Abstract
The nutritional requirements of stem cells have not been determined; in particular, the amino acid metabolism of stem cells is largely unknown. In this study, we investigated the amino acid metabolism of human mesenchymal stem cells (hMSCs), with focus on two questions: Which amino acids are consumed and/or secreted by hMSCs and at what rates? To answer these questions, hMSCs were cultured on tissue culture plastic and in a bioreactor, and their amino acid profile was analyzed. The results showed that the kinetics of hMSCs growth and amino acid metabolism were significantly higher for hMSCs in tissue culture plastic than in the bioreactor. Despite differences in culture conditions, 8 essential and 6 nonessential amino acids were consumed by hMSCs in both tissue culture plastic and bioreactor cultures. Glutamine was the most consumed amino acid with significantly higher rates than for any other amino acid. The metabolism of nonessential amino acids by hMSCs deviated significantly from that of other cell lines. The secretion of alanine, glycine, glutamate, and ornithine by hMSCs showed that there is a strong overflow metabolism that can be due to the high concentrations of amino acids provided in the medium. In addition, the data showed that there is a metabolic pattern for proliferating hMSCs, which can contribute to the design of medium without animal serum for stem cells. Further, this study shows how to implement amino acid rates and metabolic principles in three-dimensional stem cell biology.
Introduction
In this study, the amino acid metabolism of adult stem cells was explored. The study of amino acid metabolism has contributed to successfully place mammalian cell cultures at the service of biotechnology and medicine.1,2 More specific knowledge on amino acid metabolism has promoted robust cell cultures3 and design of serum-free media, which, in practice, have made possible the routine production of vaccines and antibodies.4–6 Amino acids owe their relevance and importance to the fact that they are fundamental factors in nutrition as building blocks for biomass components such as, DNA, RNA, and proteins.7
The understanding of amino acid metabolism has important implications for harnessing the therapeutic potential of stem cells. However, little is known about how stem cells use resources such as amino acids8 and other extracellular compounds.9 Stem cells from the bone marrow are used in gene, cell, and tissue therapies10–13 because of their potential to differentiate into multiple cell lineages and form bone, cartilage, muscle, fat, ligament, and tendon tissues.14–17 As essential building blocks of nature, amino acids could provide information necessary to achieve optimal (e.g., serum free) and robust (e.g., reproducible) stem cell cultures and phenotypes in three dimensions (3D).
Due to the dynamic nature of amino acid metabolism, it has been necessary to develop analytical tools, such as algorithms18 and metabolic libraries,4 to cope with the complexity. Not surprisingly, studies have indicated that when one cell type is compared with another, they differ in their amino acid metabolism.4,19 More importantly, it has become evident that amino acid metabolism reflects the internal equilibrium between using resources for energy generation and building blocks. For example, restricting the amino acid consumption leads to less overflow metabolism and higher product yields. Thus, changes in culture conditions, such as concentrations or reactor configurations, significantly upset the balance and the production or consumption of amino acids.3,5,6,19
Through amino acid metabolism, metabolic pathways are analyzed and defined by components, rates, and time constants. For stem cells, the challenges are still numerous to be able to qualitatively and quantitatively characterize all intracellular and extracellular components due to the sheer complexity of the pathways20,21 that can result in proliferation, differentiation, and 3D biology. In this study, we are making the effort to organize this complexity with the help of physical laws. First, the law of conservation of mass can be applied to determine the kinetics of amino acid transport as has been shown earlier.4,22,23 Second, the kinetics of amino acid metabolism are connected to the transport of amino acids by diffusion to determine molecular gradients in 3D. As a consequence, the benefit is that the kinetics reveal how the micro-environment of human mesenchymal stem cells (hMSCs) is shaped in time.
So far, stem cell proliferation has been the primal focus of many studies in tissue culture plastic24–26 and in bioreactors,27–30 because stem cell expansion in vitro is necessary due to their low numbers in biopsies.31 In this study, we analyzed amino acid metabolism during stem cell proliferation in tissue culture plastic and a stirred vessel bioreactor.32 In each one of those conditions, we cultured hMSCs to quantitatively and qualitatively elucidate which amino acids are consumed and/or secreted. We hypothesized that essential amino acids would be consumed by hMSCs, but nonessential amino acids could be either consumed or secreted. By determining the amino acid metabolism by hMSCs, we intend to reveal quantities necessary to support 3D stem cell biology and tissue regeneration.
Materials and Methods
Isolation and cryopreservation of hMSCs
Isolation and cryopreservation of hMSCs were performed as previously described.33 After informed consent and approval by the medical ethics committee, we obtained hMSCs from the acetabulum of five human donors who were undergoing total hip replacement surgery. Gender and donor age were as follows—Donor 1: female, 81 years. Donor 2: male, 65 years. Donor 3: female, 76 years. Donor 4: female, 68 years. Donor 5: female, 38 years. The human MSC population was isolated from the aspirates via adhesion selection. Cells were cultured up to passage 1 and cryopreserved. Experiments were performed according to protocols previously described for tissue culture plastic (Static)33 or for microcarriers in a stirred vessel bioreactor (Dynamic).30 Culture medium was prepared according to static or dynamic conditions as follows:
Static
The minimal essential medium (αMEM) proliferation medium (GIBCO) contained, 10% fetal bovine serum of a selected batch (fetal bovine serum [FBS] lot:4SB0010; Biowhittaker), 1 ng/mL basic fibroblast growth factor (bFGF, Instruchemie), penicillin G (100 Units/mL, Invitrogen), 0.2 mM l-ascorbic-acid-2-phosphate (Sigma), streptomycin (100 μg/mL, Invitrogen), and 2 mM l-glutamine (Sigma).
Dynamic
Culture medium used consisted of α-MEM (Invitrogen) supplemented with 15% FBS (FBS, Cambrex), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine (Invitrogen), 0.2 mM L-ascorbic acid-2-phosphate, 1 ng/mL bFGF (AbD Serotec), and 10 nM dexamethasone (Sigma).
Proliferation
Static
Cells from donors 1 to 3 were cultured. Proliferation of hMSCs was performed on tissue culture plastic (T-Flasks) in a standard incubator with controlled temperature (37°C) and humidified atmosphere with 5% CO2. Cryopreserved hMSCs were thawed, counted, and plated (passage 2) at 100 cells/cm2 in αMEM proliferation media in tissue culture flasks (T-flasks). αMEM proliferation medium was not refreshed to maintain a batch culture configuration. Every day, three replicates were sacrificed to obtain cell numbers and the conditioned medium.
