Abstract
The parasite Trypanosoma brucei, the causative agent of sleeping sickness across sub-Saharan Africa, depends on a remarkable U-insertion/deletion RNA editing process in its mitochondrion. A approximately 20 S multi-protein complex, called the editosome, is an essential machinery for editing pre-mRNA molecules encoding the majority of mitochondrial proteins. Editosomes contain a common core of twelve proteins where six OB-fold interaction proteins, called A1–A6, play a crucial role. Here, we report the structure of two single-strand nucleic acid-binding OB-folds from interaction proteins A3 and A6 that surprisingly, form a heterodimer. Crystal growth required the assistance of an anti-A3 nanobody as a crystallization chaperone. Unexpectedly, this anti-A3 nanobody binds to both A3OB and A6, despite only ∼40% amino acid sequence identity between the OB-folds of A3 and A6. The A3OB-A6 heterodimer buries 35% more surface area than the A6 homodimer. This is attributed mainly to the presence of a conserved Pro-rich loop in A3OB. The implications of the A3OB–A6 heterodimer, and of a dimer of heterodimers observed in the crystals, for the architecture of the editosome are profound, resulting in a proposal of a ‘five OB-fold center’ in the core of the editosome.
INTRODUCTION
Trypanosomatids form an ancient group of unicellular eukaryotes which contain several pathogens causing major global diseases. These include sleeping sickness, Chagas disease and various forms of leishmaniasis caused by, Trypanosoma brucei, Trypanosoma cruzi and a group of Leishmania species, respectively. The current treatments of infected patients needs major improvement due to serious shortcomings of the available drugs and increased drug resistance (1–4). In particular since antigenic variation makes vaccine development for these organisms exceedingly challenging, there is an urgent need to develop new drugs for treatment of patients suffering from the global diseases caused by these devastating protozoa.
Trypanosomatids harbor many unusual features (5–7), several of which are promising drug targets. Particularly interesting is a unique and extensive mRNA editing process in mitochondria. This U-insertion/deletion process edits ‘pre-mRNA’ transcribed from approximately 12 ‘cryptogenes’ located on the maxicircles of the mitochondrial DNA. Most remarkably, the information for the transformation of the pre-mRNA into mature mRNA is distributed over hundreds of guide RNAs (gRNAs), typically ∼60 nt long, which are mainly encoded by the kinetoplast minicircles (8–13). In a complex process, numerous U's are inserted and several U's are deleted from the pre-message to produce the complete and correct message. The editing process can, in some cases, lead to more than a doubling of the size of the pre-message (11). A cascade of enzymatic reactions is responsible for the insertion of many U's and the deletion of a few U's according to the information in the gRNAs (14–16). This fascinating process is found only in trypanosomes and is essential for the life stage of the parasites in humans (16–19), making it a unique target for drug design.
Several multi-protein complexes are involved in U-insertion/deletion RNA editing. One of these is called the ∼20 S editosome, hereafter, called the editosome for simplicity (15,20). Recent electron microscopy studies have revealed that the editosome has an elongated shape with dimensions of ∼80 by ∼140 by ∼200 Å (21,22). A total of about 20 proteins are incorporated into editosomes (for editosome protein and domain nomenclature, see Supplementary Figure S1) with probably one copy of each protein in this multi-protein complex (21,22). Evidence has been provided for the presence of three different types of editosomes that share a common core of 12 proteins (16,23–28). Crystal structures of two key enzymes from this core have been reported so far: the RNA-editing ligase L1 in complex with Mg-ATP (29), and the 3′-terminal uridylyl transferase (TUTase) T2 in complex with Mg-UTP (17).
In the editosome core, six OB-fold interaction proteins (A1–A6) occur and have been shown to be essential for the functioning of the editosome (24,30–41). The three large interaction proteins (A1–A3) contain two Zn-finger motifs followed by a C-terminal domain which belongs to the large superfamily of single-strand nucleic acid-binding OB-folds, also called SSB domains or SSB proteins, involved in DNA repair, recombination, replication and RNA transcription (36,42–50). The three smaller interaction proteins (A4, A5, A6) have no Zn-finger motifs but contain also a C-terminal single-strand nucleic acid-binding OB-fold (36) (Supplementary Figure S1). OB-fold proteins form homodimers and often two such dimers form homotetramers with aligned 2-fold axes and D2 symmetry.
Each OB-fold interaction protein has its own characteristics and interaction partners in the editosome. The main proteins studied in this article are A3 and A6. The interaction protein A6 is a remarkable multi-functional OB-fold protein, central to the integrity of the entire editosome. With ∼17–23 kDa in size dependent on species, it not only interacts with four interaction proteins (20,51,52) but also binds poly-U single-stranded RNA (53). In three crystal structures solved recently, essentially the same dimer of A6 was observed in all the three cases (54). The 42 kDa interaction protein A3 binds ssRNA, as well as dsRNA (30) and its C-terminal OB-fold interacts with A6 and with the editosome protein B5 (51).
Since crystal growth of A6-containing multi-protein complexes turned out to be difficult, we explored nanobodies as crystallization chaperones. Nanobodies are the variable domains of the single heavy chain antibodies occurring in cameloids (55). Nanobodies have been used with great benefit as crystallization chaperones in several previous instances in our collaborating laboratories (54,56,57). In the current work, one out of the two available anti-A3OB nanobodies was successful in promoting the growth of well-diffracting crystals.
