Abstract
Pdx1 (pancreatic and duodenal homeobox 1), Ngn3 (neurogenin 3) and MafA (v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A) have been reported to bring about the transdifferentiation of pancreatic exocrine cells to beta (β) cells in vivo. We have investigated the mechanism of this process using a standard in vitro model of pancreatic exocrine cells, the rat AR42j-B13 cell line. We constructed a new adenoviral vector encoding all three genes, called Ad-PNM (adenoviral Pdx1, Ngn3, MafA construct). When introduced into AR42j-B13 cells, Ad-PNM caused a rapid change to a flattened morphology and a cessation of cell division. The expression of exocrine markers is suppressed. Both insulin genes are up-regulated as well as a number of transcription factors normally characteristic of beta cells. At the chromatin level, histone tail modifications of the Pdx1, Ins1 (insulin 1) and Ins2 (insulin 2) gene promoters are shifted in a direction associated with gene activity, and the level of DNA CpG methylation is reduced at the Ins1 promoter. The transformed cells secrete insulin and are capable of relieving diabetes in streptozotocin-treated NOD-SCID (non-obese diabetic severe combined immunodeficiency) mice. However the transformation is not complete. The cells lack expression of several genes important for beta cell function and they do not show glucose-sensitive insulin secretion. We conclude that, for this exocrine cell model, although the transformation is dramatic, the reprogramming is not complete and lacks critical aspects of the beta cell phenotype.
Keywords: AR42j-B13 cells, beta (β) cell, diabetes, direct reprogramming, insulin
Abbreviations: Abcc8, ATP-binding cassette C8; Ad-PNM, adenoviral Pdx1, Ngn3, MafA construct; anti-anti, antibiotic-antimycotic; araC, cytosine arabinofuranoside; Arx, aristaless related homeobox; ChIP, chromatin immunoprecipitation; Cpa, carboxypeptidase A1; Cpe, carboxypeptidase E; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagles medium; Edu, 5′-ethynl-2′-deoxyuridine; FBS, fetal bovine serum; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Gcg, glucagon; GFP, green fluorescent protein; H3Ac, acetylated histone H3; Hes1, hairy and enhancer of split 1; H3K4Me3, trimethylated Lys4 of histone 3; H3K9Me2, dimethylated Lys9 of histone 3; H3K27Me3, trimethylated Lys27 of histone 3; Iapp, islet amyloid peptide; Ins, insulin; IRPT, immortalized rat kidney proximal tubular; Isl1, ISL1 transcription factor, LIM/homeodomain; Kcnj11, potassium inwardly rectifying channel; KRB, Krebs–Ringer buffer; LB, Luria–Bertani; mAb, monoclonal antibody; Mafa, v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A; Mnx1, motor neuron and pancreas homeobox 1; MOI, multiplicity of infection; Neurod1, neurogenic differentiation 1; Ngn3, neurogenin 3; Nkx, natural killer 2 transcription factor related; NOD-SCID, non-obese diabetic severe combined immunodeficiency; Pax, paired box gene; PBS-T, 0.1% Tween 20 in PBS; Pcsk2, proprotein convertase subtilisin/Kexin type 2; Pdx1, pancreatic and duodenal homeobox; Ppy, pancreatic polypeptide; Prss1, trypsin 1; Ptf1A, pancreas-specific transcription factor 1a; qPCR, quantitative PCR; Rbpj1, recombination signal binding protein for Igκ; RT–PCR, reverse transcription–PCR; qRT–PCR, quantitative RT–PCR; Slc2a2, solute carrier family 2; Sst, somatostatin; STZ, streptozotocin; TAE, Tris/acetate/EDTA
INTRODUCTION
During embryogenesis, cells undergo a sequence of developmental commitment decisions, controlled by extracellular inducing factors, and eventually become one of about 200 different cell types of the vertebrate body [1]. Differentiated cell types tend to be relatively stable and to persist long term, with or without cell division. However it is sometimes possible to reprogramme one differentiated cell type to another by overexpression of specific transcription factors which are responsible for the relevant commitment processes in normal development [2]. The first example of this was the ability of the myogenic factor MyoD to reprogram a variety of tissue culture cell lines to a myogenic phenotype [3]. More recently, several other examples have been described, including the conversion of pancreatic exocrine cells into hepatocytes [4], B-lymphocytes into macrophages [5], pancreatic exocrine cells into endocrine cells [6], fibroblasts into neurons [7] or fibroblasts into cardiomyocytes [8]. Between one and three transcription factors need to be overexpressed to achieve these transformations, typically ones that are involved in the normal embryonic development of the cell type in question [9,10]. Their function is not simply to activate direct target genes, but to shift the cell into a new stable state of gene expression. Most strikingly, it is also possible to use this approach to reprogramme a variety of normal tissue cells to a state resembling that of embryonic stem cells (induced pluripotent stem cells, [11,12]).
In defining a cell type, one important aspect is the configuration of active genes for the specific differentiation products. Another is the configuration of regulatory genes that maintain the state in a stable manner. For both it is clear that an essential feature of the differentiated cell state is the chromatin state of key genes. Two particular types of epigenetic modification have been shown to be important in this context. The modification of histone tails, particularly of histone 3, can cause both positive and negative effects on gene expression. In general acetylation is thought to be associated with an active chromatin configuration [13], whereas histone tail methylation may have either positive or negative effects, depending on the specific site in the protein [14]. The other type of modification is DNA base methylation. This occurs on the 5 position of cytosine in CpG dinucleotides. Active genes are usually associated with a relative undermethylation of CpGs in their regulatory regions (CpG islands [15]), and repressed genes are associated with a high level of methylation in their regulatory regions. DNA methylation is well established to be heritable on cell division, and there is also some evidence that histone modifications can be maintained through cell division [16]. Thus both of these general mechanisms are likely to be basic to the maintenance of differentiated cell types.
Of the various examples of direct cell type reprogramming with transcription factors, the ability to make pancreatic beta (β) cells is of particular interest from the point of view of potential cell transplantation therapy for diabetes. This is because the clinical procedure of islet cell transplantation therapy is well established and the limiting factor is entirely the lack of suitable human beta cells [17]. The study of Zhou et al. [6], in which pancreatic exocrine cells were converted into insulin-positive cells, was conducted in vivo using immunodeficient mice. Because it is difficult to investigate molecular mechanisms in vivo, we felt it important to establish an in vitro model for the process which is more amenable to study.
We selected the AR42j-B13 cell line for this purpose, referred to here as B13 cells. This is a rat cell line with a pancreatic exocrine phenotype, originally derived from a chemically induced pancreatic tumour [18]. It is easy to grow in culture and expresses amylase and other typical exocrine cell products. It has a stable phenotype, in contrast with primary cultures of pancreatic exocrine cells which undergo a rapid ductal transformation in adherent culture [19], or de-differentiation in suspension culture [20,21]. Unlike samples of pancreatic tissue from animals, which contain many cell types such as connective tissue cells and blood vessels in addition to the epithelium, a culture of B13 cells contains only one cell type, making biochemical measurements more meaningful. Although there are reports in the literature of insulin-positive cells arising from this line following culture on Matrigel and treatment with various growth factors [22,23], we have not used such conditions in the present study and see no spontaneous endocrine differentiation of the cells under the conditions used.
In the present study we describe the effects of Pdx1 (pancreatic and duodenal homeobox 1)+Ngn3 (neurogenin 3)+MafA (v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A), the gene combination used by Zhou et al. [6], on the B13 cells. Pdx1 is a major pancreatic transcription factor belonging to the ParaHox family, which is necessary both for formation of the pancreatic buds in the embryo and subsequently for beta cell formation and function [24,25]. Ngn3 is a member of basic helix-loop-helix transcription factor family and it is essential for the formation of endocrine progenitor cells during pancreatic development [26]. MafA is a member of the basic leucine zipper transcription factor family and it is needed for maturation of beta cells [27]. In addition to their developmental functions, both Pdx1 and MafA positively regulate insulin gene expression in beta cells.