Dynamic
Cells from donors 4 and 5 were cultured. Proliferation of human mesenchymal stem cells (hMSCs) was performed in 1-liter round-bottomed stirred vessel bioreactors (Applikon Biotechnology BV) on Cytodex type 1 microcarriers (GE Healthcare) at a density of 20 cm2/mL (4.5 g/l).30 During cultivation, the following conditions were regulated with an ez-Control (Applikon Biotechnology):
Temperature controlled at 37°C.
Agitation speed controlled at 50 rpm during seeding and at 60–70 rpm during expansion using a marine impeller.
pH regulated at 7.3 using 0.25 M NaOH and CO2 gas.
Dissolved oxygen concentration controlled at 4% O2 saturation by adjusting the oxygen fraction (using N2 and/or air gasses) blown over the surface of the cultures.
The hMSCs were seeded on the microcarriers at 3000 cells/cm2 in 400 mL proliferation medium without FBS.30 The suspension was stirred at 50 rpm for 4 h. After the seeding period, the medium was refreshed for 50%, and FBS was added to obtain a final concentration of 15% FBS. During 7 days of cell expansion, the culture was stirred at 70 rpm. Daily samples were taken for cell analysis and medium analysis. Since the medium level decreased over time due to sampling, the stirring rate was decreased stepwise to 60 rpm to maintain a homogeneous microcarrier suspension with comparable shear forces. Two separate experiments were performed (n=2) for each of the donors cultured in the bioreactor system.
Cell numbers
Static
To harvest cells, the T-flasks were washed with PBS (Sigma), and hMSCs were enzymatically detached from T-flasks with 0.25% trypsin/EDTA. From the cell suspension, 200 μl was diluted in 10 mL of Isoton II diluent (Beckman Coulter), and three drops of Zap-OGlobin II lytic reagent (Beckman Coulter) were added. The solutions were incubated for 30 min to maximize the effect of the lytic reagent; subsequently, the nuclei of cells were counted in a particle count and size analyzer (Z2, Beckman Coulter). The size range of counted nuclei was set to between 6 and 10.5 μm according to the 95% confidence interval of hMSCs nuclei size.
Dynamic
Sampling of microcarriers was performed during the bioreactor runs. From these samples, viable cell numbers were measured using the CellTiter-Glo assay (Promega), which is based on metabolic activity of the cell. To visualize the cell load and cell distribution on the microcarriers, cells were stained with 1% 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (Merck).
Metabolites and volume
Glucose and lactate were measured in a Vitros DT60 II chemistry system (Ortho-Clinical Diagnostics). Daily measurements were performed for every donor from independent medium samples in both static and dynamic cultures.
Static
The medium evaporated at a rate of 2.1×10−6 l/h (±1.9×10−7, n=3) in incubators at 37°C with 60% relative humidity.
Dynamic
Daily, 10–20 mL microcarrier-cell and medium samples were aseptically taken from the cell cultures in the stirred vessels. This resulted in the working volume changing from 400 to 300 mL during culture.
Amino acid analysis
Free amino acid concentrations in the medium samples were determined by high performance liquid chromatography and performed by Ansynth Service B.V. (Roosendaal).
Static
From each donor, three T-flasks were sacrificed to obtain medium samples every day. Medium samples were filtered (0.2 μm pore size, Millipore, The Netherlands). Each amino acid measurement is representative of mixing medium samples from three T-flasks. Immediately afterward, vials were stored at −80°C.
Dynamic
Medium samples from each bioreactor were filtered and stored at −80°C.
Estimation of degradation kinetics
Degradation experiments included medium incubated without cells in static and dynamic culture. Metabolites and amino acids were measured in the medium samples with their respective methods. First-order degradation kinetics were assumed during analysis. Amino acids with significant degradation are shown as Supplementary Table S2; Supplementary data available online at www.liebertonline.com/tea.
Statistical analysis
Each data point of cell numbers, glucose, and lactate concentrations was obtained from three replicates (three T-Flasks/Donor for three donors) or two parallel runs (two stirring vessels for two donors). Error bars in graphs with experimental data represent the standard deviation of measurements.
Analysis of growth and metabolism kinetics was performed and evaluated on the basis of 95% confidence intervals with the growth and kinetics model found next. Standard error of estimates (SEEs) was calculated to determine the quality of the kinetic model fit to experimental measurements.
Growth and metabolism kinetics model description
To determine amino acid metabolism by hMSCs in static and dynamic culture, the mass balances of the variables measured were defined and analyzed through the differential equations described next:
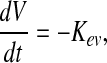 |
(1) |
where the differential equation reflects the change of medium volume (V) in time during batch culture. For static experiments, the volume is changing in time due to water evaporation; thus, Kev [l/h] is the evaporation rate constant of medium, which for the conditions tested was equal to 2.1×10−6 [l/h]. For dynamic experiments, the vessel is a closed system, where the volume is maintained constant and does not change in time.
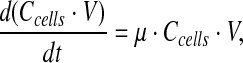 |
(2) |
where
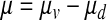 |
Equation 2 describes the change of viable cells in time, where Ccells [cells/l] is the viable cell number, μ [h−1] is the apparent specific growth rate, μv [h−1] is the specific growth rate of viable cells, and μd [h−1] is the death rate of viable cells. μv and μd cannot be estimated as independent variables, and, therefore, μ was estimated and used to predict viable cell numbers.
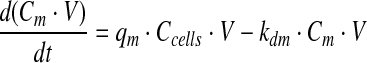 |
(3) |
Equation 3 presents the change of amino acids, glucose, and lactate in time, where Cm [mM] represents the concentration of m, the subscript m refers to metabolites or amino acids, and qm [mMoles/cell/h] refers to the specific consumption/production rate. qm is constant and is negative for consumed molecules and positive for produced molecules, and kdm [h−1] represents the first-order spontaneous degradation constant.
Parameter estimation
The metabolite concentrations depend on both production/consumption and degradation. By using the kinetics model equations 1 through 3, the contributions of consumption/production and degradation to the metabolite concentrations can be separated.
The ordinary differential equations 1 through 3 were solved with a standard differential equation solver in Matlab (ode45 in version 7.0.4 release 2007a; Mathworks) on a windows-based system. The initial values for solving the differential equations were set to the experimentally determined average seeding densities and amounts of molecules in fresh αMEM proliferation medium from five samples.