Here, we describe the 2.5 Å crystal structure of a heterotetramer formed by the OB-fold domains of the interaction proteins A3 and A6 and two copies of the same nanobody. This structure is most unusual for two reasons. First, the heterotetramer contains one copy of the anti-A3 nanobody A3Nb14 which interacts with A3OB and a second copy of A3Nb14 interacts with A6. The interaction of the A3Nb14 nanobody with A3OB is expected but the interaction with A6 is entirely unexpected since A6 shares only ∼40% sequence identity with A3OB. Second, this heterotetramer contains a tight heterodimer of the OB-folds of A3 and A6, the first heterodimer of single strand nucleic acid-binding OB-folds reported so far, to the best of our knowledge. In addition, two A3OB-A6 heterodimers form a heterotetramer in our crystals. These key features of our crystal structure have important implications for the architecture of the core of the editosome.
MATERIALS AND METHODS
Cloning, expression and purification of A2OB–A3OB–A6
Co-expressing full-length A3 with A6 did result in soluble protein which was homogeneous after purification but never crystallized, even when anti-A3 or anti-A6 nanobodies were utilized. Ternary complexes of T. brucei editosome proteins A2OB, A3OB and A6 were produced using a similar co-expression strategy as described previously (51). The gene-encoding residues 474–587 of T. brucei A2 (A2OB), preceded by an N-terminal 6xHistidine tag and a tobacco etch virus (TEV) protease cleavage site, was cloned into a pRSF vector (Novagen). The gene for T. brucei A6 (residues 20–164, i.e. with the mitochondrial signal sequence removed) and the gene-encoding residues 245–393 of T. brucei A3 (A3OB) were cloned into the bi-cistronic expression vector pACYC (Novagen) without His-tag. The pRSF vector containing the gene for A2OB was co-transformed with the A6-A3OB containing pACYC vector into Escherichia coli BL21-Gold (DE3) cells. Cells were grown to an OD600 of ∼0.6 at 37°C in Luria broth and induced with 0.5 mM IPTG at 20°C for 4 h. For SeMet substituted protein, expression was carried out in M9 minimal medium supplemented with amino acids and selenomethionine (58). Cells were lysed by sonication on ice. Soluble proteins were separated by centrifugation (30 min, 60 000g, at 4°C) from the cell pellet. The protein in the supernatant was purified on a Ni–NTA column using 250 mM imidazole, treated with TEV protease, purified further by a second Ni–NTA affinity step, and subjected to a final gel filtration run on a Superdex 200 gel filtration column using 20 mM Tris–HCl pH 8.0, 300 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and 10% glycerol.
Production of anti-A3 nanobodies
A3 protein with the import signal removed, spanning residues 47–393, was expressed from a pRSF plasmid in E. coli BL21-Gold (DE3) cells and purified similarly as the ternary protein complex described above. This protein was used for llama immunization, and subsequent selection of A3-specific nanobody clones using procedures essentially as described previously (56,57). Out of the 14 nanobodies obtained, two nanobodies, A3Nb14 and A3Nb8, appeared to bind to the ternary complex A2OB–A3OB–A6.
Crystallization and data collection
For the crystallization experiments, recombinant T. brucei A2OB–A3OB–A6 and anti-A3 nanobodies were purified separately, mixed at equimolar concentrations and incubated for 1 h at 4°C. Next, the protein mixtures were concentrated to 5 mg/ml and chromatographed on a Superdex 200 gel filtration column (GE Healthcare) in 20 mM Tris (pH 7.5), 300 mM NaCl, 1 mM TCEP and 10% glycerol. Crystals from the peak fraction containing A2OB–A3OB–A6 and A3Nb14 were obtained at room temperature by vapor diffusion experiments after mixing 200 nl of protein solution with 200 nl of a reservoir solution of 0.1 M HEPES pH 7.5, 10% 2-propanol, 22% w/v polyethylene glycol (PEG) 4000 by a Phoenix crystallization robot. A2OB–A3OB–A6 protein obtained from SeMet containing medium was also mixed with A3Nb14 protein solution and crystallized in essentially identical conditions. All crystals were soaked stepwise in cryo-protective solutions containing the mother liquor plus 20% (v/v) glycerol before being flash-cooled in liquid nitrogen for data collection. A 2.5 Å native data set and a 2.9 Å multiple-wavelength anomalous diffraction (MAD) data set were collected on beamline BL12-1 at the Stanford Synchrotron Radiation Lightsource (SSRL). The X-ray diffraction data were integrated and scaled with HKL2000 (59).
Structure determination and refinement
An initial structure was solved by MAD phasing using data collected from a crystal containing SeMet substituted protein. The program SOLVE (60) was used to locate the positions of the three Se atom sites. Initial phases were calculated using the SHELX programs at 2.9 Å resolution. After density modification by RESOLVE (60), most of the residues were automatically built by Buccaneer (61). An unrefined and partial poly-alanine model was placed successfully by PHASER (62) in the 2.5 Å resolution native crystals. The final model was improved and completed manually using the program COOT (63) and refined with the programs REFMAC5 (64) and Phenix (65), with 8 Translation/Libration/Screw (TLS) groups per asymmetric unit (66). Crystallographic data collection and refinement statistics are shown in Table 1. In the final model, there are four chains in the asymmetric unit, which comprise residues 283–393 of A3OB, residues 20–132 of A6 and residues 3–128 of both A3Nb14 molecules. The residues with weak density in A3OB are 283–287, and in A6 are 129–132. Representative electron densities in interface regions are shown in Supplementary Figure S8. The quality of the crystal structure was analyzed using MolProbity (67).
