Abstract
Breast cancers are comprised of molecularly distinct subtypes that may respond differently to pathway-targeted therapies now under development. Collections of breast cancer cell lines mirror many of the molecular subtypes and pathways found in tumors, suggesting that treatment of cell lines with candidate therapeutic compounds can guide identification of associations between molecular subtypes, pathways, and drug response. In a test of 77 therapeutic compounds, nearly all drugs showed differential responses across these cell lines, and approximately one third showed subtype-, pathway-, and/or genomic aberration-specific responses. These observations suggest mechanisms of response and resistance and may inform efforts to develop molecular assays that predict clinical response.
Keywords: genomics, therapeutics, predictor
Genomic, epigenomic, transcriptional, and proteomic analyses of breast cancers reveal subtypes that may differ in pathway activity, progression, and response to therapy. In recognition of these variances, more than 800 small molecule and biological inhibitors are now under development for the treatment of human malignancies (1). These inhibitors vary in strength of subset and pathway specificity, with molecularly targeted experimental agents tending toward stronger specificity (2). To test experimental agents more efficiently, sensitivity-enrichment schemes are needed to identify potentially responsive patient subpopulations early during clinical development. Responsive subsets can be identified during the course of molecular marker-based clinical trials; however, this approach is expensive and does not allow early evaluation of experimental compounds in subpopulations most likely to respond. As a consequence, the probability is high that compounds that are very effective only in subpopulations of patients will be missed. An alternative approach is to identify candidate predictors of response by testing compounds in well-characterized preclinical models so that early clinical trials can be powered to detect responding subpopulations predicted by these studies. This approach would reduce development costs and increase the probability of finding drugs that may be particularly effective in subsets of patients.
Several studies support the utility of preclinical testing in cell-line panels for early and efficient identification of responsive molecular subtypes to guide early clinical trials. For example, cell-line panels predict lung cancers with EGFR mutations as responsive to gefitinib (3), breast cancers with human epidermal growth factor receptor 2/erythroblastic leukemia viral oncogene homolog 2 (HER2/ERBB2) amplification as responsive to trastuzumab (4) and/or lapatinib (5), and tumors with mutated or amplified BCR-ABL (breakpoint cluster region - c-abl oncogene 1, non-receptor tyrosine kinase) as sensitive to imatinib mesylate (6). The Discovery Therapeutic Program of the National Cancer Institute (NCI) has pursued this approach on a large scale, identifying associations between molecular features and responses to >100,000 compounds in a collection of ∼60 cancer cell lines (7). Although useful for detecting compounds with diverse responses, the NCI panel has limited power to detect subtype-specific responses because of the relatively sparse representation of specific cancers and subtypes thereof. We and others therefore have promoted the use of panels of well-characterized breast cancer cell lines for statistically robust identification of associations between in vitro therapeutic compound response and molecular subtypes and activated signaling pathways (4, 5). Here we report associations between quantitative proliferation measurements and molecular features defining subtypes and activated pathways for 77 Food and Drug Administration-approved and investigational compounds in a panel of ∼50 breast cancer cell lines. Approximately one third show aberration or subtype specificity. We also show via integrative analysis of gene expression and copy number data that some of the observed subtype-associated responses can be explained by specific pathway activities. The quantitative response and cell-line characterizations are available via the Stand Up to Cancer (SU2C)/University of California, Santa Cruz Cancer Genome Browser at https://genome-cancer.soe.ucsc.edu/.
Results
Cell Lines Model Many Important Tumor Subtypes and Features.
The utility of cell-line panels for identification of clinically relevant molecular predictors of response depends on the extent to which the diverse molecular mechanisms that determine response in tumors are operative in the cell lines. We reported previously on similarities between cell-line models and primary tumors at both the transcript and genome copy-number levels (4), and we refine that comparison here by using higher-resolution platforms and enhanced analysis techniques. Hierarchical consensus clustering of gene-expression profiles for 49 breast cancer cell lines and five nonmalignant breast cell lines shows that the cell-line collection models the luminal, basal, and claudin-low subtypes defined in primary tumor samples (SI Appendix, Fig. S1A) (8, 9). The basal and claudin-low subtypes map to the previously designated basal A and basal B subtypes, respectively (Dataset S1). A high-resolution SNP copy-number analysis (SI Appendix, Fig. S1B) confirms that the cell-line panel models regions of recurrent amplification at 8q24 [v-myc myelocytomatosis viral oncogene homolog (avian) (MYC)], 11q13 [cyclin D1 (CCND1)], 17q12 (ERBB2), 20q13 [serine/threonine kinase 15 (STK15)/aurora kinase A (AURKA)], and homozygous deletion at 9p21 [cyclin dependent kinase inhibitor 2A (CDKN2A)] that are found in primary tumors. Given the clinical relevance of the ERBB2 tumor subtype, we assigned cell lines with DNA amplification of ERBB2 to a special subtype designated “ERBB2AMP.”