Growth and metabolism kinetics parameters were obtained by nonlinear least squares regression that minimizes the sum of the quadratic error over an experiment (Matlab function nlinfit). The 95% confidence intervals for the parameters were calculated (Matlab function nlparci).
Computational fluid dynamics
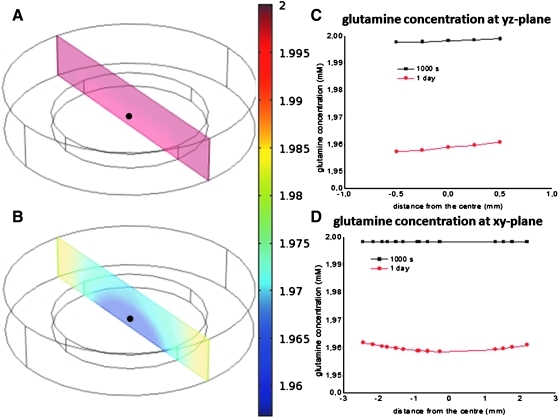
Two cylinders were representative of the 3D environment. The outer cylinder represented the medium volume, whereas the inner cylinder represented the volume, where hMSCs proliferate (Fig. 7). For the simulations, the parameters calculated from the bioreactor cultures, such as glutamine consumption rates and initial concentrations, were assumed, because bioreactor cultures are considered 3D cell culture platforms and are more representative of other 3D environments. Further, consumption rates of metabolites may be variable, instead of constant, due to environmental parameters, such as oxygen availability. However, a constant consumption rate for glutamine was a reasonable assumption, as changes in oxygen availability have not been reported to significantly change metabolite consumption rates.22 To obtain the glutamine gradients for the hMSCs volume, equations 4 and 5 were solved:
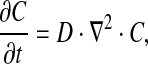 |
(4) |
FIG. 7.
Glutamine concentration changes in time and space, with gradients in 3D structures. Inner cylinder (1 mm height×5 mm diameter) is representative of hMSCs in a tissue-size volume, whereas outer cylinder represents the surrounding medium. The calculated glutamine consumption rate was used to simulate the highly consumed glutamine in time and space. (A) Cross section of a Comsol model of the glutamine gradients after 1000s. (B) Cross section of a Comsol model of the glutamine gradients after 1 day. (C) Concentration gradients across the height of the hMSCs cylinder. (D) Concentration gradients across the length of the hMSCs cylinder. Black dot indicates the center of the cylinder.
where C is the concentration of glutamine (mol/m3), t is the time (s), D is the diffusion constant of glutamine (m2/s), and ∇ is the del operator.
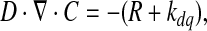 |
(5) |
where R is the glutamine consumed by the total hMSCs population in the hMSCs volume (5 mm diameter×1 mm height), and kdq is the spontaneous degradation of glutamine.
An initial concentration of glutamine (C0=2 mmol/l) was obtained from the average bulk concentration of glutamine at the beginning of culture. Glutamine diffusion constant (D) was 2.2×10−6 cm2/s.34 Glutamine spontaneous degradation (kdq) was estimated to be 3×10−3 [h−1] (Table S2).
To obtain R, glutamine consumption (qq) [pmoles/cell/h] was multiplied by 2.5×107 cells/L, the cell concentration on day 0 in dynamic culture. It was assumed that hMSCs were homogeneously organized in the 3D space.
Walls in different conditions were considered rigid and impermeable. No-slip boundary conditions were applied to surfaces.
Computational fluid dynamics models of the fluid flow in culture conditions were set up and solved in the MEMS module (microfluidics–flow with species transport–Incrompressible-Navier Stokes) in Comsol Multiphysics version 4 software (Comsol).
Results
To determine the amino acid metabolism of hMSCs without biasing the results for one culture system, hMSCs were cultured in static and dynamic platforms. All data are presented as a comparison between static and dynamic culture, with the purpose of ultimately determining transport rates across the stem cell membrane. Further, all data was collected and analyzed for the lag and logarithmic phases of hMSCs culture as defined by the viable cell numbers.
Growth and kinetics
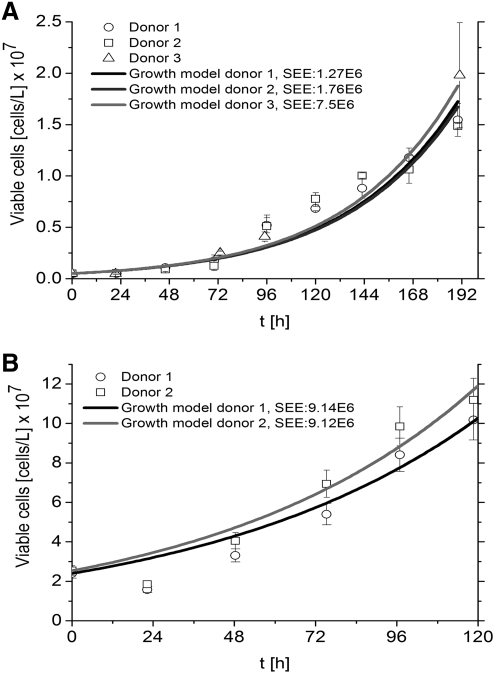
To be able to unfold consumption and production rates of amino acids for a cell, viable cell numbers were obtained for each condition (Fig. 1) and analyzed with growth model equations 1 and 2. Viable cell numbers in dynamic cultures (Fig. 1B) were significantly higher than in static cultures (Fig. 1A), which is mainly due to the higher starting cell concentration in the bioreactors. The duration of the logarithmic phase in hMSCs cultures were described as the time needed to reach the highest number of viable cells that was measured. These were determined to be 192 h for static and 120 h for dynamic cultures. According to the SEE, the estimation of viable cell numbers was more accurate in static (Fig. 1A) than in dynamic cultures (Fig. 1B).
FIG. 1.
Viable cell numbers and growth model fit for the exponential phase of hMSCs in two culture systems. (A) Static culture: Tissue culture plastic (B) Dynamic: Stirred vessel bioreactor. hMSCs have a longer exponential phase in static than in dynamic culture. Data are represented as means of three measurements±standard deviations. For the growth models, the SEE was calculated. hMSCs, human mesenchymal stem cells; SEEs, standard error of estimates.
From the growth model equations 1 and 2, the specific growth rate was estimated for every donor under every condition (Supplementary Table S1). Specific growth rates were significantly higher for hMSCs in static cultures (1.86±0.01×10−2 [h−1], mean value of n=3 donors) than in dynamic cultures (1.25±0.06×10−2 [h−1], mean value of n=2 donors).