Table 1.
Crystallographic data collection and refinement statistics
| Item | Native | Se | Se | Se |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P43212 | P43212 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 74.6, 74.6, 239.9 | 74.6, 74.6, 239.3 | ||
| Solvent content (%) | 54.9 | |||
| Peak | Inflection | Remote | ||
| Wavelength | 0.9792 | 0.9792 | 0.9794 | 0.9116 |
| Resolution (Å) | 50–2.5 | 50–2.9 | 50–2.9 | 50–2.9 |
| Rsym or Rmerge (%) | 6.5 (60.2) | 0.169 (0.381) | 0.103 (0.363) | 0.156 (0.376) |
| I / σ(I) | 19.8 (3.8) | 13.1 (3.1) | 20.8 (3.1) | 14.7 (2.6) |
| Completeness (%) | 99.9 (100) | 97.8 (77.4) | 98 (79.1) | 98.7 (86.8) |
| Redundancy | 11.5 (11.9) | 9.7 (5.9) | 10.5 (6.1) | 9.9 (5.9) |
| Refinement | ||||
| Resolution (Å) | 37–2.5 | |||
| No. of reflections | 23 086 | |||
| Rwork/Rfree (%) | 19.9/23.4 | |||
| No. atoms | ||||
| Protein | 3593 | |||
| Ligand/ion | 0 | |||
| Water | 59 | |||
| B-factors (Å2) | ||||
| Protein | 72.7 | |||
| A3OB | 77.2 | |||
| A6 | 78.2 | |||
| A3Nb14(A) | 74.3 | |||
| A3Nb14(B) | 61.1 | |||
| Ligand/ion | ||||
| Water | 66.2 | |||
| R.m.s.d. | ||||
| Bond lengths (Å) | 0.0094 | |||
| Bond angles (°) | 1.3275 | |||
| Ramachandran plota | ||||
| Preferred (%) | 96.09 | |||
| Allowed (%) | 3.48 | |||
| Outliers (%) | 0.43 | |||
aRamachandran plot analysis by Molprobity (67). Values in parentheses are for the highest resolution shell.
Figure preparation
Least squares analysis to determine the structural similarity between A3OB and A6 was carried out using LSQKAB and DaliLite (68). Protein quaternary-structure analysis was performed with the PISA server (69). Sequence alignment figures were made with Espript (70). Electrostatic potential surfaces were calculated using APBS (71). All other figures of molecular structures were prepared using PyMOL (DeLano Scientific Research LLC).
RESULTS
Protein preparation and structure determination
Obtaining a complex of multiple OB-folds from editosome interaction proteins which is homogeneous in size and composition appeared to be challenging. Eventually, a complex of A2OB–A3OB–A6 could be prepared (Supplementary Figure S2) which had the unfortunate property that the A2OB content decreased over time. Perhaps not too surprisingly therefore, no crystals of this A2OB–A3OB–A6 complex could be obtained. Given the obstacles encountered, the power of nanobodies as crystallization chaperones was evaluated. Initially, 14 nanobodies were isolated from an immune library generated against essentially full-length A3. From this set of nanobodies, A3Nb14 and A3Nb8 (Supplementary Figure S3) appeared to bind to the A3OB–A6 complex according to electromobility shift analysis (EMSA) (data not shown). After mixing the A2OB–A3OB–A6 complex with A3Nb14, well-diffracting crystals grew eventually from which a preliminary structure could be obtained by MAD phasing (See ‘Materials and Methods’ section).
Surprisingly, the asymmetric unit contained only two OB-fold proteins which formed a tight dimer. One of these OB-folds did not contain any SeMet sites and therefore, was the A6 monomer. From the distribution of SeMet sites along the chain of the second OB-fold it was evident that it was A3OB (Supplementary Figure S4). Hence, a heterodimer of A3OB–A6 is present in the crystals. Remarkably, no density representing the A2OB monomer was discernable. The absence of this monomer was confirmed by sodium dodecyl sulfate – polyacrylamide gel electrophoresis (SDS–PAGE) analysis of dissolved crystals which showed the presence of A3OB, A6 and A3Nb14, but not of A2OB (Supplementary Figure S2). In addition to containing a heterodimer of OB-folds, a second surprise was that A3OB and A6 were each interacting with an A3Nb14 nanobody, yielding an A3OB–A6-(A3Nb14)2 heterotetramer (Figure 1). The same immunoglobulin chain apparently interacts with two proteins which share only ∼40% sequence identity.