We introduced the three transcription factor genes using a single adenoviral vector, Ad-PNM (adenoviral Pdx1, Ngn3, MafA construct), ensuring that all transduced cells receive the same genes. We find that a high proportion of transduced cells alter morphology, down-regulate expression of exocrine products, such as amylase, trypsin and carboxypeptidase A, and express insulin from both of their insulin genes. The chromatin state of insulin genes is moved toward the beta-cell state in respect both of histone tail modifications and DNA methylation. The cells acquire various additional properties of pancreatic beta cells and are capable of relieving diabetes following transplantation into an experimental mouse model of diabetes. However, we also find that some important beta cell transcription factors are not up-regulated and that the vital glucose-sensing mechanism of beta cells is not present in these cells. We therefore conclude that the reprogramming achievable with these three genes, although dramatic, is not complete.
EXPERIMENTAL
Recombinant adenovirus preparation
The Pdx1, Ngn3 and MafA genes from the mouse were used to construct a single adenovirus encoding all three transcription factors, called Ad-PNM. Full-length mouse Pdx1 and Ngn3 cDNAs were amplified by PCR to replace their translational termination codons with specific restriction sites and cloned into pBluescript (KS+/−) as a XbaI/BamHI fragments. The Pdx1 cDNA and Ngn3 cDNA were then ligated to the coding region for the 18-amino-acid peptide 2A from FMDV (Foot and Mouth Disease virus) to generate pBS–Pdx1–2A and pBS–Ngn3–2A. They were then joined such that the coding region of the 2A was followed by the first amino acid codon of the Ngn3 coding region to make the construct pBS–Pdx1–2A–Ngn3–2A. This was then ligated to the MafA cDNA including the translational termination codon and cloned as BglII/EcoRV fragment in pBluescript (KS+/−) in the same way to generate the polycistronic construct PNM (pBS–Pdx1–2A–Ngn3–2A—MafA). The PNM construct was taken out as a single SalI fragment using the sites present internally upstream of Pdx1 and downstream of MafA, and cloned into the shuttle vector pShuttleT2ALCAG5PL4–7. Recombinant adenovirus containing the PNM construct was produced by homologous recombination after electroporating the linearized shuttle vector into BJ5183 bacterial cells. This was done using the pAdEasy adenoviral vector system (Agilent Technologies) according to the manufacturer's instructions.
Ad-GFP (green fluorescent protein) was used for various control experiments. Its preparation is described in [28].
Cell culture and virus administration
The rat pancreatic exocrine cell line (AR42J-B13, referred to here just as B13) was obtained from Dr David Tosh (University of Bath, Bath, U.K.) and grown in low-glucose DMEM (Dulbecco's modified Eagle's medium; Gibco) supplied with 10% (v/v) FBS (fetal bovine serum; Hyclone) and 1×anti-anti (antibiotic-antimycotic; Gibco) solution. The rat insulinoma cell line (RIN-m5F) (A.T.C.C. Manassas, VA, U.S.A) was grown in high-glucose DMEM (Gibco) supplied with 10% (v/v) FBS and 1×anti-anti solution. IRPT (immortalized rat kidney proximal tubular) cells were obtained from Dr Shunan Li (University of Minnesota, Minneapolis, MN, U.S.A.) and were grown in low-glucose DMEM supplied with 5% (v/v) FBS and 1×anti-anti solution. Media were replaced every 2 days.
B13 cells were transduced with a dose of 25 MOI (multiplicity of infection) Ad-PNM and the next day the virus including medium was replaced.
RT–PCR (reverse transcription–PCR)
Total RNA was isolated using the RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions. RNA samples were then treated with DNase (Promega) to remove genomic DNA. A PCR was run with the RNA template at 60°C for 35 cycles to confirm the absence of genomic DNA contamination. cDNAs were then synthesized by reverse transcription from 2 μg of total RNA using SuperScript III Reverse Transcriptase, oligo(dT)20 and dNTP (10 mM) (Invitrogen). The gene expression pattern between B13 cells with and without Ad-PNM was then compared by performing PCR for rat Ins (insulin) 1, Ins2 (insulin 2), Gcg (glucagon), Ppy (pancreatic polypeptide), Sst (somatostatin), Iapp (islet amyloid polypeptide), Pax (paired box gene) 4, Neurod1 (neurogenic differentiation 1), Nkx [NK (natural killer) 2 transcription factor related] 2.2, Arx (aristaless related homeobox), Pdx1, Ngn3, MafA, Nkx6.1, Isl1 (ISL1 transcription factor, LIM/homeodomain), Mnx1 (motor neuron and pancreas homeobox 1 also known as Hlxb9), Pax6, Hes1 (hairy and enhancer of split 1), Slc2a2 [solute carrier family 2 (facilitated glucose transporter), member 2 also known as Glut2], Kcnj11 (potassium inwardly rectifying channel, subfamily J, member 11 also known as Kir6.2), Abcc8 (ATP-binding cassette C8 also known as Sur1), Amy (amylase), Ptf1a (pancreas-specific transcription factor 1a), Cpe (carboxypeptidase E), Pcsk2 (proprotein convertase subtilisin/kexin type 2), Prss1 (trypsin 1), Cpa (carboxypeptidase A1; 27 cycles), Ctrb1 (chymotrypsingoen B1; 25 cycles), Rbpj1 (recombination signal binding protein for Igκ J region; 26 cycles) and Actb (β-actin), in addition to mouse Pdx1, Ngn3 and MafA. Rat pancreas cDNA was used as a positive control for Gcg, Sst and Ppy. cDNA from Ad-PNM-infected HEK (human embryonic kidney)-293 cells was used as positive control for the input genes mouse Pdx1, mouse Ngn3 and mouse MafA. cDNA from RIN-m5F cells was used as positive control for the rest of the genes. The PCR conditions were: 94°C for 3 min initial denaturation, 94°C for 30 s denaturation, 60°C for 30 s annealing, 72°C for 1 min extension and 72°C for 5 min final extension, and overall 30 cycles were performed unless otherwise indicated. Primers used for RT–PCR are listed in Supplementary Table S1 (at http://www.BiochemJ.org/bj/442/bj4420539add.htm). PCR products were run on a 1% (w/v) agarose gel (Invitrogen) in TAE (Tris/acetate/EDTA) buffer (Bio-Rad Laboratories) with ethidium bromide by electrophoresis at 100V. Gels were visualized with a Bio-Rad Laboratories gel documentation system model 2000.
qRT–PCR (quantitative RT–PCR)
Ad-PNM-treated B13 cells were kept in DMEM supplemented with 200 μM of the anti-mitotic agent araC (cytosine arabinofuranoside; Sigma) to suppress the continued proliferation of untransformed B13 cells. RNA isolation and cDNA synthesis from these cells were performed as described above. The gene expression pattern between B13 cells with and without Ad-PNM was then compared by performing qRT–PCR for rat Ins1, Ins2, Iapp, Pax4, Neurod1, Nkx2.2, Arx, endogenous Pdx, endogenous Ngn3, endogenous MafA, Isl1, Mnx1, Hes1, Slc2a2, Kcnj11, Abcc8, Amy, Ptf1a, Cpe, Pcsk2, Pcsk1, Prss, Cpa, Ctrb1, Rbpj1 and Gapdh (glyceraldehyde-3-phosphate dehydrogenase). qRT–PCR conditions were 95°C for 30 sec initial denaturation, 95°C for 5 sec denaturation, and 60°C for 10 sec annealing and extension, and overall 40 cycles were performed. The primers used for the qRT–PCR are listed in Supplementary Table S1.