Glucose and lactate
Glucose and lactate concentrations were obtained for every condition to assess the hMSCs. Glucose was consumed, and lactate was produced by hMSCs as reflected by the qgluc and qlac with negative and positive signs, respectively (Supplementary Table S1). Notably, the glucose and lactate consumption and production rates, respectively, were comparable in static and dynamic cultures. Further, the Ylac/gluc, which expresses the level of glycolysis, showed higher variability in static cultures (1.2–3), where the measurement accuracy is lower due to the larger differences in concentration at the end and the start of the experiment. On the other hand, hMSCs donors in dynamic culture showed smaller variability in the glycolysis level (1.9–2.2) (Supplementary information, Table S1). Lastly, the initial concentration of glucose was 7.5 mM in dynamic cultures and 4.5 mM in static cultures. The initial concentration of lactate was 1.7 mM in both static and dynamic cultures.
Amino acid metabolism
Essential amino acids for vertebrates are defined as those that cannot be synthesized from any other ingredients of the diet7: Histidine, isoleucine, leucine, lysine, methionine, phenylalanine, tryptophan, threonine, and valine. Nonessential amino acids are as defined by Alberts (2008)7: Alanine, arginine, asparagine, Aspartate, cysteine, glutamate, glutamine, glycine, ornithine, proline, serine, and tyrosine. Amino acid data were obtained from all donors and analyzed through equations 1 to 3.
Are essential amino acids necessary to human MSCs?
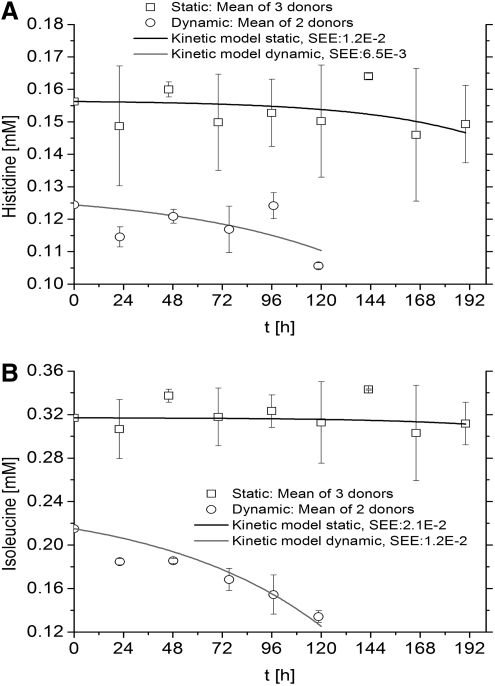
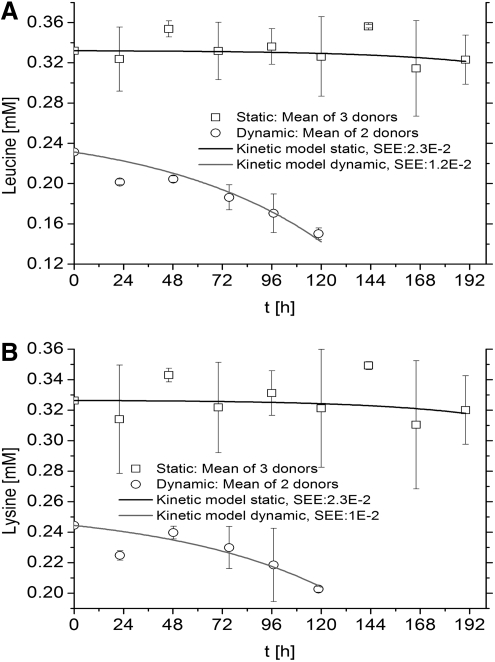
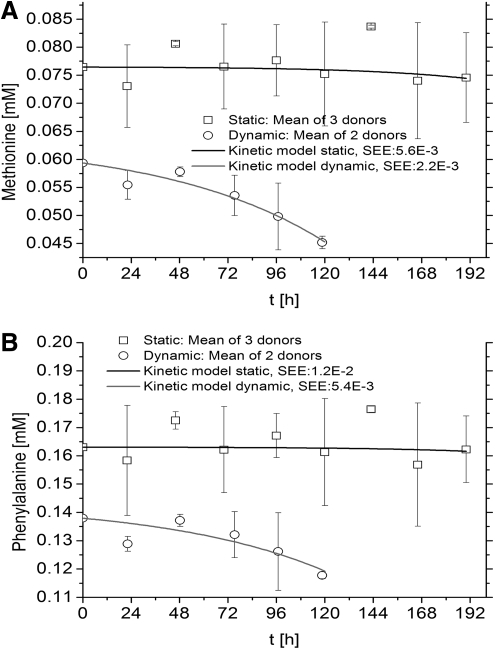
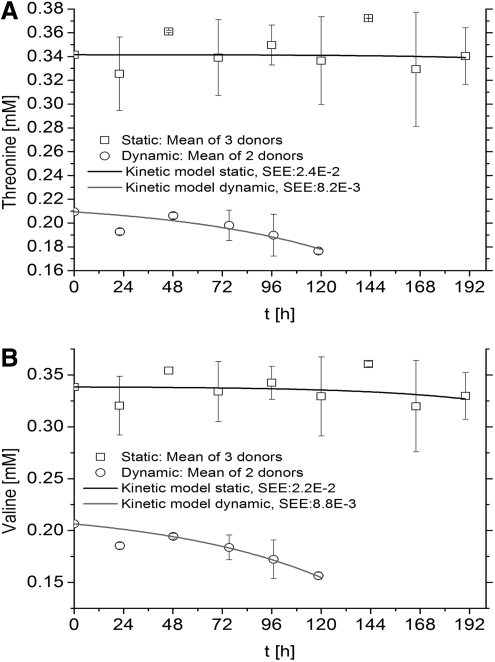
Essential amino acids were expected to be consumed by hMSCs in both static and dynamic cultures and are presented as follows; Histidine (Fig. 2A), isoleucine (Fig. 2B), leucine (Fig. 3A), lysine (Fig. 3B), methionine (Fig. 4A), phenylalanine (Fig. 4B), threonine (Fig. 5A), and valine (Fig. 5B) (Tryptophan—not measured). As measured and estimated in these figures, essential amino acids were consumed based on the model as expected beforehand. Further, the dynamic cultures were fitted more accurately as judged from the lower SEE values.
