocrinology. 1997;22:S103–108. doi: 10.1016/s0306-4530(97)00005-x. [DOI] [PubMed] [Google Scholar]
- 6.Malik A, Khaldoyanidi S, Auci D, Miller S, Ahlem C, et al. 5-Androstene-3β,7β,17 β-triol (β-AET) slows thermal-injury-induced osteopenia in mice: relation to aging and osteoporosis. PLOS1. 2010;5:e13566. doi: 10.1371/journal.pone.0013566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller C, Pompon D, Urban P, Morfin R. Inter-conversion of 7alpha- and 7beta-hydroxy-dehydroepiandrosterone by the human 11beta-hydroxysteroid dehydrogenase type 1. J Steroid Biochem Mol Biol. 2006;99:215–222. doi: 10.1016/j.jsbmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Auci DL, Ahlem CN, Kennedy MR, Page TM, Reading CL, et al. A Potential Role for 5-Androstene-3beta,7beta,17beta-triol in Obesity and Metabolic Syndrome. Obesity (Silver Spring) 2010 doi: 10.1038/oby.2010.204. doi: 10.1038/oby.2010.1204. [DOI] [PubMed] [Google Scholar]
- 9.Ahlem CN, White SK, Page TM, Frincke JM. Differential metabolism of androst-5-ene-3beta,17beta-diol between rats, canines, monkeys and humans. Steroids. 76:669–674. doi: 10.1016/j.steroids.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Villegas S, Huang Y, White SK, Ahlem C, et al. Amelioration of glucose intolerance by the synthetic androstene HE3286: link to inflammatory pathways. J Pharmacol Exp Ther. 2010;333:70–80. doi: 10.1124/jpet.109.161182. [DOI] [PubMed] [Google Scholar]
- 11.Kosiewicz M, Auci D, Fagone P, Mangano K, Caponnetto S, et al. HE3286, an orally bioavailable synthetic analogue of an active DHEA metabolite suppresses spontaneous autoimmune diabetes in the nod-obese diabetic (NOD) mouse. Eur J Pharmacol. 2011 doi: 10.1016/j.ejphar.2011.02.016. in press. [DOI] [PubMed] [Google Scholar]
- 12.Conrad D, Wang A, Pieters R, Nicoletti F, Mangano K, et al. HE3286, an oral synthetic steroid, treats lung inflammation in mice without immune suppression. J Inflamm (Lond) 2010;7:52. doi: 10.1186/1476-9255-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auci D, Kaler L, Subramanian S, Huang Y, Frincke J, et al. A new orally bioavailable synthetic androstene inhibits collagen-induced arthritis in the mouse. Ann N Y Acad Sci. 2007;1110:630–640. doi: 10.1196/annals.1423.066. [DOI] [PubMed] [Google Scholar]
- 14.Auci D, Mangano K, Destiche D, White SK, Haung Y, et al. Oral treatment with HE3286 ameliorates disease in rodent models of rheumatoid arthritis. Int J Mol Med. 2010;25:625–633. doi: 10.3892/ijmm_00000385. [DOI] [PubMed] [Google Scholar]
- 15.Offner H, Firestein GS, Boyle DL, Pieters R, Frincke JM, et al. An orally bioavailable synthetic analog of an active dehydroepiandrosterone metabolite reduces established disease in rodent models of rheumatoid arthritis. J Pharmacol Exp Ther. 2009;329:1100–1109. doi: 10.1124/jpet.108.145086. [DOI] [PubMed] [Google Scholar]
- 16.Lu M, Patsouris D, Li P, Flores-Riveros J, Frincke JM, et al. A new antidiabetic compound attenuates inflammation and insulin resistance in Zucker diabetic fatty rats. Am J Physiol Endocrinol Metab. 2010;298:E1036–1048. doi: 10.1152/ajpendo.00668.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reading C, Stickney D, Flores-Riveros J, Destiche D, Ahlem C, et al. Improved insulin sensitivity by HE3286 depends on insulin resistance, inflammation and body mass index. Submitted for publication to Obesity 2011 [Google Scholar]
- 18.Ahlem CN, Kennedy MR, Page TM, Reading CL, White SK, et al. Studies of the pharmacology of 17alpha-ethynyl-androst-5-ene-3beta,7beta,17beta-triol, a synthetic anti-inflammatory androstene. Int J Clin Exp Med. 4:119–135. [PMC free article] [PubMed] [Google Scholar]
- 19.Mann M. Comparative analysis to guide quality improvements in proteomics. Nat Methods. 2009;6:717–719. doi: 10.1038/nmeth1009-717. [DOI] [PubMed] [Google Scholar]
- 20.Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleuren WW, Verhoeven S, Frijters R, Heupers B, Polman J, et al. CoPub update: CoPub 5.0 a text mining system to answer biological questions. Nucleic Acids Res. 39:W450–454. doi: 10.1093/nar/gkr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. BioGrid_Database v (3.1.74) Available: http://thebiogrid.org/. Accessed 2011 Nov 30.