Immunostaining
B13 cells were fixed with 4% (w/v) paraformaldehyde (Sigma) in PBS (Sigma) for 20 min. After fixation cells were washed three times with PBS-T [0.1% Tween 20 (Bio-Rad Laboratories) in PBS] for 5 min in each wash. Cells were then permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 15 min. After 1 h of blocking with PBS-T containing 1% (w/v) BSA (Sigma), cells were incubated overnight with primary antibodies at 4°C and then for 1 h with secondary antibodies at room temperature (20°C). The primary antibodies used were: guinea pig anti-insulin (1:200 dilution; Sigma); rabbit anti-Pdx1 (1:2000 dilution; Upstate Biotechnology); rabbit anti-C-peptide (1:100 dilution; Cell Signaling Technology), rabbit anti-Ngn3 (1:100 dilution; Santa Cruz Biotechnology); and rabbit anti-MafA (1:100 dilution; Santa Cruz Biotechnology). The secondary antibodies used are as follows: Alexa Fluor® 480 goat anti-rabbit IgG (1:500 dilution; Invitrogen) and Alexa Fluor® 594 goat anti-guinea pig IgG (1:500 dilution; Invitrogen). Images were taken using a Leica DMI6000 B inverted microscope.
EdU (5′-ethynl-2′-deoxyuridine) labelling and staining
B13 cells (5×105) were plated in each well of six-well plates. EdU (10 μM) from the Click-iT EdU 594 Imaging Kit (Invitrogen) was used for labelling. After the incubation cells were fixed and stained for EdU, according to the manufacturer's instructions to detect the number of cells in cycle during the EdU treatment. Images were taken using a Leica DMI6000 B inverted microscope.
ELISA for total insulin content and glucose-stimulated insulin release
B23 cells (5×105) were plated in each well of a six-well plate. Cells in three wells were kept without Ad-PNM as controls. The cells in the remaining three wells were transduced with Ad-PNM overnight, after which the medium was changed. At 3 days after Ad-PNM administration, DMEM was removed and cells were incubated either with 2.8 mM or 20 mM glucose in KRB (Krebs–Ringer buffer) for 1 h at 37°C. In some experiments 30 mM KCl was included to effect depolarization of the cells. The KRB was then removed and kept at −20°C for measurement of insulin release into the medium. After the removal of KRB, the cells were scraped into 0.18 M HCl/35% ethanol. The cells were homogenized by sonication with a Branson Sonifier 450 (6×10 s on ice with 1 min waiting on ice between pulses) and rotated overnight at 4°C. The next day, the cells in acid/ethanol were subjected to centrifugation (10000 g for 30 min at 4°C). The supernatant was kept at −20°C to measure the total amount of insulin in the cells. The remaining cell pellets were resuspended in 0.1 M NaOH to measure the total amount of protein in the cells. The amount of the insulin released into the medium and the total insulin in the cells were measured using an Ultrasensitive Rat Insulin ELISA kit (Mercodia), according to the manufacturer's instructions. The total protein amount in the cells was measured using the BCA (bicinchoninic acid) protein assay kit (Thermo Scientific), according to manufacturer's instructions.
ChIP (chromatin immunoprecipitation) for histone tail modifications and qPCR (quantitative PCR)
B13 cells (107) with no Ad-PNM, B13 cells 3 days after administration of Ad-PNM, RIN-m5F cells or IRPT cells were plated on 20 cm tissue culture dishes. Formaldehyde (36.5%; Sigma) was added to the dishes to final concentration of 1% in each dish. After 10 min incubation, the cross-linking reaction was quenched by addition of 2 ml of 1.25 M glycine (Sigma). The cells were subjected to centrifugation (10000 g for 15 min at 4°C) and the supernatant was discarded. Cell and nucleus lysis buffers (Magna ChIP™ G Kit, Millipore) were added to the cell pellets. The cells/DNA were sheared by sonication using a Branson Sonifier 450 (15×10 s on ice with 1 min waiting on ice between pulses). Then ChIP was performed for H3K4Me3 (trimethylated Lys4 of histone 3), H3K9Me2 (dimethylated Lys9 of histone 3), H3K27Me3 (trimethylated Lys27 of histone 3) and H3Ac (acetylated histone H3) by using Magna ChIP™ G Kit (Millipore), according to the manufacturer's instructions. Antibodies used for ChIP were: a rabbit anti-H3K4Me3 mAb (monoclonal antibody; Cell Signaling Technology), mouse anti-H3K9Me2 mAb (Abcam) and mouse anti-H3K27Me3 mAb (Abcam) and rabbit anti-H3Ac polyclonal antibody (Millipore). For each immunoprecipitation reaction 5 μg of antibody was used. After immunoprecipitation of DNA fragments bearing the histone modifications mentioned above, qPCR was performed by using specific primer pairs (Supplementary Table S1) spanning approximately −200 bp to +50 bp of the Ins1, Ins2 and Pdx1 genes. This region includes a number of important regulatory sites as shown in Supplementary Figure S1 (at http://www.BiochemJ.org/bj/442/bj4420539add.htm) [29–33]. qPCR conditions were: 50°C for 2 min, 95°C for 10 min initial denaturation, 95°C for 15 s denaturation, 60°C for 30 s annealing and 60°C for 30 s extension (overall 40 cycles were run) using a Eppendorf Realplex4 mastercycler. To normalize the ChIP data, the fold enrichment method was used in which ChIP signals were divided by non-specific antibody signals.
DNA methylation assay with bisulfite sequencing
Genomic DNA isolation from B13 cells with no Ad-PNM and from B13 cells with 3 days after Ad-PNM, RIN-m5F cells or IRPT cells was performed with QIAamp DNA Mini Kit (Qiagen), according to the manufacturer's instructions. Bisulfite conversion of isolated genomic DNA was performed with the EZ DNA methylation-gold kit (Zymo Research), according to the manufacturer's instructions. Bisulfite-treated DNA-specific primer pairs (Supplementary Table S1) which span the proximal promoters and first exons of Ins1, Ins2 and Pdx1 genes (Supplementary Figure S1) were designed by using MethPrimer software [34]. To amplify the bisulfite-treated genomic DNA, PCR was performed as follows: 95°C for 10 min initial denaturation, 95°C for 30 s denaturation, 50°C for 40 s annealing, 72°C for 1 min extension and 72°C for 7 min final extension, and overall 40 cycles were performed. Hot Taq DNA Polymerase (Zymo Research) was used in these PCRs. PCR products were run on 1% (w/v) agarose gel in 1×TAE buffer by electrophoresis at 100 V. DNA fragments of interest were purified from the gel with Wizard SV Gel and PCR Clean-Up System (Promega), according to the manufacturer's instructions. Purified PCR products were cloned into the pCR4-TOPO vector with TOPO TA Cloning Kit (Invitrogen) for subsequent sequencing. Plasmids including the DNA fragments of interest were transformed into the chemically competent One Shot TOP10 cells provided by the kit, according to the manufacturer's instructions. Cells were grown on LB (Luria–Bertani) agar plates containing kanamycin. From each plate ten colonies were picked up randomly and grown in kanamaycin LB broth overnight at 37°C. The next day plasmid DNA isolation was performed with Wizard Plus SV Minipreps DNA Purification System (Promega), according to the manufacturer's instructions. Each collected clone was restriction-digested with EcoRI (Promega) to verify that they contain the DNA fragments of interest. Then ten different clones for each DNA fragment were sequenced by the DNA Sequencing and Analysis Facility, Biomedical Genomics Center, University of Minnesota, MN, U.S.A.