FIG. 2.
Essential amino acids are consumed by hMSCs in both culture systems. (A) Histidine. (B) Isoleucine. Experimental concentrations, kinetics model, and SEE are shown.
FIG. 3.
Essential amino acids are consumed by hMSCs in both culture systems. (A) Leucine. (B) Lysine. Experimental concentrations, kinetics model, and SEE are shown.
FIG. 4.
Essential amino acids are consumed by hMSCs in both culture systems. (A) Methionine. (B) Phenylalanine. Experimental concentrations, kinetics model, and SEE are shown.
FIG. 5.
Essential amino acids are consumed by hMSCs in both culture systems. (A) Threonine. (B) Valine. Experimental concentrations, kinetics model, and SEE are shown.
Are nonessential amino acids unnecessary to human MSCs?
Nonessential amino acids can be produced and/or consumed. The metabolism of these amino acids by hMSCs revealed that: (1) Alanine (Supplementary Fig. S1A), glutamate (Supplementary Fig. S3B), glycine (Supplementary Fig. S4B), and ornithine (Supplementary Fig. S5A) were produced by hMSCs in both static and dynamic hMSCs cultures. (2) hMSCs consumed arginine (Supplementary Fig. S1B), asparagine (Figure S2A), aspartate (Supplementary Fig. S2B), glutamine (Supplementary Fig. S4A), serine (Supplementary Fig. S6A), and tyrosine (Supplementary Fig. S6B) in both static and dynamic cultures. (3) Cysteine (Supplementary Fig. S3A) and proline (Supplementary Fig. S5B) were produced by hMSCs in static culture and consumed by hMSCs in dynamic culture. According to SEE values, the kinetic model fits were more accurate for data in dynamic than static cultures, except for glutamate.
Arginine, asparagine, aspartate, glutamine, and ornithine showed significant spontaneous degradation in static and dynamic cultures (Supplementary Table S2). Thus, the kinetic model fit for each of these amino acids was corrected for their degradation through degradation constants.
How much transport of amino acids in a human MSC is there?
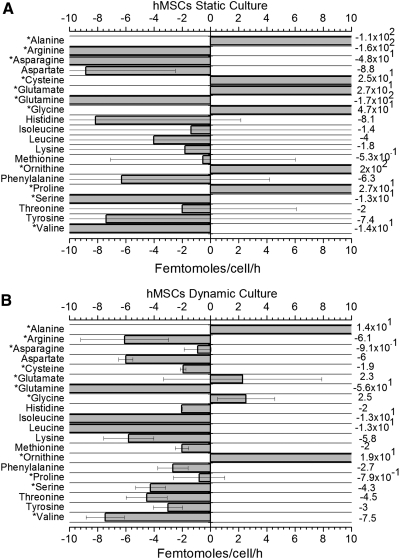
The amount of transport for each amino acid was obtained through the growth and kinetic models (Equations 1 to 3). The mean metabolic rates are presented in Figure 6 for static and dynamic cultures. Amino acids left of the zero were consumed, and right of the zero were secreted out of hMSCs, the standard deviations reflect the donor variation for hMSCs metabolism in each condition. Figure 6 displays that (1) mean metabolic rates are higher for amino acids in static than in dynamic culture. This difference was statistically significant for alanine, arginine, asparagine, cysteine, glutamate, glutamine, glycine, ornithine, proline, serine, and valine. (2) The most consumed amino acid by hMSCs was glutamine. (3) Standard deviations of metabolic rates were higher in static cultures than in dynamic cultures.
FIG. 6.
hMSCs metabolic rates for essential and nonessential amino acids. (A) Static culture. Mean of three donors±standard deviations. (B) Dynamic culture. Mean of two donors±standard deviations. Parameter estimation was used for calculation (See Materials and Methods section). All numbers are reported in femtomoles/cell/h. Consumed amino acids are negative, and produced amino acids are positive. Due to the difference in magnitude of values, some values are not graphically available. The precise metabolic rates for each amino acid are located on the right of the diagram. *Statistically significant values (p<0.05) between static and dynamic cultures.
Three-dimensional biology of stem cells: molecular gradients
The amino acid rates have important applications in the study of stem cell biology in 3D. With the specific consumption rates for stem cells, the gradients in 3D structures can be depicted. To show this, the specific glutamine consumption rate 5.6×101 [femtomol/cell/h] (Fig. 6) in dynamic culture, the most consumed amino acid, was introduced into equation 5. Figure 7 shows the gradients in space and time in a 1 mm height×5 mm diameter cylinder, which is the inner cylinder where hMSCs are homogeneously distributed. The outer cylinder represents the volume of medium. Figure 7A represents the 3D graphical distribution of glutamine gradients at t=1000s in the hMSCs and medium 3D cylinders. Figure 7B displays the graphical representation of glutamine concentration at t=1 day with glutamine being more abundant on the medium volume (outer) than in the hMSCs volume (inner). Figure 7C–D reveal the glutamine concentrations and their change in time and space across the height (Fig. 7C) and length (Fig. 7D) of the hMSCs cylinder. In a time lapse of 1 day, glutamine concentrations vary by 2% across the height and length of the hMSCs volume. The same cell number was implemented in the calculations for both time lapses, because the cell numbers would not significantly change during the first 24 h, where only glutamine consumption accounts for concentration changes; However, after 1 day, as the cell numbers exponentially increase, the glutamine concentration decreases faster.
Discussion
To unravel the amino acids required by hMSCs, the direction and amount of amino acid transport through the plasma membrane were considered. First, the numbers of viable cells were analyzed. Specific growth rates, glucose, and lactate metabolic rates in static and dynamic cultures were consistent with values found in literature,22,23,32,35 where average values are 2×10−2 h−1; 0.5 picomoles glucose/cell/h and 1 picomoles lactate/cell hour, respectively. Our results were consistent with previous reports that specific growth rates are significantly lower in dynamic than in static culture of hMSCs.32 This is possibly because shear stress36 can have a significant effect on proliferation through the formation of turbulent flow.37,38 In addition, micro-carrier and tissue culture plastic also present important surface differences39 that account for different specific growth rates between static and dynamic cultures. Further, glycolysis levels (Ylac/glac) were maintained and were comparable to previous reports of static22,23,35 and dynamic30 cultures, where glycolysis and proliferation data suggest that the Warburg effect40 is the rule rather than the exception in hMSCs metabolism.