Cell Lines Exhibit Differential Sensitivities to Most Therapeutic Compounds.
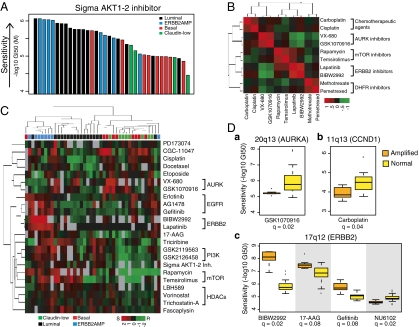
We quantified the sensitivity of our cell-line panel to 77 therapeutic compounds by measuring the concentration needed for each compound to inhibit proliferation by 50% (designated the GI50, where GI indicates “growth inhibition”) after 72 h of continuous exposure (Dataset S2). The anticancer compounds tested included both conventional cytotoxic agents (e.g., taxanes, platinum compounds, anthracylines) and targeted agents (e.g., hormonal and kinase inhibitors). Replicate analyses of the responses to several compounds showed that the median absolute deviation of −log10(GI50) values was 0.15 in log10 space (SI Appendix, Fig. S2). Waterfall plots showing responses to the 77 compounds are shown in SI Appendix. Most compounds showed high variation in response across the cell-line panel; however, three did not and were excluded from further analysis. The representative waterfall plot in Fig. 1A shows relatively higher efficacy in luminal and ERBB2AMP cells for the Sigma AKT1-2 inhibitor. Overall, responses to drugs with similar mechanisms and targets were highly correlated across the cell lines, as illustrated in Fig. 1B (also see Dataset S3).
Fig. 1.
Cell lines show a broad range of responses to therapeutic compounds. (A) Luminal and ERBB2AMP cell lines respond preferentially to AKT inhibition. Each bar represents the response of a single breast cancer cell line to the Sigma AKT1-2 inhibitor and is colored according to subtype. Cell lines are ordered by decreasing sensitivity [−log10(GI50)]. (B) Drug-response profiles for compounds with similar mechanisms and targets are highly correlated. Heatmap shows hierarchical clustering of pairwise correlations between responses of breast cancer cell lines treated with one of eight compounds. Red indicates positively correlated sensitivity across the panel of cell lines. Green indicates anticorrelated drug-response profiles. (C) Many compounds are significantly associated with subtype. Each column represents one cell line, and each row represents the median-centered −log10(GI50) for a particular compound. Both rows and columns are clustered hierarchically. Red represents sensitivity, green represents resistance, and gray represents missing values. Colored boxes below the dendogram identify sample subtype. Overall, cell lines of similar subtype tend to cluster together, as do compounds with similar targets or mechanisms. (D) CNAs are associated with compound response. Boxplots show distribution of response sensitivity for cell lines with amplified (A) and normal (N) copy number at the noted genomic locus. (a) 20q13 (STK15/AURKA) amplification is associated with GSK1070916 response (A = 7; N = 26 samples). (b) Amplification at 11q13 (CCND1) is associated with response to carboplatin (A = 9; N = 28 samples). (c) 17q12 (ERBB2) amplification is associated with sensitivity to BIBW2992 (A = 6; N = 19 samples), 17-AAG (A = 7; N = 27 samples), and gefitinib (A = 7; N = 18 samples), as well as resistance to NU6102 (A = 6; N = 21 samples).
Many Compounds Were Preferentially Effective in Subsets of the Cell Lines.
We assessed response–subtype associations with nonparametric Kruskal–Wallis ANOVAs to compare −log10(GI50) values across the clinically relevant transcriptional and genomic subtypes. We tested three comparisons: (i) luminal vs. basal vs. claudin-low; (ii) luminal vs. basal and claudin-low; and (iii) ERBB2AMP vs. non-ERBB2AMP.
Overall, 23 of 74 compounds showed transcriptional subtype-specific responses [Benjamini–Hochberg false discovery rate (q) <0.1] (Table 1 and Dataset S4). Although doubling times (DT) vary according to subtype, these subtype associations cannot be explained by differences in DT (SI Appendix, Fig. S3). Fig. 1C shows a hierarchical clustering of the quantitative responses of 23 agents with significant subtype associations. The 10 agents most strongly associated with subtype include inhibitors of receptor tyrosine kinase signaling and histone deacetylase. Several of these compounds showed preferential sensitivity in both the luminal and ERBB2AMP cell lines [e.g., lapatinib and GSK2126458 (pan phosphoinositide-3-kinase, catalytic; PIK3C inhibitor)], although the degree of specificity could vary. For instance, lapatinib was associated more strongly with the ERBB2AMP than with the luminal subtype, whereas the opposite pattern was observed for GSK2126458. Other compounds were specific for only one of these subtypes, as exemplified by the preferential sensitivity of luminal cells to vorinostat. The ERBB2AMP cell lines alone were preferentially responsive to AG1478, BIBW2992, and gefitinib, all of which target EGFR and/or ERBB2. VX-680 (AURK A/B/C inhibitor) was negatively associated with ERBB2 amplification. Etoposide, docetaxel, and cisplatin showed preferential activity in basal or claudin-low cell lines, as observed clinically (10, 11). Agents targeting the mitotic apparatus, including GSK1070916 (AURK B/C inhibitor), also were more active against basal and/or claudin-low cell lines. Interestingly, some compounds targeting the mitotic apparatus (e.g., ispinesib and GSK923295) showed no significant subtype specificity, indicating the complexity of signaling through this network.