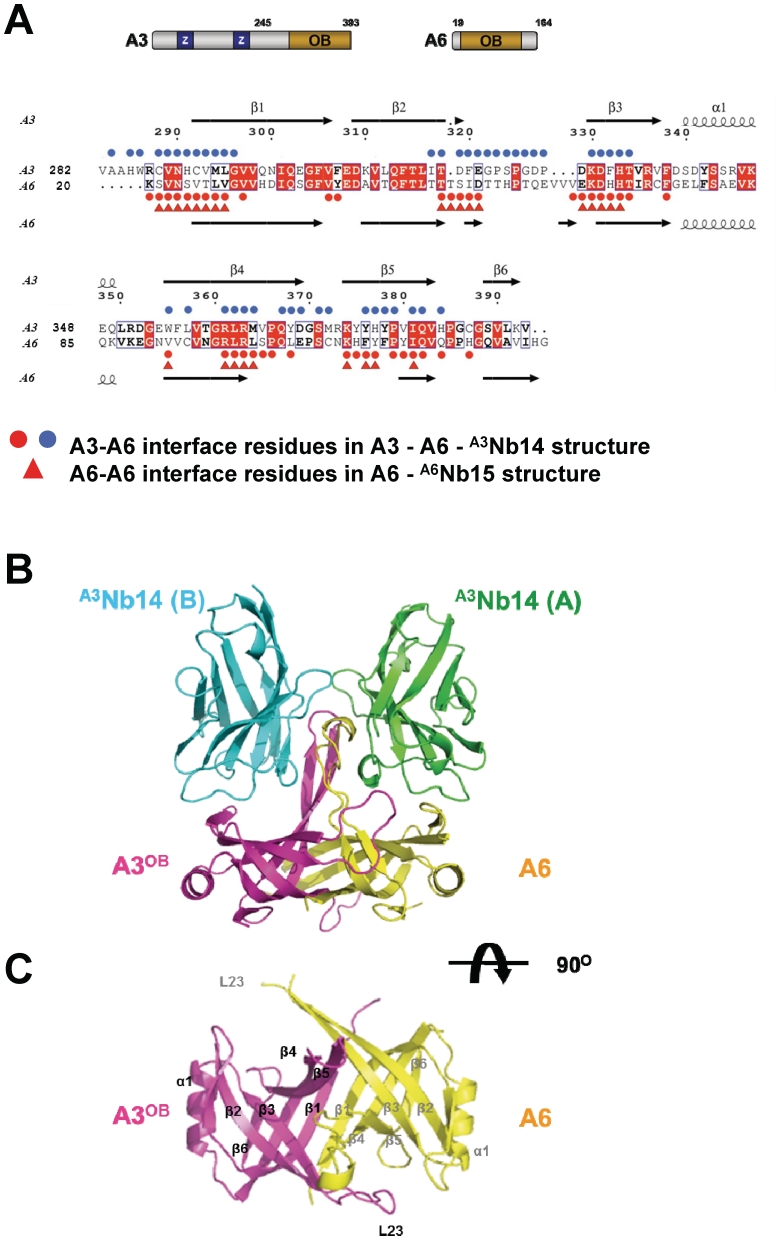
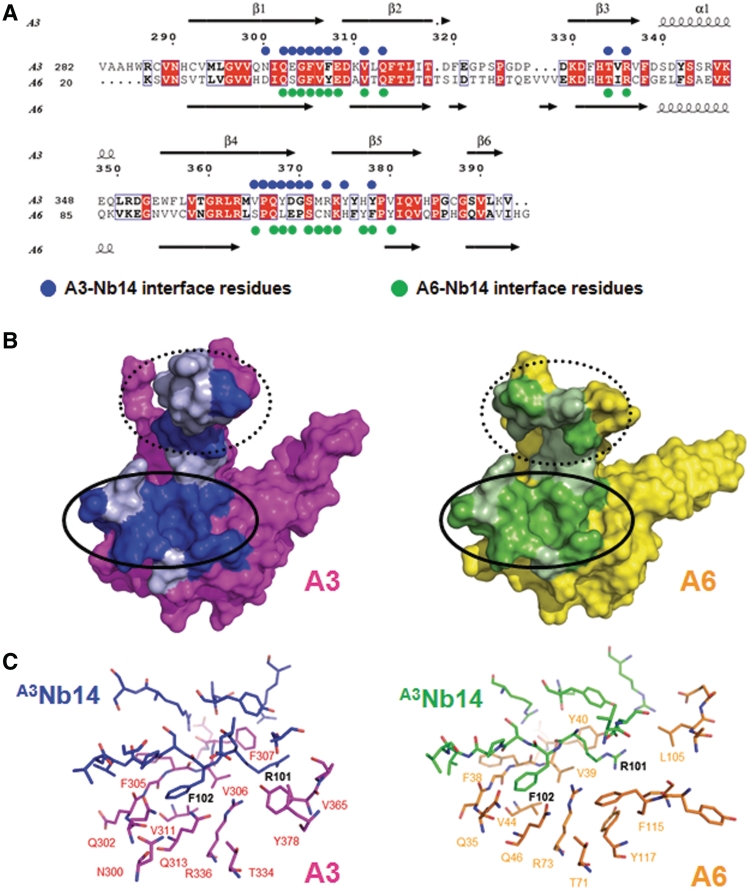
Figure 1.
Structure of heterotetramer and heterodimer. (A) Sequence alignment of A3OB and A6. A schematic representation of full-length A3 and full-length A6 with their OB-fold domain and the zinc finger domains (Z) shown in the upper panel. A3OB and A6 sequences (acronyms provided on the left) are aligned in the lower panel. Strictly conserved residues in both sequences are in red boxes. Secondary structure elements are indicated on top and below. Red and blue spheres indicate residues forming the A3OB–A6 interface; red triangles the interface residues of the A6 dimer (PDB-ID: 3K7U) (54). (B) The A3OB–A6-(A3Nb14)2 heterotetramer. The heterotetramer is shown perpendicular to the pseudo-2-fold axis relating the OB-folds of A3OB and A6. A3OB is depicted in magenta; A6 in yellow; A3Nb14 in complex with A3OB in blue; A3Nb14 in complex with A6 in green. The heterotetramer depicted is part of an [A3OB–A6–(A3Nb14)2]2 heterooctamer occurring in the crystal lattice (Supplementary Figure S7). (C) The A3OB–A6-heterodimer. A view along the pseudo-2-fold of the A3OB–A6 dimer of the heterotetramer shown in (B) is in the same color code. The N-terminal β-strands β1 of each monomer run anti-parallel to each other in the center of an extended β-sheet.