In our study, there were significant differences between static and dynamic culture with regard to shear stress, cell-attachment material, seeding density, and donor variation. Either one of these factors could have an effect on hMSCs proliferation.23,36,39 Further, the standard deviations in the data reflect the variability associated with hMSCs in static culture that can be explained by three factors. First, lower cell numbers in static cultures induce smaller changes of concentrations than higher cell numbers in dynamic cultures. Second, the differences lie in samples that are taken from different culture flasks, whereas in dynamic cultures, a homogenous representative sample can be taken. Lastly, environmental and operational conditions are not as closely monitored and controlled as in dynamic cultures.28 In other cell types, whenever the bioreactor configuration is changed, the transport of amino acids changes direction.4,19 Despite these differences, the number of amino acids that have similar kinetics in both static and dynamic culture is remarkable. Our results indicated that hMSCs consumed arginine, asparagine, aspartate, glutamine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, serine, threonine, tyrosine, and valine in both static and dynamic cultures. This metabolic pattern is different from nonstem cells, particularly in the nonessential amino acids. For example, in cell lines such as HEK-293 and CHO (4), asparagine can be secreted and alanine, glutamate, and glycine are consumed, which is the opposite from the pattern observed here for these amino acids. In addition, under various reactor configurations and media compositions, the Vero41 and MDCK42 cell lines consume glycine, whereas hMSCs do not. These differences emphasize that metabolic principles cannot be generalized or extrapolated to hMSCs. Instead, metabolic principles, experimentally and theoretically explored in hMSCs, can shed light on processes such as the production of extracellular matrix, which is crucial to hMSCs proliferation and differentiation.
The metabolic pattern described here was obtained from the concentration data and mean metabolic rates, which are proposed to be representative of the basic nutritional requirement for hMSCs in culture. The practical implications of this statement are that the amino acids that enter the cell (Consumed) are required for hMSCs proliferation in both tissue culture plastic, stirring vessels and perhaps other culture configurations. Despite the fact that standard deviations in metabolic rates suggest that some of the amino acids on this list may be consumed and/or produced during the logarithmic phase by hMSCs, it is unlikely that the amino acids showing large standard deviations will be produced in static culture, because essential amino acids should never be produced, whereas nonessential amino acids can be produced or consumed.
With regard to amino acid metabolism in the cell, it is possible that isoleucine, leucine, serine, and valine are converted to derivates of the Krebs cycle, because these have been shown to be indirect to the generation of energy, and directly associated with protein biosynthesis.43 In addition, arginine and histidine could be converted to glutamate for energy generation. Leucine,44 methionine,45 and threonine46 have also been shown to be vital for cell and tissue development, where the amount of consumption could depend on the differentiation lineage that hMSCs follow. Consequently, consumption of these 14 amino acids by hMSCs could be essential not only for hMSCs proliferation but also in differentiation.
Amino acids transport through the plasma membrane is a balance, which is highly dependent on concentrations.47 It is important to note that no amino acid was depleted during the static or dynamic culture period, which means that amino acid concentrations are unnecessarily high in the prepared medium. This is particularly evident in the group of amino acids that were produced (secreted) by hMSCs, where kinetic rates were significantly higher in static culture for alanine, glutamate, glycine, and ornithine. In literature, alanine secretion has been known to occur when there is excess of carbon sources in culture media.48 Further, arginine, histidine, proline, and glutamine are converted into glutamate before being fed into the TCA cycle.7 In addition, glycine is a by-product of serine metabolism; thus, glycine secreted in hMSCs cultures could be the result of serine consumption.43 Lastly, ornithine is not present in the basic medium; thus, its production may be due to the fact that it is the in-between product from arginine to glutamate.49 The production of amino acids is important for hMSCs medium design, because it signals excessive concentrations of carbon and energy sources, thus leading to pathway overflow. Consequently, alanine, glutamate, and glycine addition to the proliferating medium of hMSCs may be unnecessary.
Cysteine and proline showed transport across the plasma membrane dependent on culture conditions where they were both produced by hMSCs in static culture and consumed by hMSCs in dynamic culture. Considering the attachment surface39 and shear stress38 differences in static and dynamic cultures, these amino acids could indicate the activation of mechanosensitive pathways.50
Amino acid rates are important to stem cell biology both intracellularly and extracellularly. Intracellularly, amino acid rates can yield useful understanding of mass and energy management as has been successfully shown in nonstem cells.4,41,42 Extracellularly, metabolic rates have been useful in showing the gradients across 3D cartilage constructs and experimentally exploring the effects of gradients on cartilage biology.51 As shown in this study, numbers such as the specific glutamine consumption rate can be used to identify the expected molecular gradients in time and space. The cylindrical shape depicted here is representative of an in vitro 3D environment, which shows that amino acids can quickly change concentration for hMSCs found inside a 3D structure. The size of the bone marrow52 suggests that there can be many biological effects on hMSCs biology directly caused by molecular gradients. The amino acid metabolic rates reported here can provide a better understanding on how the amino acids support the 3D environment in vitro and in vivo, where cell culture parameters such as culture volume and medium composition can be tailored according to amino acid metabolism, nutrient depletion, and other basic stem cell needs instead of being defined by standard and broader cell culture methods. Similarly, from building blocks, such as amino acids, the theoretical and experimental analysis of these and other extracellular components can increase our understanding of hMSCs 3D organization, proliferation, migration, and differentiation.
In this study, it is shown that there are amino acids which are consistently consumed by hMSCs despite differences in culture conditions. The culture conditions affect the rates themselves rather than the direction of transport, as consistently revealed by hMSCs consumption of: arginine, asparagine, aspartate, glutamine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, serine, threonine, tyrosine, and valine. Consequently, these amino acids indicate the minimal nutritional requirement by hMSCs in various culture configurations. However, the rates are not high enough to deplete amino acids and thereby limit growth. In addition the amino acids produced (alanine, glutamate, glycine, and ornithine) could be omitted, because these imply a high degree of mass overflow in amino acid metabolism, which can increase the amount of toxic species (e.g., Through oxidation) and wear of the molecular machinery of hMSCs. In the scheme of amino acid metabolism, a realistic illustration of molecular gradients in 3D can be obtained to develop, control, and optimize culture systems specific for stem cells. This offers a unique insight into the intracellular and extracellular mass and energy dynamics of human MSCs and musculoskeletal tissues.
Supplementary Material
Acknowledgment
This work was financially supported by the Dutch SenterNovem research grant 15044112.