- 23. KEGG Pathway Available: http://www.genome.jp/kegg/pathway.html Accessed 2011 Jun 14.
- 24.Ceschin DG, Sanchez MC, Chiabrando GA. Insulin induces the low density lipoprotein receptor-related protein 1 (LRP1) degradation by the proteasomal system in J774 macrophage-derived cells. J Cell Biochem. 2009;106:372–380. doi: 10.1002/jcb.22014. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Links PH, Ngsee JK, Tran K, Cui Z, et al. Localization of low density lipoprotein receptor-related protein 1 to caveolae in 3T3-L1 adipocytes in response to insulin treatment. J Biol Chem. 2004;279:2221–2230. doi: 10.1074/jbc.M310679200. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann SM, Zhou L, Perez-Tilve D, Greer T, Grant E, et al. Adipocyte LDL receptor-related protein-1 expression modulates postprandial lipid transport and glucose homeostasis in mice. J Clin Invest. 2007;117:3271–3282. doi: 10.1172/JCI31929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordts PL, Reekmans S, Lauwers A, Van Dongen A, Verbeek L, et al. Inactivation of the LRP1 intracellular NPxYxxL motif in LDLR-deficient mice enhances postprandial dyslipidemia and atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:1258–1264. doi: 10.1161/ATVBAHA.109.192211. [DOI] [PubMed] [Google Scholar]
- 28.Gaultier A, Arandjelovic S, Niessen S, Overton CD, Linton MF, et al. Regulation of tumor necrosis factor receptor-1 and the IKK-NF-kappaB pathway by LDL receptor-related protein explains the antiinflammatory activity of this receptor. Blood. 2008;111:5316–5325. doi: 10.1182/blood-2007-12-127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croy JE, Brandon T, Komives EA. Two apolipoprotein E mimetic peptides, ApoE(130–149) and ApoE(141–155)2, bind to LRP1. Biochemistry. 2004;43:7328–7335. doi: 10.1021/bi036208p. [DOI] [PubMed] [Google Scholar]
- 30.Baitsch D, Bock HH, Engel T, Telgmann R, Muller-Tidow C, et al. Apolipoprotein E induces antiinflammatory phenotype in macrophages. Arterioscler Thromb Vasc Biol. 31:1160–1168. doi: 10.1161/ATVBAHA.111.222745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boucher P, Herz J. Signaling through LRP1: Protection from atherosclerosis and beyond. Biochem Pharmacol. 81:1–5. doi: 10.1016/j.bcp.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, et al. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Pfluger PT, Kim JY, Nogueiras R, Duran A, et al. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010;11:226–232. doi: 10.1038/embor.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung S, Brown JM, Provo JN, Hopkins R, McIntosh MK. Conjugated linoleic acid promotes human adipocyte insulin resistance through NFkappaB-dependent cytokine production. J Biol Chem. 2005;280:38445–38456. doi: 10.1074/jbc.M508159200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo LH, Tsai PJ, Jiang MJ, Chuang YL, Yu L, et al. Toll-like receptor 2 deficiency improves insulin sensitivity and hepatic insulin signalling in the mouse. Diabetologia. 2011;54:168–179. doi: 10.1007/s00125-010-1931-5. [DOI] [PubMed] [Google Scholar]
- 36.Jager J, Gremeaux T, Gonzalez T, Bonnafous S, Debard C, et al. Tpl2 kinase is upregulated in adipose tissue in obesity and may mediate interleukin-1beta and tumor necrosis factor-{alpha} effects on extracellular signal-regulated kinase activation and lipolysis. Diabetes. 2010;59:61–70. doi: 10.2337/db09-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li JH, Liang Y, Xie YX. Efficient palladium-catalyzed homocoupling reaction and Sonogashira cross-coupling reaction of terminal alkynes under aerobic conditions. J Org Chem. 2005;70:4393–4396. doi: 10.1021/jo0503310. [DOI] [PubMed] [Google Scholar]
- 38. Pierce Scientific http://www.piercenet.com/pisearch/?prodnum=26200 Accessed 2011 Jun 22.
- 39.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 40.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR., 3rd Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 41.Ong SE, Schenone M, Margolin AA, Li X, Do K, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Brief description of the 26 gene products bound by HE3286.
(XLS)
Interactions from the BioGRID protein-protein interaction database (v. 3.1.74) suggest 1,569 possible proteins under the influence of HE3286. Using this expanded set of proteins, multiple nodes within pathways found in the KEGG database suggest that HE3286 may be involved in various pathways linked to inflammation. In the included three example pathways, Adipocytokine Signaling, Insulin Signaling, and Type II Diabetes Mellitus, nodes are highlighted in green for proteins found in this SILAC study and nodes are highlighted in red if the targets are involved in a protein-protein interaction. The two column tables below each pathway list the HE3286 bound proteins in the first column (black) while the partnered protein (listed in BioGRID) is shown in the second column, highlighted in the same color as in the pathway above.
(PDF)