The preliminary model derived from the SeMet MAD electron density was used as a starting point for refinement against the 2.5 Å data from a native crystal that belonged to the same space group. This eventually resulted in a refined structure with Rwork and Rfree values of 20 and 23.4%, respectively, with good statistics (Table 1).
Monomers, dimer and tetramer
The main structural elements of both the A3OB and A6 monomers are a six-stranded anti-parallel β-barrel with a Greek key motif, helix α1 linking β3 and β4, and three β-hairpin loops (L12, L23 and L45) extending out of a globular core β-barrel (Figure 1). Superimposing A3OB onto A6 results in an r.m.s.d. of 1.2 Å for 91 equivalent Cα atoms. The main structural differences between the A3OB and A6 monomers are localized in loop L23 and the β4–β5 hairpin (Figure 2A). The two monomers are related by a pseudodyad which is perpendicular to the extended β-sheet formed by an anti-parallel pair of N-terminal β-strands of each monomer. The binding of the nanobodies to these two different proteins is such that the two A3Nb14 molecules are related by the same pseudo-2-fold which relates the OB-folds (Figure 1B).
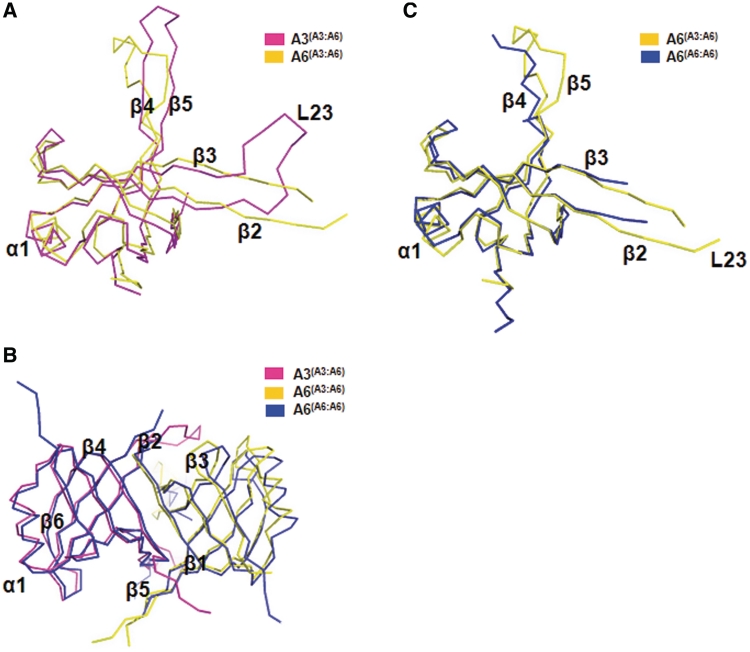
Figure 2.
Comparison of A3OB and A6. (A) Superposition of the A3OB and A6 chains from the A3OB-A6 heterodimer. The A3OB chain is shown in magenta; A6 from the A3OB–A6 heterodimer in yellow. Note the large conformational differences in the L23 loop, as well as in the β4–β5 hairpin. (B) Superposition of the A3OB–A6 and A6–A6 dimers. A view along the pseudodyad of the heterodimer. The A3OB chain is shown in magenta; A6 from the A3OB–A6 heterodimer in yellow; both A6 subunits from the A6–A6 homodimer in blue (PDB-ID: 3K7U) (54). (C) Superposition of A6 in the A3OB-A6 heterodimer and A6 in the A6 homodimer. A6 from the heterodimer is depicted in yellow; A6 from the homodimer in blue (PDB-ID: 3K7U) (54). Note that the L23 loop, as well as the β4–β5 hairpin of A6 is better ordered in the A3OB-A6 heterodimer than in the A6 dimer.
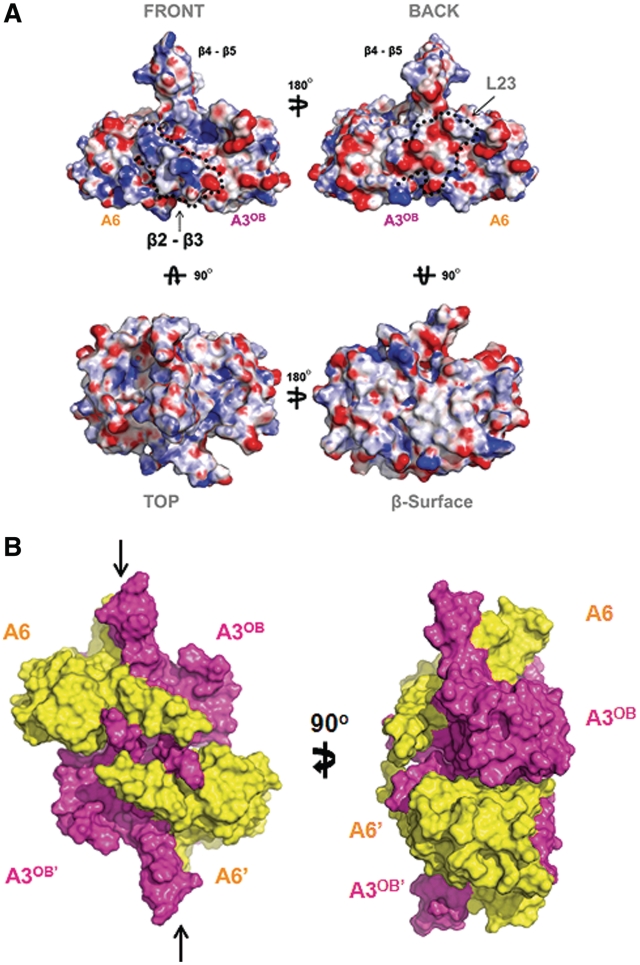
The distribution of charged residues over the surface of the A3OB-A6 heterodimer is highly irregular (Figure 3A). The ‘front’ surface has a preponderance of positive charges and the ‘back’ surface an excess of negative charges. The ‘top’ surface is relatively abundant in positively charged side chains and aromatic residues that are typically involved in interactions with single-strand RNA or ssDNA by this class of proteins (47,72–74). In contrast, the ‘bottom’ surface contains few charged residues and a large number of branched non-polar residues resulting in a quite hydrophobic ‘β-surface’ of the heterodimer (Figure 3A).