Mouse experiments
For the cell transplantation experiments, adult male NOD-SCID (non-obese diabetic severe combined immunodeficiency) mice weighting 22–27 g (up to 42 days old) were injected intraperitoneally with STZ (streptozotocin; Sigma) at a dose of 120 mg/kg of body mass. Mice were considered diabetic if their blood glucose level reached a stable value over 400 mg/dl. The diabetic mice were transplanted with Ad-PNM-treated B13 cells or with Ad-GFP-transduced cells as a control. Under the kidney membrane of each mouse, 2×106 cells were transplanted. Blood was drawn from the tail vein and blood glucose levels were measured with a glucose meter every 2 days. At 10 days after cell transplantation, the kidneys with transplanted cells were removed and blood glucose levels of mice were monitored for two more days. The blood sugar changes were quantified by measuring the area under the curve, and comparisons were made using a Student's t test. The excised kidneys were fixed in 10% (v/v) formalin (Fisher) and processed for tissue sectioning. After antigen retrieval, sections were stained for insulin, E-cadherin and DAPI (4′,6-diamidino-2-phenylindole) as described above. The mouse anti-E-cadherin (1:500 dilution; BD Bioscience Pharmingen) primary antibody and Alexa Fluor® 488 goat anti-mouse IgG (1:500 dilution; Invitrogen) secondary antibody were used to stain E-cadherin protein.
For expression of Pdx1, Ngn3 and MafA in the exocrine pancreas, adult CD1 mice were anaesthetized with avertin. A portion of skin was shaved and wiped with betadine. A small incision was then made and the pancreas everted from the wound. Ad-GFP or Ad-PNM (100 μl; 1011 plaque-forming units/ml) were injected into the splenic lobe of the pancreas using an insulin needle. This caused formation of a visible bubble of fluid within the pancreas. The pancreas was reinternalized and the wound repaired in layers. The mice were killed 7 days later. The pancreases were removed, fixed in 10% (v/v) formalin (Fisher) and processed for tissue sectioning (frozen for GFP and paraffin for immunostaining). Sections were stained for insulin and Ngn3 as described above.
The animal experiments were carried out under IACUC (Institutional Animal Care and Use Committee) protocol 1007A85640 of the University of Minnesota.
RESULTS
Effects of Pdx1, Ngn3 and MafA on B13 cells
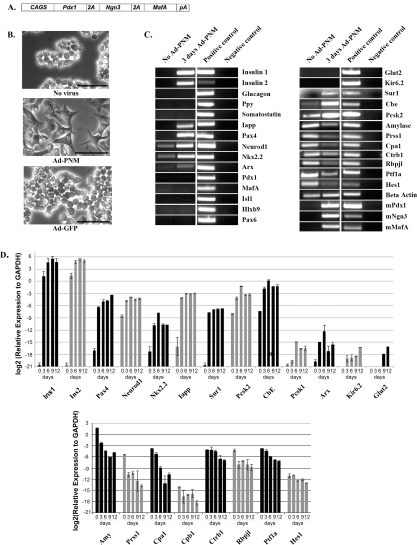
When three adenoviruses carrying different genes are used to transduce cells, they do so at random such that individual cells may receive one, two or three genes. To remove this source of variation we made an adenoviral vector carrying all three genes in order that all transduced cells would receive all three (Figure 1A). The three coding regions are joined by 2A sequences, which cause autocleavage of the extending polypeptide during protein synthesis [35]. The whole is driven by the highly active cags (cytomegalovirus-chicken β-actin) promoter [36]. We confirmed that this vector would infect cells efficiently and that all three proteins could be detected by immunostaining (shown in Figure 2). Trials of different viral doses indicated that for B13 cells the optimum MOI was 25 and this dose was used in the experiments to be described. The day after virus administration, cells changed their shape from an oval structure to a more flattened fibroblast-like shape (Figure 1B). A control virus encoding GFP did not have this effect, showing that it is not a consequence of virus infection itself.
Figure 1. Effect of PNM on cell shape and gene expression profile of B13 cells.
(A) Diagram of the Ad-PNM construct. CAGS, cytomegalovirus-chicken β-actin. (B) Shape of the B13 cells with no treatment, Ad-PNM or Ad-GFP. Images were taken 3 days after virus transduction. Scale bars=100 μm. (C) Changes in the gene expression profile of B13 cells, with and without Ad-PNM, by RT–PCR. From left to right the lanes are as follows: untreated B13 cells, B13 cells 3 days after Ad-PNM, positive control sample and water (negative control). Input gene expression is labelled as mPdx1, mNgn3 and mMafA. Each horizontal strip comes from a single gel but different strips may come from different gels. The results for Abcc8/Sur1, Prss1, Cpa1, Ctrb1 and Rbpjl were obtained with the qRT–PCR primers. (D) Changes in the gene expression profile of B13 cells, with and without Ad-PNM, by qRT–PCR. From left to right the bars are as follows: untreated B13 cells as day 0, Ad-PNM-treated B13 cells as day 3, day 6, day 9 and day 12. Cultures were kept in araC to remove untransduced B13 cells. The heights of bars shows the expression relative to the housekeeping gene Gapdh. Results are means±S.E.M. (n=3).
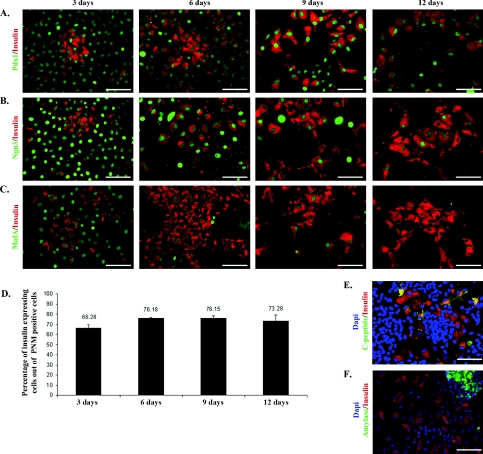
Figure 2. Expression of insulin and exogenous Pdx1, Ngn3, MafA.
B13 cells were fixed and stained 3, 6, 9, 12 days after Ad-PNM transduction. Cells were then co-stained for insulin (red) and Pdx1 (green) (A), insulin (red) and Ngn3 (green) (B), insulin (red) and MafA (green) (C). (D) Percentage of insulin-positive cells out of the population of PNM-positive cells. Data are means±S.E.M. (n=3). (E) C-peptide (green), insulin (red) and DAPI (blue) at day 12. (F) Amylase (green), insulin (red) and DAPI (blue) at day 6. Scale bars=100 μm.
Ad-PNM also altered the gene expression pattern. RT–PCR results are shown at 3 days and qRT–PCR over a time course of 12 days (Figures 1C and 1D). For the 12 day studies cells were cultured in the presence of araC. This kills dividing cells and thus removes the minority of untransduced cells that continue to divide and would otherwise overgrow the cultures. These results therefore reflect just the transduced cells. They started to express Ins1 and Ins2 genes (rodents have two insulin genes encoding similar products). Expression of Iapp and Pax4, which are not normally expressed in B13 cells, were also up-regulated. Moreover, these cells elevated the expression level of genes encoding the beta cell transcription factors Neurod1, Nkx2.2 and the pro-insulin processing enzymes cpe and Pcsk2 which are all expressed to some extent in the untreated cells. One of the genes for the ATP-sensitive potassium channel Sur1 (Abcc8) was up-regulated, but the gene for another component of this channel, Kir6.2 (Kcnj11), was not. In contrast with the beta cell markers, the expression of all the exocrine markers (Amy, Prss1, Cpa1, Ctrb1, Rbpj1 and Ptf1a) was down-regulated, as was the centroacinar marker Hes1. The fall in Amy, Prss1 and Cpa1 transcripts was approximately 100-fold, whereas that of Ctrb1, Rbpj1 and Ptf1a was approximately 10-fold. There was no up-regulation of the genes encoding the hormones produced by the non-beta endocrine cell types found in the pancreas: Gcg, Ppy, Sst and Ghrl (ghrelin).