Table 1.
Therapeutic compounds that show significant subtype specificity
| Compound | Target | Basal vs. claudin-low vs. luminal | Basal and claudin-low vs. luminal | ERBB2AMP vs. not ERBB2AMP | Subtype specificity |
| Lapatinib | EGFR, ERBB2 | 7.23E-02 | 3.34E-02 | 2.26E-06 | Luminal/ERBB2AMP |
| Sigma AKT1-2 inh. | AKT1, AKT2 | 1.17E-03 | 2.63E-04 | 1.29E-01 | Luminal |
| GSK2126458 | PIK3C A/B/D/G | 1.27E-03 | 1.27E-03 | 8.67E-02 | Luminal/ERBB2AMP |
| Gefitinib | EGFR | 4.89E-01 | 3.35E-01 | 4.14E-03 | ERBB2AMP |
| BIBW 2992 | EGFR, ERBB2 | 6.93E-01 | 8.08E-01 | 6.39E-03 | ERBB2AMP |
| GSK2119563 | PIK3CA | 2.85E-02 | 8.11E-03 | 8.67E-02 | Luminal/ERBB2AMP |
| Rapamycin | MTOR | 1.45E-02 | 8.11E-03 | 3.84E-01 | Luminal |
| AG1478 | EGFR | 9.34E-01 | 9.34E-01 | 2.60E-02 | ERBB2AMP |
| Etoposide | TOP2A | 3.34E-02 | 5.13E-02 | 8.89E-01 | Claudin-low |
| LBH589 | HDAC | 5.14E-02 | 3.34E-02 | 3.22E-01 | Luminal |
| Vorinostat | HDAC | 7.23E-02 | 3.34E-02 | 6.89E-01 | Luminal |
| Cisplatin | DNA cross-linker | 8.45E-02 | 4.31E-02 | 8.52E-01 | Basal/Claudin-low |
| Fascaplysin | CDK4 | 4.83E-02 | 4.31E-02 | 3.70E-01 | Luminal |
| Docetaxel | TUBB1, BCL2 | 8.67E-02 | 4.83E-02 | 8.44E-01 | Basal/Claudin-low |
| GSK1070916 | AURK B/C | 5.13E-02 | 4.83E-02 | 4.82E-01 | Claudin-low |
| PD173074 | FGFR3 | 5.13E-02 | 3.68E-01 | 5.06E-01 | Claudin-low |
| Trichostatin A | HDAC | 1.22E-01 | 5.13E-02 | 7.10E-01 | Luminal |
| Triciribine | AKT, ZNF217 | 8.67E-02 | 5.91E-02 | 3.56E-01 | Luminal |
| CGC-11047 | Polyamine analog | 6.51E-02 | 1.25E-01 | 8.08E-01 | Basal |
| Temsirolimus | MTOR | 1.64E-01 | 7.25E-02 | 1.29E-01 | Luminal |
| VX-680 | AURK A/B/C | 2.95E-01 | 4.02E-01 | 7.77E-02 | not ERBB2AMP |
| 17-AAG | HSP90AA1 | 1.83E-01 | 1.10E-01 | 8.67E-02 | ERBB2AMP |
| Erlotinib | EGFR | 9.48E-02 | 2.83E-01 | 2.33E-01 | Basal |
Each column represents q-values for one ANOVA. Compounds are ranked by the minimum q-value achieved across the three tests. BCL2, B-cell CLL/lymphoma 2; CDK4, cyclin-dependent kinase 4; HDAC, histone deacetylase; MTOR, mechanistic target of rapamycin; TOP2A, topoisomerase (DNA) II alpha 170kDa; TUBB1, tubulin β1; ZNF217, zinc finger protein 217.
Fig. 1D shows responses for six compounds that were significantly associated with recurrent focal, high-level copy-number aberrations (CNAs) (t-test, q ≤ 0.1). A χ2 test showed only nonsignificant trends in transcriptional subtype associations for the CNAs. Amplification at 20q13, encoding AURKA was associated with resistance to the AURK B/C inhibitor GSK1070916 (12). This resistance suggests that amplification of AURKA provides a bypass mechanism for AURK B/C inhibitors. Amplification at 11q13, encoding CCND1, was associated with resistance to carboplatin. CCND1 is a G1/S cell-cycle checkpoint gene that monitors for unrepaired DNA damage, and whose overexpression is known to be associated with cisplatin resistance in other tumor types (13, 14). Amplification at 17q12 (ERBB2) was associated with sensitivity to BIBW2992 and gefitinib, inhibitors of ERBB2 and/or EGFR, as well as 17-AAG [heat shock protein 90kDa alpha (cytosolic), class A member 1 (HSP90AA1) inhibitor]. 17q12 amplification also was associated with resistance to the cyclin-dependent kinase 1 (CDK1)/CCNB1 inhibitor, NU6102, perhaps reflecting the fact that ERBB2 negatively regulates CDK1 (15, 16), thereby diminishing the impact of the CDK1 inhibitor.