Disclosure Statement
No competing financial interests exist.
References
- 1.Masters J.R. Stacey G.N. Changing medium and passaging cell lines. Nature Protocols. 2007;2:2276. doi: 10.1038/nprot.2007.319. [DOI] [PubMed] [Google Scholar]
- 2.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122:501. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 3.Simpson N.H. Singh R.P. Perani A. Goldenzon C. Al-Rubeai M. In hybridoma cultures, deprivation of any single amino acid leads to apoptotic death, which is suppressed by the expression of the bcl-2 gene. Biotechnol Bioeng. 1998;59:90. doi: 10.1002/(sici)1097-0290(19980705)59:1<90::aid-bit12>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 4.Kontoravdi C. Wong D. Lam C. Lee Y.Y. Yap M.G.S. Pistikopoulos E.N., et al. Modeling amino acid metabolism in mammalian cells toward the development of a model library. Biotechnol Prog. 2007;23:1261. doi: 10.1021/bp070106z. [DOI] [PubMed] [Google Scholar]
- 5.Duval D. Demangel C. Munierjolain K. Miossec S. Geahel I. Factors controlling cell-proliferation and antibody-production in mouse hybridoma cells .1. Influence of the amino-acid supply. Biotechnol Bioeng. 1991;38:561. doi: 10.1002/bit.260380602. [DOI] [PubMed] [Google Scholar]
- 6.Sanfeliu A. Cairo J.J. Cases C. Sola C. Godia F. Analysis of nutritional factors and physical conditions affecting growth and monoclonal antibody production of the hybridoma KB-26.5 cell line. Biotechnol Prog. 1996;12:209. doi: 10.1021/bp950078x. [DOI] [PubMed] [Google Scholar]
- 7.Alberts B. Johnson A. Lewis J. Raff M. Roberts K. Walter P. Molecular Biology of the Cell. 5th. New York: Garland Science; 2008. [Google Scholar]
- 8.Choi K.M. Yoon H.H. Seo Y.K. Song K.Y. Kwon S.Y. Lee H.S., et al. Effect of essential and nonessential amino acid compositions on the in vitro behavior of human mesenchymal stem cells. Korean J Chem Eng. 2007;24:1058. [Google Scholar]
- 9.Caplan A. Why are MSCs therapeutic? New data: new insight. J Pathol. 2009;217:318. doi: 10.1002/path.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prockop D.J. Marrow stromal cells for nonhematopoietic tissues. Science. 1997;276:71. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 11.Owen M. Friedenstein A.J. Stromal stem-cells—marrow-derived osteogenic precursors. Ciba Foundation Symp. 1988;136:42. doi: 10.1002/9780470513637.ch4. [DOI] [PubMed] [Google Scholar]
- 12.Caplan A.I. Mesenchymal stem cells and gene therapy. Clin Orthop Relat Res. 2000;379:S67. doi: 10.1097/00003086-200010001-00010. [DOI] [PubMed] [Google Scholar]
- 13.Piersma A.H. Brockbank K.G.M. Ploemacher R.E. Vanvliet E. Brakelvanpeer K.M.J. Visser P.J. Characterization of fibroblastic stromal cells from murine bone-marrow. Exp Hematol. 1985;13:237. [PubMed] [Google Scholar]
- 14.Jaiswal N. Haynesworth S.E. Caplan A.I. Bruder S.P. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem. 1997;64:295. [PubMed] [Google Scholar]
- 15.Johnstone B. Hering T.M. Caplan A.I. Goldberg V.M. Yoo J.U. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 16.Wakitani S. Saito T. Caplan A.I. Myogenic cells derived from rat bone-marrow mesenchymal stem-cells exposed to 5-Azacytidine. Muscle Nerve. 1995;18:1417. doi: 10.1002/mus.880181212. [DOI] [PubMed] [Google Scholar]
- 17.Pittenger M.F. Mackay A.M. Beck S.C. Jaiswal R.K. Douglas R. Mosca J.D., et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 18.Neeleman R. Boxtel A. Estimation of specific growth rate from cell density measurements. Bioprocess Biosystems Eng. 2001;24:179. [Google Scholar]
- 19.Doverskog M. Ljunggren J. Ohman L. Haggstrom L. Physiology of cultured animal cells. J Biotechnol. 1997;59:103. doi: 10.1016/s0168-1656(97)00172-7. [DOI] [PubMed] [Google Scholar]
- 20.Larson B.L. Ylostalo J. Prockop D.J. Human multipotent stromal cells undergo sharp transition from division to development in culture. Stem Cells. 2008;26:193. doi: 10.1634/stemcells.2007-0524. [DOI] [PubMed] [Google Scholar]
- 21.Gregory C.A. Gunn W.G. Reyes E. Smolarz A.J. Munoz J. Spees J.L., et al. How wnt signaling affects bone repair by mesenchymal stem cells from the bone marrow. Stem Cell Biol Dev Plast. 2005;1049:97. doi: 10.1196/annals.1334.010. [DOI] [PubMed] [Google Scholar]
- 22.Santos F.D. Andrade P.Z. Boura J.S. Abecasis M.M. Silva C.L.D. Cabral J.M.S. Ex Vivo expansion of human mesenchymal stem cells: a more effective cell proliferation kinetics and metabolism under hypoxia. J Cell Physiol. 2010;223:27. doi: 10.1002/jcp.21987. [DOI] [PubMed] [Google Scholar]
- 23.Higuera G. Schop D. Janssen F. van Dijkhuizen-Radersma R. van Boxtel T. van Blitterswijk C.A. Quantifying in vitro growth and metabolism kinetics of human mesenchymal stem cells using a mathematical model. Tissue Eng Part A. 2009;15:2653. doi: 10.1089/ten.TEA.2008.0328. [DOI] [PubMed] [Google Scholar]
- 24.Sekiya I. Larson B.L. Smith J.R. Pochampally R. Cui J.-G. Prockop D.J. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 25.Colter D.C. Class R. DiGirolamo C.M. Prockop D.J. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci U S A. 2000;97:3113. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colter D.C. Sekiya I. Prockop D.J. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci U S A. 2001;98:7841. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frauenschuh S. Reichmann E. Ibold Y. Goetz P.M. Sittinger M. Ringe J. A microcarrier-based cultivation system for expansion of primary mesenchymal stem cells. Biotechnol Prog. 2007;23:187. doi: 10.1021/bp060155w. [DOI] [PubMed] [Google Scholar]