Thus in a number of respects the Ad-PNM-treated B13 cells appeared to have a phenotype more resembling that of pancreatic beta cells than of pancreatic exocrine cells. On the other hand, apart from Abcc8 (Sur1), other genes for beta cell membrane channel proteins, such as those encoding Slc2a2/(Glut2) and Kcnj11 (Kir6.2), were not significantly up-regulated (Figures 1C and 1D). The ability to secrete insulin in response to a rise in external glucose concentration is a key attribute of the beta cell, and the essential components of this mechanism are not all induced, indicating that the reprogramming of cell type is incomplete. Also, the reduction of exocrine gene expression, although striking, is not a complete extinction. Furthermore, expression is not induced from the endogenous counterparts of the three input genes. The viral-encoded Pdx1, Ngn3 and MafA are mouse sequences and so their mRNA can be distinguished from those of the rat B13 cells using suitable primers. Although the expression of Ngn3 might be expected to be confined to the period of formation of endocrine precursors, we would expect to see Pdx1 and MafA expressed long-term in genuine beta cells, and it is not seen.
We also performed immunostaining for insulin, its side product C-peptide, the major exocrine protein amylase, and the three input proteins Pdx1, Ngn3 and MafA. The majority of Ad-PNM-transduced cells started to express insulin protein by 3 days (Figure 2) and these insulin-positive cells can persist for at least 4 weeks. Not all of the virus-transduced cells became insulin-positive, but about 70% did so and this proportion remains similar over at least 12 days (Figure 2D). C-peptide was found to be present and to be co-localized with insulin. C-peptide is normally excised during the maturation of pro-insulin to the mature two-chain structure and its presence in insulin-positive cells indicates that the pro-insulin protein is properly processed after expression (Figure 2E). Moreover, as viewed by immunostaining, the insulin-positive cells completely lost amylase protein which is one of the major products of pancreatic exocrine cells (Figure 2F). Note that araC was not used in these experiments so there are still untransduced B13 cells present which are growing rapidly. This provides the internal positive control for amylase staining in Figure 2(F).
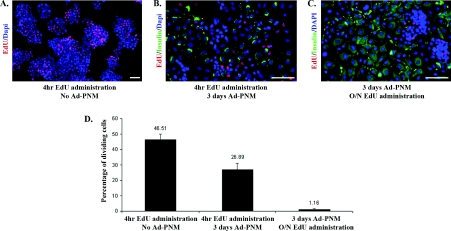
Next we looked at the proliferation ability of B13 cells before and after Ad-PNM transduction (Figure 3). A 4 h pulse of EdU was found to label about half of the untreated B13 cells as expected for a rapidly dividing population (Figures 3A and 3D). If Ad-PNM is added after such a 4 h pulse and insulin-positive cells are examined 3 days later, a high proportion are labelled, although somewhat less than 50%, indicating that the transformation may occur either in cells in S-phase or not in S-phase (Figures 3B and 3D). However, if the cells were transduced with Ad-PNM, left for 3 days and then given an overnight label with EdU, the proportion of insulin-positive cells that are also EdU-labelled is extremely small (Figures 3C and 3D). This shows that cell division ceases almost entirely following the transformation. If overgrowth by the untransformed B13 cells is suppressed with araC, then the fibroblast-shaped insulin-positive cells can persist for a long time. Cultures have been maintained for 4 weeks, although there is some cell death of the insulin-positive cells over this period.
Figure 3. Cell proliferation assay with EdU.
(A) B13 cells were given EdU for 4 h. Then they were fixed and stained for EdU (red) and DAPI (blue). (B) B13 cells were given EduU for 4 h. Then the cells were transduced with Ad-PNM. At 3 days later, cells were fixed and stained for EdU (red), insulin (green) and DAPI (blue). (C). B13 cells were transduced with Ad-PNM. At 3 days later these cells were given EdU overnight. Next day they were fixed and stained for EdU (red), insulin (green) and DAPI (blue). Scale bars=100 μm. The red colour is slightly enhanced in (B and C) to be consistent with (A). (D) Left-hand bar: percentage of proliferating cells out of total population. Middle and right-hand bars: percentage of proliferating insulin-positive cells out of total insulin-positive cells. Data are means±S.E.M. (n=3).
Insulin content and secretion in transformed cells
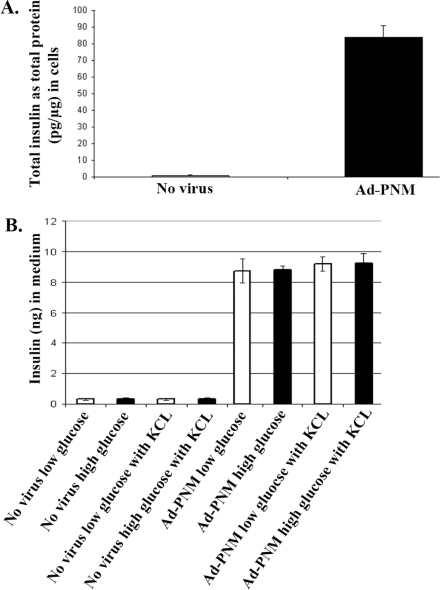
To detect the amount of the insulin protein produced we measured the total insulin amount by ELISA in the B13 cells with or without Ad-PNM. This showed a substantial increase, although it did not reach the level of mature beta cells which can contain as much as 5% of insulin protein as a proportion of total cell protein (Figure 4A). Because glucose-stimulated insulin release is the key physiological property of beta cells we examined this by treating the cells with 2.8 or 20 mM glucose in KRB for 1 h and then analysing the insulin content of the medium by ELISA (Figure 4B). We also tested KCl which produces complete depolarization of beta cells and causes them to emit a maximal amount of insulin. As expected, there was no insulin secretion from untreated B13 cells. The Ad-PNM-transduced cells did secrete insulin, but in a completely unregulated manner, showing no effect of glucose or of KCl. We conclude that the transformed cells do secrete insulin in a constitutive, but not a regulated, manner.
Figure 4. Total insulin and secreted insulin amount by ELISA.
(A) The total insulin amount in the B13 cells with and without Ad-PNM was measured by ELISA. (B) The amount of insulin in the medium released from B13 cells with and without Ad–PNM transduction was measured by ELISA. Cells were stimulated either with low-glucose (open bars) or high-glucose (closed bars) and KCl. Experiments with Ad-PNM-treated cells were performed 3 days after transduction. Data are means±S.E.M. (n=3).
Effects on histone tail modifications at Pdx1 and insulin genes
Histone tail modifications such as methylation and acetylation can be functionally important either for up-regulation or repression of a gene [13,14]. H3K4Me3 and H3Ac are two of the indicators of the active transcription when found in histones at the promoter region of a gene, whereas H3K9Me2 and H3K27Me3 are two of the indicators of the repression of transcription.
Reprogramming can be expected to activate regulatory genes controlling the cell phenotype as well as the genes encoding the differentiation products for the gene in question. For this reason we examined these four histone tail modifications at the promoter of the endogenous Pdx1 gene as well as at the promoters of the Ins1 and Ins2 genes [29–33] (Figure 5). The Gapdh gene, which is active in all cell types, was used as positive control (Figure 5A). We compared RIN-m5F cells (a rat insulinoma line which is Ins1+, Ins2+ and Pdx1+), B13 cells without Ad-PNM (which are Ins1−, Ins2− and Pdx1−), the B13 cells with Ad-PNM (which are Ins1+, Ins2+ and Pdx1−) and IRPT cells (a rat kidney line which is Ins1−, Ins2− and Pdx1−). ChIP was performed for H3K4Me3, H3K9Me2, H3K27Me3 and H3Ac modifications followed by qRT–PCR for the promoter region under study. For Ad-PNM-transduced cells, ChIP was conducted 3 days after viral transduction.