Integration of Copy-Number and Transcription Measurements Identifies Biologically Relevant SuperPathways.
We used the network analysis tool PARADIGM (17) to identify pathway-based mechanisms that underlie subtype-specific responses. PARADIGM uses copy number and transcription data to calculate integrated pathway levels (IPLs) for 1,441 curated signal transduction, transcriptional, and metabolic pathways (18). We compared IPLs for cell lines and primary breast tumors using data from The Cancer Genome Atlas (TCGA) project (http://cancergenome.nih.gov) and found a general concordance between transcriptional subtype and pathway activity across the two cohorts (SI Appendix, Figs. S4 and S5 and Dataset S5). This subtype-specific pathway activity likely explains much of the observed subtype specific responses.
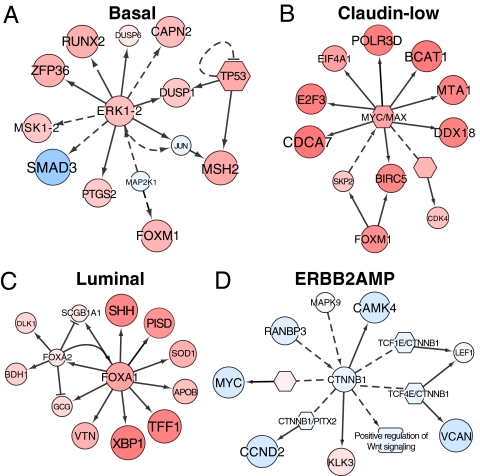
Mechanistic interpretation of IPLs for 1,441 pathways is complicated by the overlapping elements in many of the curated pathways. We overcame this complication by merging the 1,441 curated pathways into a single “SuperPathway” in which redundant pathway elements are eliminated. This approach enabled us to identify SuperPathway subnets that differed in activity between transcriptional subtypes (SI Appendix, Fig. S6). As an example, comparison of subnet activities between basal cell lines and all others in the collection identified a network comprised of 1,104 nodes (e.g., proteins, protein complexes, or cellular processes) connected by 1,242 edges (e.g., protein–protein interactions) between these elements. Several subnetworks were up- or down-regulated in the SuperPathway networks. Fig. 2A, for example, shows up-regulation of an ERK1/2 subnetwork controlling cell cycle, adhesion, invasion, and macrophage activation (19). The forkhead box M1 and DNA-damage subnetworks also were up-regulated markedly in the basal cell lines. The claudin-low network showed up-regulation of many of the same subnetworks, as well as up-regulation of a MYC/Myc-associated factor X (MAX) subnetwork (Fig. 2B) associated with metabolism, proliferation, angiogenesis, and oncogenesis (20). Comparison of the luminal cell lines with all others showed down-regulation of an activating transcription factor 2 network, which inhibits tumorigenicity in melanoma (21), as well as up-regulation of forkhead box A1 (FOXA1)/forkhead box A2 (FOXA2) networks that control transcription of estrogen receptor-regulated genes (Fig. 2C) and are associated with good-prognosis luminal breast cancers (22, 23). ERBB2AMP subnetworks were similar to those for luminal cells; this similarity is not surprising because most ERBB2AMP cells also can be classified as luminal. However, Fig. 2D shows down-regulation of a β-catenin (CTNNB1) network in ERBB2AMP cell lines; up-regulation of this network has been implicated in tumorigenesis and is associated with poor prognosis (24, 25).
Fig. 2.