- 28.Martin I. Wendt D. Heberer M. The role of bioreactors in tissue engineering. Trends Biotechnol. 2004;22:80. doi: 10.1016/j.tibtech.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y. Rossi F.M.V. Putnins E.E. Ex vivo expansion of rat bone marrow mesenchymal stromal cells on microcarrier beads in spin culture. Biomaterials. 2007;28:3110. doi: 10.1016/j.biomaterials.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Schop D. Janssen F.W. Borgart E. de Bruijn J.D. van Dijkhuizen-Radersma R. Expansion of mesenchymal stem cells using a microcarrier based cultivation system: growth and metabolism. J Tissue Eng Regen Med. 2008;2:126. doi: 10.1002/term.73. [DOI] [PubMed] [Google Scholar]
- 31.Aubin J.E. Bone stem cells. J Cell Biochem. 1998;30:73. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<73::AID-JCB11>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Schop D. van Dijkhuizen-Radersma R. Borgart E. Janssen F.W. Rozemuller H. Prins H.J., et al. Expansion of human mesenchymal stromal cells on microcarriers: growth and metabolism. J Tissue Eng Regen Med. 2010;4:131. doi: 10.1002/term.224. [DOI] [PubMed] [Google Scholar]
- 33.Both S.K. van der Muijsenberg A.J. van Blitterswijk C.A. de Boer J. de Bruijn J.D. A rapid and efficient method for expansion of human mesenchymal stem cells. Tissue Eng. 2007;13:3. doi: 10.1089/ten.2005.0513. [DOI] [PubMed] [Google Scholar]
- 34.Kallinowski F. Runkel S. Fortmeyer H.P. Forster H. Vaupel P. L-Glutamine: a major substrate for tumor cells in vivo? J Cancer Res Clin Oncol. 1987;113:209. doi: 10.1007/BF00396375. [DOI] [PubMed] [Google Scholar]
- 35.Schop D. Janssen F.W. van Rijn L.D. Fernandes H. dev Bruijn J.D. van Dijkhuizen-Radersma R. Growth, metabolism and growth inhibitors of mesenchymal stem cells. Tissue Eng Part A. 2009;15:1877. doi: 10.1089/ten.tea.2008.0345. [DOI] [PubMed] [Google Scholar]
- 36.Zhao F. Chella R. Ma T. Effects of shear stress on 3-D human mesenchymal stem cell construct development in a perfusion bioreactor system: experiments and hydrodynamic modeling. Biotechnol Bioeng. 2007;96:584. doi: 10.1002/bit.21184. [DOI] [PubMed] [Google Scholar]
- 37.van ’tRiet K. Tramper J. Basic Bioreactor Design. 1st. New York: Basel: Marcel Dekker, Inc.; 1991. [Google Scholar]
- 38.Meinel L. Karageorgiou V. Fajardo R. Snyder B. Shinde-Patil V. Zichner L., et al. Bone tissue engineering using human mesenchymal stem cells: effects of scaffold material and medium flow. Ann Biomed Eng. 2004;32:112. doi: 10.1023/b:abme.0000007796.48329.b4. [DOI] [PubMed] [Google Scholar]
- 39.Wei J. Heo S.J. Kim D.H. Kim S.E. Hyun Y.T. Shin J.W. Comparison of physical, chemical and cellular responses to nano- and micro-sized calcium silicate/poly(epsilon-caprolactone) bioactive composites. J R Soc Interface. 2008;5:617. doi: 10.1098/rsif.2007.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almeida A. Bolanos J.P. Moncada S. E3 Ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci U S A. 2010;107:738. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quesney S. Marc A. Gerdil C. Gimenez C. Marvel J. Richard Y., et al. Kinetics and metabolic specificities of Vero cells in bioreactor cultures with serum-free medium. Cytotechnology. 2003;42:1. doi: 10.1023/A:1026185615650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahl A. Sidorenko Y. Dauner M. Genzel Y. Reichl U. Metabolic flux model for an anchorage-dependent MDCK cell line: characteristic growth phases and minimum substrate consumption flux distribution. Biotechnol Bioeng. 2008;101:135. doi: 10.1002/bit.21873. [DOI] [PubMed] [Google Scholar]
- 43.Nelson D. Cox M. Lehninger Principles of Biochemistry. New York: Worth Publishers; 2000. [Google Scholar]
- 44.Zhang P.C. McGrath B.C. Reinert J. Olsen D.S. Lei L. Gill S., et al. The GCN2 eIF2 alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681. doi: 10.1128/MCB.22.19.6681-6688.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rees W.D. Manipulating the sulfur amino acid content of the early diet and its implications for long-term health. Proc Nutr Soc. 2002;61:71. doi: 10.1079/pns2001137. [DOI] [PubMed] [Google Scholar]
- 46.Wang J. Alexander P. Wu L.J. Hammer R. Cleaver O. McKnight S.L. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bendall S.C. Hughes C. Stewart M.H. Doble B. Bhatia M. Lajoie G.A. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol Cell Proteomics. 2008;7:1587. doi: 10.1074/mcp.M800113-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drews M. Doverskog M. Ohman L. Chapman B.E. Jacobsson U. Kuchel P.W., et al. Pathways of glutamine metabolism in Spodoptera frugiperda (Sf9) insect cells: evidence for the presence of the nitrogen assimilation system, and a metabolic switch by H-1/N-15 NMR. J Biotechnol. 2000;78:23. doi: 10.1016/s0168-1656(99)00231-x. [DOI] [PubMed] [Google Scholar]
- 49.Tritsch G.L. Moore G.E. Spontaneous decomposition of glutamine in cell culture media. Exp Cell Res. 1962;28:360. doi: 10.1016/0014-4827(62)90290-2. [DOI] [PubMed] [Google Scholar]
- 50.Reilly G.C. Engler A.J. Intrinsic extracellular matrix properties regulate stem cell differentiation. J Biomech. 2010;43:55. doi: 10.1016/j.jbiomech.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Zhou S. Cui Z. Urban J.P. Nutrient gradients in engineered cartilage: metabolic kinetics measurement and mass transfer modeling. Biotechnol Bioeng. 2008;101:408. doi: 10.1002/bit.21887. [DOI] [PubMed] [Google Scholar]
- 52.Gurkan U.A. Akkus O. The mechanical environment of bone marrow: a review. Ann Biomed Eng. 2008;36:1978. doi: 10.1007/s10439-008-9577-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.