Figure 3.
Characteristics of A3OB–A6 heterodimer and heterotetramer. (A) Surface charge distribution. Four views of the surface charge distribution of the A3OB–A6 heterodimer. The electrostatic potential surface of the A3OB and A6 dimer is calculated using APBS (71) and displayed as blue for a positively charged surface potential, red for negative and gray for neutral. The ‘front surface’ (upper left) is predominantly positively charged and the ‘back surface’ (upper right) predominantly negative. The ‘top’ view (lower left) shows pockets of positively charged surface areas next to the protruding β4–β5 hairpin. The ‘β-surface’ (lower right), formed mainly by the extended β-sheet of the dimer, contains large hydrophobic areas (gray). (B) The ‘shifted tetramer’ formed by two A3OB-A6 heterodimers in the crystal. The A3OB and A6 proteins in the complex are shown in surface mode, with the two A3OB chains in magenta and the two A6 domains in yellow. The β-surfaces of two A3OB–A6 heterodimers contact each other in the crystals. The ∼2000 Å2 total solvent accessible surface buried in this dimer–dimer interface is composed of ∼800 Å2 from A3OB–A3OB′ contacts, ∼600 Å2 contributed by A3OB–A6′, ∼600 Å2 by A3OB′–A6. The A6 and A6′ subunits do not interact, while A3OB interacts with three different OB-folds in this assembly. This (A3OB–A6)2 heterotetramer is distinct from canonical OB-fold homotetramers (78) in that the upper and lower dimer pseudo-dyads are not coinciding, but are shifted with respect to each other parallel to the interacting β-surface (See arrows representing pseudo-dyads in the upper and lower heterodimers).
In the crystals, two A3OB-A6 heterodimers related by crystallographic symmetry form a tetramer, in which the hydrophobic β-surfaces of two A3OB–A6 heterodimers face each other, yielding an (A3OB–A6)2 heterotetramer (Figure 3B). The total solvent accessible buried surface in this interface is ∼2000 Å2 (69). The possible relevance of this heterotetramer of four OB-folds for the architecture of the editosome will be discussed further below.
Interactions between A3OB and A6
The interaction between A3OB and A6 buries 3105 Å2 surface area upon forming the heterodimer. Among the 47 residues from A3OB and the 43 residues of A6 engaged in intersubunit contacts, five amino acids from each subunit are the major players and each contribute 70–150 Å2 to the buried surface area (Supplementary Table S1). The interaction surface is highly convoluted but can approximately be described as the sum of three major areas of contact between the two subunits.
The first contact area includes and surrounds the two antiparallel β1 strands of both OB-folds, contributing to 30% of the total buried interface area (Figure 2B, Supplementary Tables S1 and S2). Five residues from strand β1A3 form main chain hydrogen bonds with six β1A6 residues, a feature shared with the A6 homodimer (54). Different from the A6-A6 dimer, these β1A3– β1A6 interactions are in addition stabilized by contacts of the two N-terminal residues Lys20A6 and Ser21A6 with residues from β2A3 and β4A3. The highly conserved Ser21A6 has contacts with not less than seven residues from A3OB (Supplementary Table S2).
Amino acids contributing to the second contact area include residues from β4A3–L45–β5A3 and β2A6–L23–β3A6 (Figure 4A and B). The invariant Glu66A6 side chain protrudes from strand β4 and forms a hydrogen bond with the backbone amide of Arg361A3, and a salt bridge with the side chain of the same Arg361A3. Another salt bridge occurs between Asp68A6 and Arg363A3. Residues Leu362A3, Arg363A3 and Met364A3 from β4A3 are three of the five A3 residues contributing each more than 70 Å2 to the interface, indicating the importance of this β-strand for the A3–A6 interaction.