Cell-line subtypes have unique SuperPathway network features. In all panels, each node represents a pathway “concept” corresponding to a protein (circle), a multimeric complex (hexagon), or an abstract cellular process (square). Node sizes are drawn in proportion to the DA score; larger nodes correspond to concepts more correlated with a particular subtype than with all other subtypes. Color indicates whether the concept is correlated positively (red) or negatively (blue) with the subtype of interest. Lines represent interactions, including protein–protein interactions (dashed lines) and transcriptional interactions (solid lines). Interactions are included if they connect concepts whose absolute level of DA is higher than the mean absolute level. Labels on some nodes are omitted for clarity. (A) An ERK1/2 subnet preferentially activated in basal breast cancer cell lines. (B) A MYC/MAX network activated in claudin-low cell lines. (C) A FOXA1/FOXA2 network up-regulated in the luminal subtype. (D) A CTNNB1 subnet down-regulated in the ERBB2AMP subtype. APOB, apolipoprotein B [including Ag(x) antigen]; BCAT1, branched chain amino-acid transaminase 1, cytosolic; BDH1, 3-hydroxybutyrate dehydrogenase, type 1; BIRC5, baculoviral IAP repeat containing 5; CAMK4, calcium/calmodulin-dependent protein kinase IV; CAPN2, calpain 2, (m/II) large subunit; CCND2, cyclin D2; CDCA7, cell division cycle associated 7; DDX18, DEAD (Asp-Glu-Ala-Asp) box polypeptide 18; DLK1, delta-like 1 homolog (Drosophila); DUSP1, dual specificity phosphatase 1; DUSP6, dual specificity phosphatase 6; E2F3, E2F transcription factor 3; EIF4A1, eukaryotic translation initiation factor 4A1; ERK1-2, mitogen-activated protein kinase 3-1; GCG, glucagon; JUN, jun proto-oncogene; KLK3, kallikrein-related peptidase 3; LEF1, lymphoid enhancer-binding factor 1; MAP2K1, mitogen-activated protein kinase kinase 1; MAPK9, mitogen-activated protein kinase 9; MSH2, mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli); MSK1-2, ribosomal protein S6 kinase, 90kDa, polypeptide 5; MTA1, metastasis associated 1; PISD, phosphatidylserine decarboxylase; PITX2, paired-like homeodomain 2; POLR3D, polymerase (RNA) III (DNA directed) polypeptide D, 44kDa; PTGS2, prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase); RANBP3, RAN binding protein 3; RUNX2, runt-related transcription factor 2; SCGB1A1, secretoglobin, family 1A, member 1 (uteroglobin); SKP2, S-phase kinase-associated protein 2 (p45); SMAD3, SMAD family member 3; SOD1, superoxide dismutase 1, soluble; SSH, slingshot homolog; TCF1E, HNF homeobox A; TCF4E, transcription factor 4; TFF1, trefoil factor 1; TP53, tumor protein p53; VCAN, versican; VTN, vitronectin; XBP1, X-box binding protein 1; ZFP36, zinc finger protein 36, C3H type, homolog (mouse).
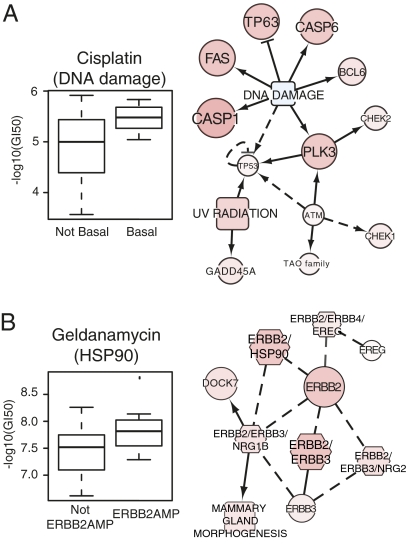
SuperPathway analysis of differential drug response among the cell lines also revealed subnet activities that provide information about mechanisms of response. For example, basal cell line sensitivity to the DNA-damaging agent cisplatin was associated with up-regulation of a DNA-damage response subnetwork that includes ataxia telangiectasia mutated and checkpoint kinase 1 homolog, key genes associated with response to cisplatin (Fig. 3A) (26). Likewise, ERBB2AMP cell line sensitivity to geldanamycin [an inhibitor of heat-shock protein 90 (HSP90)] was associated with up-regulation of an ERBB2-HSP90 subnetwork (Fig. 3B). This observation is consistent with the known ERBB2 degradation induced by geldanamycin binding (27, 28).
Fig. 3.
Pathway diagrams can be used to predict response to therapies. (A) (Left) Basal breast cancer cell lines respond preferentially to the DNA-damaging agent cisplatin. Each boxplot represents the distribution of drug response data for basal (right) and non-basal (left) cell lines. (Right) Basal cell lines show enhanced pathway levels in a subnetwork associated with the DNA-damage response, providing a possible mechanism by which cisplatin acts in these cell lines. (B) (Left) ERBB2AMP cell lines are sensitive to the HSP90 inhibitor geldanamycin. (Right) The ERBB2–HSP90 network is up-regulated in ERBBP2AMP cell lines. Conventions are as in Fig. 2. BCL6, B-cell CLL/lymphoma 6; CASP1, caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase); CASP6, caspase 6, apoptosis-related cysteine peptidase; CHEK2, CHK2 checkpoint homolog (S. pombe); DOCK7, dedicator of cytokinesis 7; ERBB3, v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian); ERBB4, v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian); EREG, epiregulin; FAS, Fas (TNF receptor superfamily, member 6); GADD45A, growth arrest and DNA-damage-inducible, alpha; NRG1B, neuregulin 1; NRG2, neuregulin 2; PLK3, polo-like kinase 3; TP53, tumor protein p53; TP63, tumor protein p63.
