Abstract
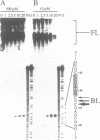
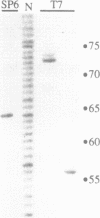
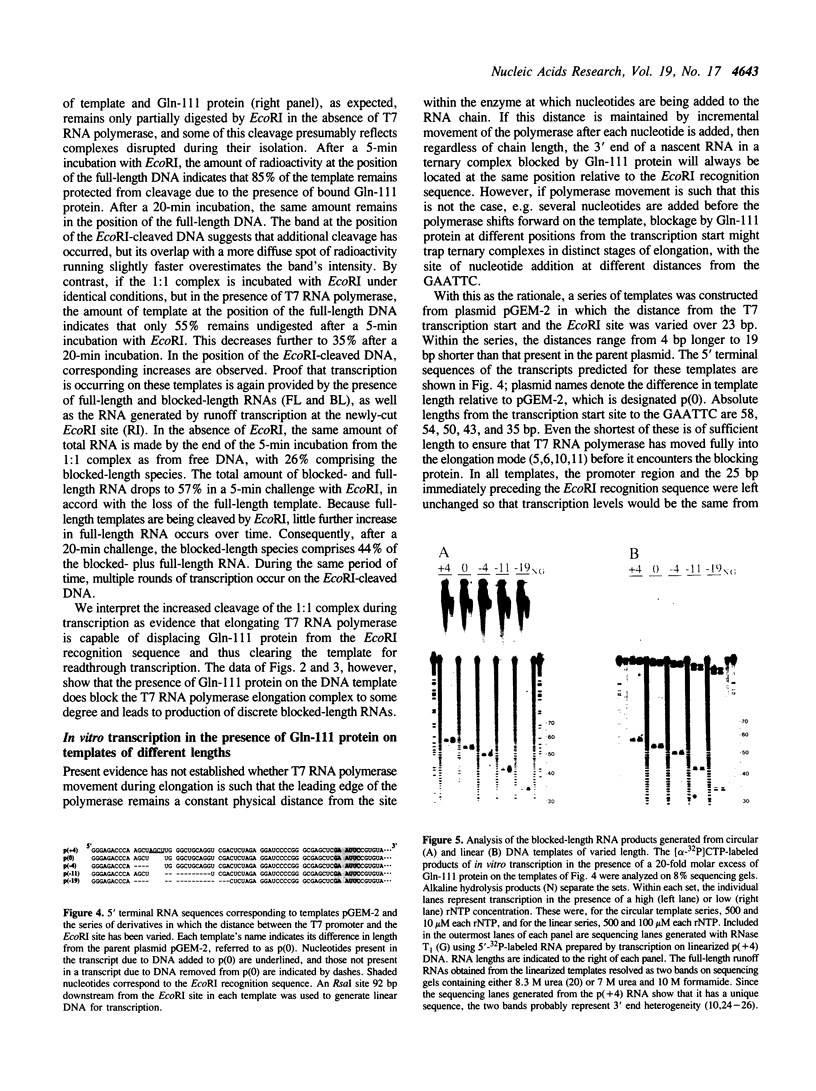
As a means of generating homogeneous populations of elongation complexes with the RNA polymerases encoded by phages T7 and SP6, transcription has been carried out in vitro on templates associated with the Gln-111 mutant of EcoRI endonuclease. The Gln-111 protein, as a result of a single amino acid substitution at position 111, lacks cleavage function yet shows higher than wild-type affinity for the EcoRI recognition sequence GAATTC. On a series of linear and circular templates associated with Gln-111 protein, blockage of the phage RNA polymerase elongation complex is observed. The 3' endpoint of the major blocked-length RNA species, just 3 bp upstream from the GAATTC, reveals an extremely close approach of polymerase's leading edge to essential contacts between Gln-111 protein and its binding site. In contrast to E. coli RNA polymerase, which is blocked stably and quantitatively by Gln-111 protein (Pavco, P.A. and Steege, D. A. (1990) J. Biol. Chem. 265, 9960-9969), the phage polymerases show substantial levels of readthrough transcription beyond the protein block.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. II. Inhibitors of the enzyme and their application to the study of the enzymatic reaction. J Biol Chem. 1973 Mar 25;248(6):2245–2250. [PubMed] [Google Scholar]
- Christianson T. W., Clayton D. A. A tridecamer DNA sequence supports human mitochondrial RNA 3'-end formation in vitro. Mol Cell Biol. 1988 Oct;8(10):4502–4509. doi: 10.1128/mcb.8.10.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S., Manley J. L. RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol Cell Biol. 1989 Nov;9(11):5254–5259. doi: 10.1128/mcb.9.11.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle U., Gentz R., Bujard H. lac Repressor blocks transcribing RNA polymerase and terminates transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4134–4137. doi: 10.1073/pnas.83.12.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubendorff J. W., Studier F. W. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J Mol Biol. 1991 May 5;219(1):45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano T. J., Deuschle U., Bujard H., McAllister W. T. Regulation of coliphage T3 and T7 RNA polymerases by the lac repressor-operator system. Gene. 1989 Dec 14;84(2):209–219. doi: 10.1016/0378-1119(89)90494-0. [DOI] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J Biol Chem. 1974 May 10;249(9):2858–2863. [PubMed] [Google Scholar]
- Gunderson S. I., Chapman K. A., Burgess R. R. Interactions of T7 RNA polymerase with T7 late promoters measured by footprinting with methidiumpropyl-EDTA-iron(II). Biochemistry. 1987 Mar 24;26(6):1539–1546. doi: 10.1021/bi00380a007. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Ikeda R. A., Richardson C. C. Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3614–3618. doi: 10.1073/pnas.83.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Jorgensen E. D., Durbin R. K., Risman S. S., McAllister W. T. Specific contacts between the bacteriophage T3, T7, and SP6 RNA polymerases and their promoters. J Biol Chem. 1991 Jan 5;266(1):645–651. [PubMed] [Google Scholar]
- Kassavetis G. A., Kaya K. M., Chamberlin M. J. Escherichia coli RNA polymerase-rifampicin complexes bound at promoter sites block RNA chain elongation by Escherichia coli RNA polymerase and T7-specific RNA polymerase. Biochemistry. 1978 Dec 26;17(26):5798–5804. doi: 10.1021/bi00619a029. [DOI] [PubMed] [Google Scholar]