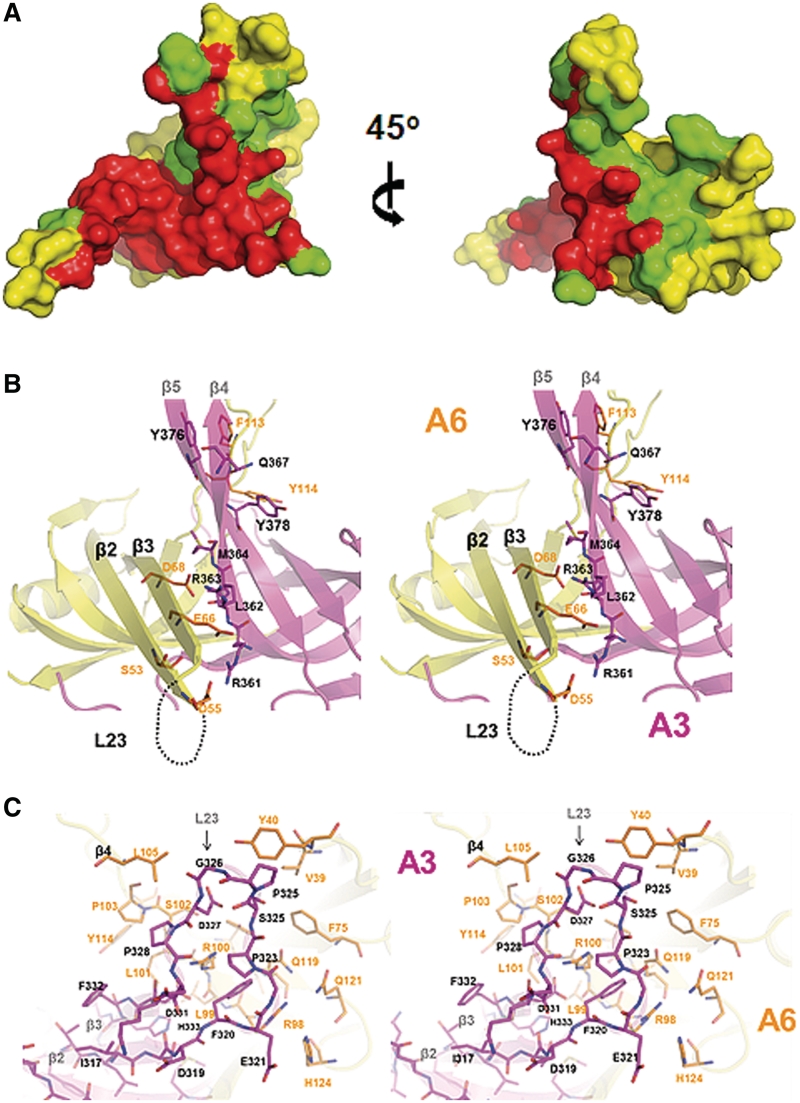
Figure 4.
Interactions in A3OB–A6 compared with those in A6–A6. (A) The fingerprint of A3OB on A6 is larger than that of A6 on A6. The interactions are ‘painted’ onto the surface of A6. Red: the contact area which is the same in the A3OB–A6 heterodimer and in the A6 homodimer. Green: the extra contact area in the heterodimer compared with the homodimer. Yellow: the surface of A6 neither in contact with A3OB in the heterodimer, nor with the second A6 subunit in the homodimer. The surface buried in the interaction in A3OB–A6 is ∼35% larger than in the A6–A6 dimer (PDB-ID: 3K7U)(54). (B) Strands β2–β3 of A6 contacting A3OB. A stereo view of the ‘second contact area’ (see text) of A3–A6 with important residues highlighted shown in ribbon representation with the same color coding as in Figure 1B. The secondary structure elements and key residues mediating the A3–A6 dimer interactions are labeled. (C) The Pro-rich L23 loop of A3OB contacting A6. A stereo view, 180° different about the vertical axis from (B), of the ‘third contact area’ (see text) in the heterodimer shown with stick representations. Selected secondary structure elements and key contact residues are labeled. A3 is colored magenta, A6 yellow-gold.
In the third contact region, residues from β2A3–L23–β3A3 and β4A6–L45–β5A6 interact with each other. The β2A6–β3A6 hairpin shows a large conformational difference compared to the equivalent region in A3 (Figure 2A). Interestingly, residues Phe320A3 and Arg100A6, which each contribute more than 100 Å2 to this interface, are in close proximity (Supplementary Figure S8A and Supplementary Tables S1 and S2). A key point is that the proline-rich loop L23 of A3OB is substantially involved in the A3OB–A6 dimer formation (Figure 4C), whereas loop L23 of A6 is not engaged in similar interactions due to a different course of the polypeptide chain (Figures 2A and 4B).
Comparison of the A3OB-A6 heterodimer and the A6 homodimer
The A3OB–A6 heterodimer and the A6–A6 homodimer (PDB-ID: 3K7U) can be superimposed with an r.m.s.d. of 1.2 Å for 179 equivalent Cα atoms, indicating that the general architecture of these dimers is similar (Figure 2B). The β4–β5 hairpins in the A6 subunits differ in conformation and loop L23 is less well defined in the A6 subunit of the homodimer (Figure 2C). The 3105 Å2 surface area buried in the A3OB–A6 heterodimer is considerably more than the 2015 Å2 buried in the formation of the A6 homodimer (54). The major additional contacts in the heterodimer interface (Figure 4A) are due to (i) extra contacts by N-terminal residues of A3 and (ii) the different conformation of loop L23 of A3OB which allows contacts with more A6 residues than occur in the A6 dimer. Clearly, according to the structure, formation of the A3OB–A6 heterodimer is more favorable than that of the A6–A6 homodimer, in agreement with the fact that during co-expression, the A3–A6 dimer is formed.
The cognate A3Nb14-A3OB and the cross-reacting A3Nb14-A6 interfaces
The interface between the A3Nb14 and its cognate antigen A3OB is formed by 29 residues contributed by A3Nb14 and 22 residues by A3OB, resulting in a total buried surface area of 1775 Å2 (Figure 5 and Supplementary Figure S3). All three complementarity defining regions (CDRs) of the nanobody make specific contacts with A3OB (Supplementary Figure S3). Additionally, nine residues participating in the interaction with A3OB are highly conserved framework residues of the nanobody.