Discussion
Efforts to personalize breast cancer treatment are aimed at identifying subsets of patients most likely to benefit from treatment and avoiding treatment-associated morbidity and mortality in patients who are unlikely to respond. We have supported this effort by testing 77 therapeutic compounds in an in vitro cell-line panel and have shown that approximately one third are preferentially effective in one or more transcriptional or genomic breast cancer subtypes. We also have shown that integration of the transcriptional and genomic data for the cell lines reveals SuperPathway subnetworks that provide mechanistic information about the observed subtype-specific responses. Comparative analysis of pathways between cell lines and tumors shows that the majority of subtype-specific subnetworks are conserved between cell lines and tumors. This similarity is important, given the very different environments between a cell line growing in a standard 2D culture and a primary or metastatic tumor, and supports the clinical relevance of the in vitro studies.
The potential clinical utility of these findings is supported by concordance of in vitro-derived molecular predictors of response to therapeutic compounds and clinical results. For example, ERBB2-amplified cell lines are preferentially sensitive to ERBB2-targeted agents, and basal subtype cell lines are preferentially sensitive to platinum salts, as observed clinically. That said, additional work remains before the signatures reported in this study can be used to select patients for clinical trials. Such future work would include the development of robust and reliable molecular assays that can be applied to clinical samples, establishment of predictive algorithms with decision-making thresholds optimized for clinical use, and validation of predictive power in multiple independent studies. To initiate this process, we suggest that the response-associated signatures identified in this study be developed into standardized assays that can be assessed for clinical predictive power in early-stage clinical trials and used to design trials that are properly powered to detect the responses in the clinical subsets predicted by the in vitro studies. Assays that show positive predictive power in early clinical trials then can be “locked down” and tested for predictive power in follow-on clinical trials.
We anticipate that the power of this in vitro systems approach will increase as additional molecular features, including mutations, methylation, and alternative splicing, are included in the analysis. In addition, expanding the cell-line panel will increase the power to identify low-frequency molecular patterns and to develop robust predictive models. Most important, however, is iterative refinement of the in vitro assay system based on lessons learned by comparing in vitro predictions with clinical reality.
Methods
Unless stated otherwise, analyses were performed in R (http://www.r-project.org).
Cell Proliferation Assay and Growth Rate.
The efficacy of 77 compounds in ∼50 breast cancer cell lines was assessed as described previously (29). Briefly, cells were treated in triplicate for 72 h with nine doses of each compound in 1:5 serial dilution. Cell proliferation was estimated using the Cell Titer-Glo assay (Promega). DT was estimated from the ratio of 72 h to 0 h for untreated wells. A Gompertz curve was fit to the dose-response data using a nonlinear least squares procedure with the following parameters: upper and lower asymptotes, slope, and inflection point. The fitted curve was transformed into a GI curve as described previously (30, 31).
Several quantitative response metrics were estimated including the GI50, the concentration needed to inhibit proliferation completely (TGI), and the concentration needed to reduce the population by 50% (LC50). When the underlying proliferation data were of high quality but the end point response (GI50, TGI, or LC50) was not reached, the values were set to the highest concentration tested. The drug-response data were filtered so that (i) the median SD across the nine triplicate-treated data points was <0.20; (ii) the DT was within 2 SD of the median DT for each cell line; (iii) the slope of the fitted curve was >0.25; (iv) we identified assays with no response by requiring inhibition at the maximum concentration <50%. Approximately 80% of all assays passed all filtering requirements.
Compound and Cell-line Screening.
Compounds were excluded from analysis if (i) more than 40% of GI50 values were missing across the set of cell lines and/or (ii) the GI50 was not >1.5 times the median GI50 for a given drug (mGI50) or <0.5 mGI50 for least three cell lines. Nonmalignant cell lines were not included in the molecular-response assessments.
SNP Array and DNA Copy-Number Analysis.
Genome copy number was assessed using the Affymetrix Genome-Wide Human SNP Array 6.0 analysis platform. Arrays were analyzed using aroma.affymetrix (http://aroma-project.org) (32), data were normalized as described (33), and DNA copy-number ratios at each locus were estimated relative to a set of 20 normal sample arrays. Data were segmented using circular binary segmentation from the bioconductor package DNAcopy (34). Significant DNA copy-number changes were analyzed using MATLAB-based Genomic Identification of Significant Targets in Cancer (GISTIC) (35). Nonmalignant cell lines were not included in the GISTIC analysis. GISTIC scores for one member of each isogenic cell-line pair were used to infer genomic changes in the other (Dataset S1). Raw data are available in The European Genotype Archive (accession no. EGAS00000000059).
Exon Array Analysis.
Gene expression was assessed using the Affymetrix GeneChip Human Gene 1.0 ST exon array platform. Gene-level summaries of expression were computed using aroma.affymetrix (33) with quantile normalization and a log-additive probe-level model based on the HuEx-1_0-st-v2,DCCg,Spring2008 chip definition file (CDF). The raw data are available in ArrayExpress (E-MTAB-181).
Consensus Clustering.