- King K., Benkovic S. J., Modrich P. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11807–11815. [PubMed] [Google Scholar]
- Kruse B., Narasimhan N., Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989 Jul 28;58(2):391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Bartsch I., Grummt I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990 Apr 5;344(6266):559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Krummel B., Chamberlin M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987 Jul 5;196(1):85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Jack W. E., Modrich P. DNA determinants important in sequence recognition by Eco RI endonuclease. J Biol Chem. 1981 Dec 25;256(24):13200–13206. [PubMed] [Google Scholar]
- Martin C. T., Muller D. K., Coleman J. E. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988 May 31;27(11):3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A DNA-binding protein is required for termination of transcription by RNA polymerase I in Xenopus laevis. Mol Cell Biol. 1990 Jun;10(6):2793–2800. doi: 10.1128/mcb.10.6.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D. K., Martin C. T., Coleman J. E. T7 RNA polymerase interacts with its promoter from one side of the DNA helix. Biochemistry. 1989 Apr 18;28(8):3306–3313. doi: 10.1021/bi00434a028. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Osterman H. L., Coleman J. E. T7 ribonucleic acid polymerase-promotor interactions. Biochemistry. 1981 Aug 18;20(17):4884–4892. doi: 10.1021/bi00520a013. [DOI] [PubMed] [Google Scholar]
- Pavco P. A., Steege D. A. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990 Jun 15;265(17):9960–9969. [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. A synthetic substrate for tRNA splicing. Anal Biochem. 1987 Oct;166(1):90–106. doi: 10.1016/0003-2697(87)90551-3. [DOI] [PubMed] [Google Scholar]
- Rokeach L. A., Zyskind J. W. RNA terminating within the E. coli origin of replication: stringent regulation and control by DnaA protein. Cell. 1986 Aug 29;46(5):763–771. doi: 10.1016/0092-8674(86)90352-1. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitti M. A., Pavco P. A., Steege D. A. lac repressor blocks in vivo transcription of lac control region DNA. Proc Natl Acad Sci U S A. 1987 May;84(10):3199–3203. doi: 10.1073/pnas.84.10.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. B., Gamper H., Hearst J. E. Interaction of T7 RNA polymerase with DNA in an elongation complex arrested at a specific psoralen adduct site. J Biol Chem. 1988 Jan 5;263(1):527–534. [PubMed] [Google Scholar]
- Strothkamp R. E., Oakley J. L., Coleman J. E. Promoter melting by T7 ribonucleic acid polymerase as detected by single-stranded endonuclease digestion. Biochemistry. 1980 Mar 18;19(6):1074–1080. doi: 10.1021/bi00547a005. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vournakis J. N., Celantano J., Finn M., Lockard R. E., Mitra T., Pavlakis G., Troutt A., van den Berg M., Wurst R. M. Sequence and structure analysis of end-labeled RNA with nucleases. Gene Amplif Anal. 1981;2:267–298. [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- Wright D. J., King K., Modrich P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem. 1989 Jul 15;264(20):11816–11821. [PubMed] [Google Scholar]