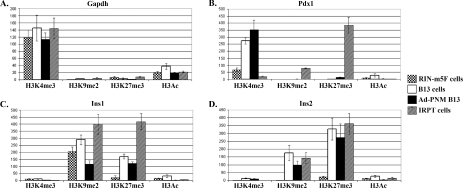
Figure 5. Changes of the histone tail modifications at the promoter regions of the Pdx1, Ins1 and Ins2 genes.
ChIP assays were carried out to compare the proportion of the histone tail modifications (H3K4Me3, H3K9Me2, H3K27Me3 and H3Ac) at the promoters of the Gapdh (A), Pdx1 (B), Ins1 (C) and Ins2 (D) genes. The cell types analysed are RIN-m5F cells (stippled bars), B13 cells without Ad-PNM (open bars), B13 cells with Ad-PNM (closed bars) and IRPT cells (diagonal hatched bars). The y-axis shows fold change of samples over negative control rat IgG. Data are means±S.E.M. (n=3).
At the Pdx1 promoter of the B13 cells, Ad-PNM increased the proportion of H3K4Me3 to a modest extent and it did not change the level of H3K9Me2, H3K27Me3 or H3Ac (Figure 5B). This is consistent with the failure to activate the endogenous gene as shown by the RT–PCR studies. At both the Ins1 and Ins2 promoters of B13 cells, Ad-PNM reduced the inhibitory H3K9Me2 and H3K27Me3 proportions to a degree (not statistically significant for Ins2 H3K27Me3) (Figures 5C and 5D). This is consistent with the up-regulation of both of these genes as shown by RT–PCR. We had hypothesized that the chromatin for regulatory genes such as Pdx1 might be ‘open’ in pancreatic exocrine cells and ‘closed’ in non-pancreatic cells such as the kidney. As shown in Figure 5(B) this is the case in relation to the H3K4Me3 and the two inhibitory indicators. However since we see no significant up-regulation of endogenous Pdx1 with Ad-PNM it may be that the chromatin is not really accessible and is rendered inactive by other modifications not examined in the present study.
Effects on promoter methylation at Pdx1 and insulin genes
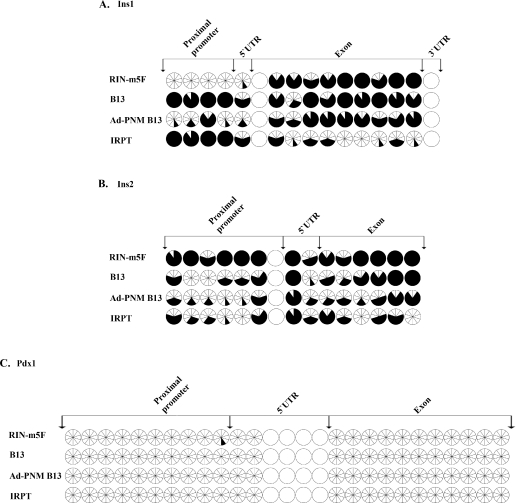
In addition to histone tail modifications, DNA base methylation at promoter regions of the genes is another important epigenetic factor affecting gene expression. The methylation of cytosines at CpG islands is usually associated with repression of gene activity [15]. To see whether Ad-PNM changes the DNA methylation pattern in treated cells, we compared the same four cell populations used for the ChIP analysis and performed bisulfite sequencing for the same promoter regions of Pdx1, Ins1 and Ins2 genes. The results indicate that the expression of the Pdx1 gene is not under the control of DNA methylation at the sites examined, as there is no CpG methylation at either proximal promoter or exons in any of the cell types. However, expression of the Ins1 gene correlated with DNA methylation at the proximal promoter (Figure 6A). In RIN-m5F cells (which are Ins1+), the CpG methylation ratio was only 2% at the proximal promoter as compared with 63% for the whole gene. In IRPT cells (which are Ins−), the CpG methylation ratio was 90% at the proximal promoter compared with 47% for the whole gene. This suggests that demethylation of the proximal promoter of the Ins1 gene is necessary for gene expression. As with IRPT cells, the proximal promoter of B13 cells (which are Ins−) is highly methylated at 90%. After addition of Ad-PNM (such that the cells become Ins1+), proximal promoter CpG methylation was reduced to 28%, with little change of methylation in the Ins1 exon. Unlike Ins1, the expression of the Ins2 gene may not be under the control of DNA methylation at the sites examined as there was no correlation with expression state. In the RIN-m5F cells (Ins2+), the methylation ratio was 88% at the proximal promoter and 89% throughout the whole gene. In IRPT cells (Ins2−), the methylation ratio was 42% at the proximal promoter and 40% throughout the whole gene. After Ad-PNM treatment the methylation ratio in B13 cells fell slightly from 40% to 35% at the proximal promoter and from 54% to 49% throughout the whole gene (Figure 6B).
Figure 6. Changes of the DNA methylation pattern of the Pdx1, Ins1 and Ins2 genes after transduction with Ad-PNM.
Comparison of the CpG methylation pattern of the Ins1 (A), Ins2 (B) and Pdx1 (C) genes in RIN-m5F cells, B13 cells with and without Ad-PNM, and IRPT cells. Each circle, divided into ten wedges, represents ten different clones for the same CpG. Open wedges represent unmethylated CpGs and closed wedges represent methylated CpGs. Intact open circles represent CpGs that were not covered by the bisulfite sequencing. UTR, untranslated region.
In summary, the DNA base methylation study indicates that there is no pre-existing competence of these genes for up-regulation in the pancreatic cells as compared with the kidney cells, and that of the genes examined Ins1 shows promoter demethylation following Ad-PNM treatment, whereas Ins2 does not.
Insulin-expressing B13 cells rescue diabetic mice
In diabetes research a standard assay for a beta cell phenotype consists of a transplantation assay to ameliorate diabetes in experimental animals. For this purpose we used immune-deficient (NOD-SCID) mice which will tolerate a graft of rat cells. Despite their name (non-obese diabetic), these mice are not diabetic, and diabetes is induced by an injection of the toxin STZ, which destroys beta cells to generate a model of Type 1 diabetes.
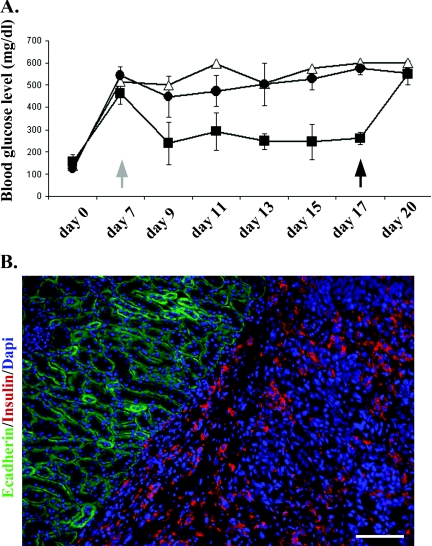
Once diabetes (blood glucose >400 mg/dl) was established, we transplanted cells under the kidney capsule and then monitored the mice every 2 days. The fall in blood glucose indicates a relief of the diabetes. Following this, the grafted kidneys were removed for analysis. The rise of blood glucose at this point indicates that the relief of diabetes was in fact due to the graft and not to a non-specific effect of surgery or to consequent effects on feeding behaviour. These experiments were terminated at 20 days because there is a risk of tumour formation from residual non-transformed B13 cells.
Figure 7(A) shows three mice that were successfully engrafted with Ad-PNM-treated cells. Immunostaining of kidney sections from the excised kidneys demonstrated the persistent presence of insulin-positive rat cells under the kidney capsule (Figure 7B). The controls consisted of one mouse receiving Ad-GFP-treated cells and three receiving Ad-PNM-treated cells, but where the graft did not take. The results were quantified by measuring the area under the curve for each mouse from days 9–17 inclusive. This gave mean values of 1285 (S.D.=426) for the active grafts, 2531 (S.D.=385) for the grafts that did not take, and 2780 (S.D.) for the diabetic control. Comparison of the active grafts with those that did not take showed a P value of 0.020. Comparison of the active grafts with diabetic control gave a P value of 0.026. By contrast, comparison of the grafts that did not take with the diabetic control showed no difference (P=0.379). This result indicates that the Ad-PNM-treated cells are capable of relieving diabetes in an animal model, even though they do not exhibit glucose-sensitive insulin secretion.