Cell-line subtypes were identified using hierarchical consensus clustering (36) of genes with an SD >1.0 on the log2 scale across all cell lines. Consensus was computed using 500 samplings of the cell lines, 80% of the cell lines per sample, agglomerative hierarchical clustering, Euclidean distance metric, and average linkage.
Associations of Subtypes and Response to Therapeutic Agents.
Associations between drug response and subtype were assessed for (i) luminal vs. basal vs. claudin-low; (ii) luminal vs. basal and claudin-low; and (iii) ERBB2AMP vs. non–ERBB2AMP. Differences between −log10(GI50) of the groups were compared with a nonparametric Kruskall–Wallis ANOVA. The P values for the three sets of tests were combined, and the Benjamini–Hochberg false discovery rate (q-value) was used to correct for multiple testing. For the three-sample test, the most sensitive group was identified by performing a post hoc analysis on the significant compounds in which we compared each group with all others. The P values for the post hoc test were adjusted together. In all cases, q <0.10 was deemed significant. If the basal and claudin-low group was significant in scheme ii, but only one of these groups was significant in scheme i, precedence was given to the three-sample case when assigning class specificity. No minimum difference in medians was required.
Association of Genomic Changes and Response to Therapeutic Agents.
A t test was used to assess the association between recurrent copy number changes at 9p21, 11q13, 17q12, and 20q13, as identified in the GISTIC analysis, and drug response. Cell lines with low or no amplification were combined into a single group and compared with cell lines with high amplification. A similar analysis was performed for regions of deletion. Cell lines for which the GI50 was equal to the maximum concentration tested were omitted from analysis (e.g., after censoring lapatinib, there were only two samples in the amplified copy number group for 17q12; Dataset S6). Compounds were omitted if the distribution deviated greatly from normality, as assessed by a quartile–quartile plot. The complete set of P values was adjusted for multiple comparisons, and q < 0.10 was deemed significant.
Integrated Pathway Analysis.
Copy number, gene expression, and pathway interaction data were integrated using the PARADIGM software (17). This procedure infers IPLs for genes, complexes, and processes using pathway interactions and genomic and functional genomic data from a single cell line or patient sample. See SI Appendix for details.
TCGA and Cell-line Clustering.
Pathway activities inferred for the cell lines were combined with activities inferred for TCGA tumor samples (Dataset S7) and analyzed using hierarchical clustering. Cell lines and tumor samples were clustered using a set of 2,351 nonredundant activities determined by a correlation analysis to avoid biases caused by highly connected hub genes and highly correlated activities. The degree to which cell lines clustered with tumor samples of the same subtype was calculated using a Kolmogorov–Smirnov test to compare a distribution of t statistics calculated from correlations between pairs of cell lines and tumor samples of the same subtype with a distribution calculated from cell line pairs of different subtypes. See SI Appendix for details.
Identification of Subtype Pathway Markers.
Interconnected genes that collectively showed differential IPLs with respect to subtype were identified by treating each subtype as a dichotomization of the cell lines into a group containing the subtype of interest and a group containing the remaining cell lines. The R implementation of the two-class significance analysis of microarrays algorithm (37) was used to compute a differential activity (DA) score for each concept in the SuperPathway. For subtypes, positive DA corresponds to higher activity in the subtype compared with the other cell lines.
The coordinated up- and down-regulation of closely connected genes in the SuperPathway reinforced the activities inferred by PARADIGM. Entire subnetworks with high DA scores were expected if the activities of neighboring genes also were correlated with a particular phenotype. Regions in the SuperPathway were identified in which concepts of high absolute DA were interconnected by retaining only links that connected two concepts that both had DA scores higher than the average absolute DA.
Supplementary Material
Acknowledgments
This work was supported by the Director, Office of Science, Office of Biological and Environmental Research, of the US Department of Energy under Contract DE-AC02-05CH11231; by the National Institutes of Health, National Cancer Institute Grants P50 CA058207 (to J.W.G.); U54 CA112970 (to J.W.G.); NHGRI U24 CA126551 (to P.T.S.), K08CA137153 (to E.A.C.), DOD BC087768 (to A.S.), and by Stand Up to Cancer-American Association for Cancer Research Dream Team Translational Cancer Research Grant SU2C-AACR-DT0409 (to J.W.G.). Additionally, this work was supported by contracts with the following companies: SmithKline Beecham Corporation (LB06002417); Millennium Pharmaceuticals, Inc. (LB09005492); Cytokinetics, Inc. (LB08004488); Celgate, Inc. (LB07003395); and Progen Pharmaceuticals Ltd. (LB08005005).
Footnotes
Conflict of interest statement: K.W.W. is an employee at Cytokinetics. P.G.S. is an employee at Millenium Pharmaceuticals. L.T.V. is an employee at Hoffman-LaRoche. J.G., K.E.B. M.A.H. and R.F.W., are employees and stockholders of GlaxoSmithKline. L.J.M. is an employee at Progen.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
Data deposition: Genome data has been deposited at the European Genome-Phenome Archive (EGA), http://www.ebi.ac.uk/ega/, hosted at the EBI (accession no.EGAS00000000059). Gene expression data for the cell lines were derived from Affymetrix GeneChip Human Gene 1.0 ST exon arrays. Raw data are available in ArrayExpress (E-MTAB-181).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018854108/-/DCSupplemental.
References
- 1.Anonymous New Medicines Database | PHRMA. 2010 Available at http://newmeds.phrma.org/. Accessed September, 2011. [Google Scholar]
- 2.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konecny GE, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66:1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 6.Scappini B, et al. Changes associated with the development of resistance to imatinib (STI571) in two leukemia cell lines expressing p210 Bcr/Abl protein. Cancer. 2004;100:1459–1471. doi: 10.1002/cncr.20131. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein JN. Spotlight on molecular profiling: “Integromic” analysis of the NCI-60 cancer cell lines. Mol Cancer Ther. 2006;5:2601–2605. doi: 10.1158/1535-7163.MCT-06-0640. [DOI] [PubMed] [Google Scholar]
- 8.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76; 1–17. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey LA, et al. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 11.Silver DP, et al. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardwicke MA, et al. GSK1070916, a potent Aurora B/C kinase inhibitor with broad antitumor activity in tissue culture cells and human tumor xenograft models. Mol Cancer Ther. 2009;8:1808–1817. doi: 10.1158/1535-7163.MCT-09-0041. [DOI] [PubMed] [Google Scholar]
- 13.Nakashima T, et al. The effect of cyclin D1 overexpression in human head and neck cancer cells. Eur Arch Otorhinolaryngol. 2005;262:379–383. doi: 10.1007/s00405-004-0831-z. [DOI] [PubMed] [Google Scholar]
- 14.Huerta S, et al. Gene expression profile of metastatic colon cancer cells resistant to cisplatin-induced apoptosis. Int J Oncol. 2003;22:663–670. [PubMed] [Google Scholar]
- 15.Yu D, et al. Overexpression of ErbB2 blocks Taxol-induced apoptosis by upregulation of p21Cip1, which inhibits p34Cdc2 kinase. Mol Cell. 1998;2:581–591. doi: 10.1016/s1097-2765(00)80157-4. [DOI] [PubMed] [Google Scholar]
- 16.Tan M, et al. Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to taxol-induced apoptosis. Mol Cell. 2002;9:993–1004. doi: 10.1016/s1097-2765(02)00510-5. [DOI] [PubMed] [Google Scholar]
- 17.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristensen VN, et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc Natl Acad Sci USA. 2012;109:2802–2807. doi: 10.1073/pnas.1108781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 20.Hynes NE, Stoelzle T. Key signalling nodes in mammary gland development and cancer: Myc. Breast Cancer Res. 2009;11:210–219. doi: 10.1186/bcr2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhoumik A, Jones N, Ronai Z. Transcriptional switch by activating transcription factor 2-derived peptide sensitizes melanoma cells to apoptosis and inhibits their tumorigenicity. Proc Natl Acad Sci USA. 2004;101:4222–4227. doi: 10.1073/pnas.0400195101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habashy HO, et al. Forkhead-box A1 (FOXA1) expression in breast cancer and its prognostic significance. Eur J Cancer. 2008;44:1541–1551. doi: 10.1016/j.ejca.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Badve S, et al. FOXA1 expression in breast cancer—correlation with luminal subtype A and survival. Clin Cancer Res. 2007;13:4415–4421. doi: 10.1158/1078-0432.CCR-07-0122. [DOI] [PubMed] [Google Scholar]
- 24.Michaelson JS, Leder P. beta-catenin is a downstream effector of Wnt-mediated tumorigenesis in the mammary gland. Oncogene. 2001;20:5093–5099. doi: 10.1038/sj.onc.1204586. [DOI] [PubMed] [Google Scholar]
- 25.Lin SY, et al. Beta-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 27.Blagosklonny MV. Hsp-90-associated oncoproteins: Multiple targets of geldanamycin and its analogs. Leukemia. 2002;16:455–462. doi: 10.1038/sj.leu.2402415. [DOI] [PubMed] [Google Scholar]
- 28.Baselga J, Swain SM. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 29.Kuo WL, et al. A systems analysis of the chemosensitivity of breast cancer cells to the polyamine analogue PG-11047. BMC Med. 2009;7:77–88. doi: 10.1186/1741-7015-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Cancer Institute Screening Services - NCI-60 DTP Human Tumor Cell Line Screen. 2011 Available at http://dtp.nci.nih.gov/branches/btb/ivclsp.html. Accessed September 2011. [Google Scholar]
- 31.Monks A, et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst. 1991;83:757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 32.Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–767. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 33.Bengtsson H, Wirapati P, Speed TP. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics. 2009;25:2149–2156. doi: 10.1093/bioinformatics/btp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 35.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monti S, Tamayo P, Mesirov JP, Golub TA. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52:91–118. [Google Scholar]
- 37.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.