Figure 7. Amelioration of diabetes by Ad-PNM-transduced B13 cells.
(A) Mice were given STZ to induce diabetes at day 0. Cells were transplanted under the kidney capsule at day 7 (grey arrow). Blood glucose levels were then measured every 2 days. At day 17 the transplanted kidneys were removed and analysed for the presence of graft cells (black arrow). Δ, Ad-GFP-treated cells (control); ■, Ad-PNM-treated cells, n=3 and one mouse died after kidney removal; ●, Ad-PNM-treated but without successful engraftment, n=3. Results are means±S.E.M. (B) Explanted kidneys were stained for insulin (red), E-cadherin (green) and DAPI (blue). Scale bar=100 μm.
Transduction of exocrine pancreas in vivo
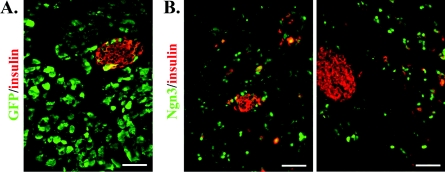
Ad-GFP or Ad-PNM were injected directly into the pancreas as described previously [6] and the results are shown in Figure 8. Ad-GFP gives a very high efficiency of transduction in exocrine tissue, with most cells expressing GFP protein. A small number of beta cells in the islets, visualized by immunostaining for insulin, were also transduced (Figure 8A). When a similar dose of Ad-PNM was given, a high level of transduction was detected by expression of the vector-encoded proteins. Figure 8(B) shows the expression of Ngn3, which is not normally found in mature exocrine or endocrine cells. The sections were stained for insulin and it was evident that a proportion of the scattered cells expressing Ngn3 were also expressing insulin. The insulin expression was not necessarily observed in those cells expressing the highest levels of Ngn3, rather it was more likely to be observed in those with intermediate levels. This experiment confirms that the basic observation of Zhou et al. [6] is correct. On the other hand it is difficult to establish for certain the phenotype of scattered cells in vivo. Consistent with the study of the B13 cells, the insulin-positive cells in vivo superficially resemble exocrine cells making insulin, rather than true beta cells.
Figure 8. Transduction of the exocrine pancreas in vivo.
(A) Injection of pancreas with Ad-GFP, visualized by GFP fluorescence, showing a high level of transduction in exocrine tissue. (B) Transduction with Ad-PNM, immunostained for Ngn3 (green) and insulin (red). Scale bars=100 μm.
DISCUSSION
The direct reprogramming of differentiated cells is of interest for two reasons. It is an important intellectual issue because it raises the question of whether the 200 or so visible cell types are the only possible stable states of vertebrate cells or whether numerous other states are potentially stable but are not accessed during normal development. This can be experimentally probed by studies in which developmental transcription factors are misexpressed in order to drive cells into a new developmental state. Because of the dense web of regulatory connections between developmental transcription factors, overexpression of a small number of factors can sometimes push the cell into a new state which involves a change in activity of thousands of genes.
In the case of transformation to a beta cell phenotype, direct reprogramming is also important from a practical point of view. The technique of islet cell transplantation has been practiced on a modest scale for the last 10 years and is used especially for cases of Type 1 diabetes showing unawareness of hypoglycaemia [17]. This is potentially life threatening as it can lead to sudden loss of consciousness. In the technique, islets from the pancreas of human cadaver donors are infused into the portal vein and lodge in the liver, where they can survive and function effectively. This is a successful type of cell transplantation therapy as the unawareness of hypoglycaemia resolves in virtually all patients and a proportion become insulin-independent for some years. However, the supply of cells is highly limited and there are many problems associated with the obligatory lifelong immunosuppression. In this context, the possibility of creating new beta cells from a tissue of an individual patient would be of great importance since it would resolve both the problem of cell supply and of alloimmunity.
We were motivated to conduct the present study following the publication by Zhou et al. [6]. This showed the induction of insulin-positive cells in the exocrine pancreas of mice following direct injection of an adenovirus encoding the three genes Pdx1, Ngn3 and MafA. As we show in Figure 8, there is no doubt that the procedure can induce ectopic insulin-positive cells, but it is less easy to prove that they are indeed beta cells with the full range of properties expected of this cell type.
In order to facilitate mechanistic studies we employed a well-known in vitro model of pancreatic exocrine cells, the cell line AR42j-B13. Our results show that there is a remarkable phenotypic change in these cells following the introduction of the three genes. They change morphology to a flat fibroblast-like shape, stop dividing, down-regulate the expression of a range of exocrine markers, express insulin from both the endogenous genes and display a number of characteristic transcription factors normally found in beta cells. However, the transformation is not complete, as the endogenous counterparts of the introduced genes are not up-regulated, and some components of the all-important glucose-sensing mechanism of beta cells are not induced. We showed this both from the lack of expression of the Slc2a2 (also known as Glut2) and Kcnj11 (also known as Kir-6.2) genes and from the lack of glucose-induced insulin secretion by these cells. Significantly, a previous study by Aldabbiat et al. [23], involving formation of insulin-positive cells from AR42j cells cultured on Matrigel and treated with growth factors, also reported no regulated insulin secretion.
During pancreatic development both exocrine and endocrine cells arise from the same endodermal cell population of the pancreatic bud [37,38]. It is possible that the endocrine and exocrine cells might share some aspects of chromatin state as a result of their developmental relatedness, and this may facilitate interconversion by overexpression of selected transcription factors. For example, in exocrine cells perhaps the genes characteristic of endocrine cells, while not expressed, are still accessible to transcription factors rather than being sequestered in inactive regions of chromatin. For this reason we made the study of the histone tail modifications and DNA base methylation of Pdx1 and the two insulin genes. In the case of Pdx1, which is a key regulatory gene in pancreatic development, the histone methylation studies did suggest an open configuration in the pancreatic, but not the non-pancreatic, cell types. Although our line of B13 cells does not express Pdx1, other lines of the original cells in use elsewhere have been reported to do so in their normal state [23,39]. With regard to the insulin genes, the results of the histone methylation and DNA methylation studies indicate that the introduction of the PNM factor combination itself modifies the chromatin to promote expression.
If cellular reprogramming to beta cells does eventually become a method for clinical use, it is likely to take the form of in vitro cell transformation followed by a graft similar to the current islet transplantation procedure. This is because direct introduction of genes into the pancreas is likely to pose risks of oncogenic transformations as seen in other types of gene therapy [40]. Thus it is necessary to show that the transformed cells can indeed cure diabetes following transformation. In our case we have shown that the Ad-PNM-treated B13 cells can cure STZ-induced diabetes. However, we have good evidence that the cells are not glucose-responsive, so their curative ability must derive from their constitutive insulin secretion. This is a reminder that the ability to rescue experimental diabetes in rodents is not a very discriminating characteristic. For treatment of human diabetes, cells must have the exact glucose responsiveness of real beta cells or they will not be significantly better than the present regimes of insulin injection. Whether beta cells are obtained by direct reprogramming or from other sources, such as directed differentiation of pluripotent stem cells [41], it will be necessary to carefully characterize the phenotype of the cells before proceeding.
Online data
AUTHOR CONTRIBUTION
Ersin Akinci did most of the experimental work and helped write the paper. Anannya Banga, Lucas Greder and James Dutton contributed to the experimental work. Jonathan Slack conceived and directed the work, and wrote the paper.
ACKNOWLEDGEMENTS
We thank Dr David Tosh (University of Bath, Bath, U.K.) for the AR42j-B13 cells, and Dr Sandeep Gupta and Dr Shunan Li for the IRPT cells. We thank Dr Nobuaki Kikyo for advice on the chromatin studies. This work was carried out by Ersin Akinci in part fulfilment of a PhD program at the University of Minnesota.
FUNDING
This work was supported by a Ministry of National Education of the Republic of Turkey Scholarship (to E.A.) and the National Institutes of Health [grant number R01DK080747 (to J.M.W.S.)].
References
- 1.Slack J. M. W. Essential Developmental Biology. Oxford: Blackwell Science; 2005. [Google Scholar]
- 2.Zhou Q., Melton D. A. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell-lines by forced expression of Myod. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen C. N., Slack J. M. W., Tosh D. Molecular basis of transdifferentiation of pancreas to liver. Nat. Cell Biol. 2000;2:879–887. doi: 10.1038/35046522. [DOI] [PubMed] [Google Scholar]
- 5.Xie H., Ye M., Feng R., Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ieda M., Fu J.-D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slack J. M. W. Homeotic transformations in Man: implications for the mechanism of embryonic development and for the organization of epithelia. J. Theor. Biol. 1985;114:463–490. doi: 10.1016/s0022-5193(85)80179-x. [DOI] [PubMed] [Google Scholar]
- 10.Slack J. M. W. Metaplasia and transdifferentiation: from pure biology to the clinic. Nat. Rev. Mol. Cell Biol. 2007;8:369–378. doi: 10.1038/nrm2146. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Nishikawa S.-I., Goldstein R. A., Nierras C. R. The promise of human induced pluripotent stem cells for research and therapy. Nat. Rev. Mol. Cell Biol. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- 13.Eberharter A., Becker P. B. Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep. 2002;31:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 15.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 16.Margueron R., Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paty B. W. Islet transplantation for type 1 diabetes: an overview. Paediatr. Child Health. 2005;10:38–40. doi: 10.1093/pch/10.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longnecker D. S., Lilja H. S., French J. I., Kuhlmann E., Noll W. Transplantation of azaserine-induced carcinomas of pancreas in rats. Cancer Lett. 1979;7:197–202. doi: 10.1016/s0304-3835(79)80080-4. [DOI] [PubMed] [Google Scholar]
- 19.Vila M. R., Lloreta J., Real F. X. Normal human pancreas cultures display functional ductal characteristics. Lab. Invest. 1994;71:423–431. [PubMed] [Google Scholar]
- 20.Rooman I., Heremans Y., Heimberg H., Bouwens L. Modulation of rat pancreatic acinoductal transdifferentiation and expression of PDX-1 in vitro. Diabetologia. 2000;43:907–914. doi: 10.1007/s001250051468. [DOI] [PubMed] [Google Scholar]
- 21.Pinho A. V., Rooman I., Reichert M., De Medts N., Bouwens L., Rustgi A. K., Real F. X. Adult pancreatic acinar cells dedifferentiate to an embryonic progenitor phenotype with concomitant activation of a senescence programme that is present in chronic pancreatitis. Gut. 2011;60:958–966. doi: 10.1136/gut.2010.225920. [DOI] [PubMed] [Google Scholar]
- 22.Mashima H., Shibata H., Mine T., Kojima I. Formation of insulin-producing cells from pancreatic acinar AR42J cells by hepatocyte growth factor. Endocrinology. 1996;137:3969–3976. doi: 10.1210/endo.137.9.8756573. [DOI] [PubMed] [Google Scholar]
- 23.Aldibbiat A., Marriott C. E., Scougall K. T., Campbell S. C., Huang G. C., Macfarlane W. M., Shaw J. A. M. Inability to process and store proinsulin in transdifferentiated pancreatic acinar cells lacking the regulated secretory pathway. J. Endocrinol. 2008;196:33–43. doi: 10.1677/JOE-07-0397. [DOI] [PubMed] [Google Scholar]
- 24.Guz Y., Montminy M. R., Stein R., Leonard J., Gamer L. W., Wright C. V. E., Teitleman G. Expression of murine STF-1, a putative insulin gene transcription factor, in cells of pancreas, duodenal epithelium and pancratic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 25.Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 26.Gradwohl G., Dierich A., LeMeur M., Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kataoka K., Han S. I., Shioda S., Hirai M., Nishizawa M., Handa H. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- 28.Dutton J. R., Daughters R. S., Chen Y., O'Neill K. E., Slack J. M. W. Use of adenovirus for ectopic gene expression in Xenopus. Dev. Dyn. 2009;238:1412–1421. doi: 10.1002/dvdy.21932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.German M., Ashcroft S., Docherty K., Edlund H., Edlund T., Goodison S., Imura H., Kennedy G., Madsen O., Melloul D., et al. The insulin gene promoter. Diabetes. 1995;44:1002–1004. doi: 10.2337/diab.44.8.1002. [DOI] [PubMed] [Google Scholar]
- 30.Campbell S. C., Macfarlane W. M. Regulation of the pdx1 gene promoter in pancreatic beta-cells. Biochem. Biophys. Res. Comm. 2002;299:277–284. doi: 10.1016/s0006-291x(02)02633-5. [DOI] [PubMed] [Google Scholar]
- 31.Gannon M., Gamer L. W., Wright C. V. E. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. Dev. Biol. 2001;238:185–201. doi: 10.1006/dbio.2001.0359. [DOI] [PubMed] [Google Scholar]
- 32.Iype T., Francis J., Garmey J. C., Schisler J. C., Nesher R., Weir G. C., Becker T. C., Newgard C. B., Griffen S. C., Mirmira R. G. Mechanism of insulin gene regulation by the pancreatic transcription factor Pdx-1: application of pre-mRNA analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes. J. Biol. Chem. 2005;280:16798–16807. doi: 10.1074/jbc.M414381200. [DOI] [PubMed] [Google Scholar]
- 33.Miyatsuka T., Matsuoka T.-A., Shiraiwa T., Yamamoto T., Kojima I., Kaneto H. Ptf1a and RBP-J cooperate in activating Pdx1 gene expression through binding to Area III. Biochem. Biophys. Res. Comm. 2007;362:905–909. doi: 10.1016/j.bbrc.2007.08.076. [DOI] [PubMed] [Google Scholar]
- 34.Li L. C., Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 35.de Felipe P. Skipping the co-expression problem: the new 2A ‘CHYSEL’ technology. Genet. Vaccines Ther. 2004;2:13. doi: 10.1186/1479-0556-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung S. M., Andersson T., Sonntag K. C., Bjorklund L., Isacson O., Kim K. S. Analysis of different promoter systems for efficient transgene expression in mouse embryonic stem cell lines. Stem Cells. 2002;20:139–145. doi: 10.1634/stemcells.20-2-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Percival A. C., Slack J. M. W. Analysis of pancreatic development using a cell lineage label. Exp. Cell Res. 1999;247:123–132. doi: 10.1006/excr.1998.4322. [DOI] [PubMed] [Google Scholar]
- 38.Gu G., Dubauskaite J., Melton D. A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from gut progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 39.Ogihara T., Fujitani Y., Uchida T., Kanno R., Choi J. B., Hirose T., Kawamori R., Watada H. Combined expression of transcription factors induces AR42J-B13 cells to differentiate into insulin-producing cells. Endocrine J. 2008;55:691–698. doi: 10.1507/endocrj.k07e-169. [DOI] [PubMed] [Google Scholar]
- 40.Flotte T. R. Gene therapy: the first two decades and the current state-of-the-art. J. Cell Physiol. 2007;213:301–305. doi: 10.1002/jcp.21173. [DOI] [PubMed] [Google Scholar]
- 41.Murry C. E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.