Figure 5.
The A3OB–A3Nb14 and A6–A3Nb14 complexes. (A) Sequences and interface residues of A3OB and A6 with A3Nb14. Structure-based alignment of the T. brucei A3 sequence (upper line) and A6 sequence (lower line). Residues highlighted in red are completely conserved. Blue spheres indicate A3 residues in contact with nanobody A3Nb14. Green sphere residues of A6 are residues in contact with the second copy of nanobody A3Nb14 in the heterotetramer. (B) Footprints of A3Nb14 onto A3OB (left) and onto A6 (right). Interaction Region 1 is outlined by an ellipse with a solid line; Interaction Region 2 with a dashed line. Left: dark blue: A3OB residues contacting A3Nb14 residues which are identical in A6; Light blue: A3OB residues contacting A3Nb14 residues which are different in A6; Magenta: A3OB atoms not contacting the nanobody. Right: dark green: A6 residues contacting A3Nb14 residues which are identical in A3OB; light green: A3Nb14 residues-contacting residues of A6 which are different in A3OB. Yellow: A6 atoms not contacting the nanobody. (C) Key interactions of A3Nb14 with A3 (left) and of A3Nb14 with A6 (right). Close-ups of Interaction Region 1 of both A3 and A6 are shown in stick representations. Residues involved in protein–protein interactions are indicated and colored according to the molecules they belong to. Colors: A3OB in magenta with its A3Nb14 bound in blue, A6 in yellow-gold with its A3Nb14 bound in green. Nitrogens blue, oxygens red.
The majority of cognate contacts occur between CDR3 of the nanobody and β1–β2 (residues 302–313) of A3OB. This interface alone accounts for 61% of the buried surface, and will hereafter be called Recognition Area 1. On the A3OB surface, Recognition Area 1 is separated from that of Recognition Area 2 which is formed mainly by residues from strands β4 and β5. The footprints of these two Recognition Areas on A3OB are approximately perpendicular to each other (Figure 5B). Recognition area 1 contains Phe307A3 which, with 157 Å2, is the A3OB residue contributing most to the contacts with the nanobody (Supplementary Table S3). Important contact residues of A3Nb14 are Arg101 and Phe102 of CDR3, the only nanobody residues contributing each more than 100 Å2 to the buried surface. The Phe102 side chain of A3Nb14 fits into a deep pocket in the center of Recognition Area 1 of A3OB (Figure 5B and C; Supplementary Figure S8).
Nanobody A3Nb14, raised against A3, interacts also with A6 (Figure 1). The unexpected ‘cross-reacting’ A6-A3Nb14 interface buries 1673 Å2 solvent accessible surface area contributed by 23 residues of A3Nb14 and 22 residues of A6. Of the amino acids of A3OB and A6 recognized by CDRs, about 71% are identical between A3OB and A6 (Figure 5 and Supplementary Table S3). Many identical or similar contact residues between A3OB and A6 are clustered in Interaction Region 1. For instance, like its counterpart Phe307A3, Tyr40A6 is the major contributor from A6 to the interface, with 118 Å2 buried surface area (Supplementary Table S3). Also, the Phe102 side chain of A3Nb14 fits into a deep pocket in the center of Recognition Area 1 of A6, very similar to the mode of binding of Phe102 of the nanobody to its cognate antigen A3 (Figure 5B and C). Interaction Region 2 is more different in the two cases. The largest variation occurs here in the β4–β5 hairpin where Asp369, Tyr368, Gly370 and Tyr375 of A3OB are replaced by Cys109, Asn110, Lys111 and Glu106 in A6 (Supplementary Table S3). Interestingly, these residues only interact with framework residues of A3Nb14 indicating that Contact Region 2 is largely involved in non-specific interactions.
DISCUSSION
Cross-reactivity of A3Nb14 with A3OB and A6
Crystal structures of the same immunoglobulin domain in complex with two different proteins appear to be rare. To the best of our knowledge, there are only two previous reports of structures with the same antibody in complex with different proteins, if we ignore the special case of idiotype:anti-idiotype complexes. The first example is provided by the structures of guinea fowl lysozyme and hen egg white lysozyme in complex with Fab F9.13.7 generated against hen egg white lysozyme (75). In this case, the two lysozymes bound to the same Fab are 81% identical in amino acid sequence. A second example is Fab F8.12.19 generated against the surface protein AMA1 from the malaria parasite Plasmodium vivax that cross-reacts with the homologous protein from several Plasmodium species. The crystal structures of complexes of Fab F8.12.19 with fragments of Plasmodium falciparum AMA1 and with P. vivax AMA1 have been elucidated. These two AMA1 proteins share 67% amino acid sequence identity (76). The monoclonal antibody Fab F8.12.19 recognizes a discontinuous epitope, with the major contact region, a loop with a cysteine knot, and interacts mainly with conserved residues in these two proteins.
Nanobody A3Nb14 interacts with two proteins which share only ∼40% amino acid sequence identity. This is a substantially lower amino acid sequence identity than in the two above-mentioned reports on structures of cross-reacting immunoglobulin domains. The two copies of A3Nb14 in the A3OB–A6-(A3Nb14)2 heterotetramer have an r.m.s.d. of only 0.45 Å for 125 residues, with the largest difference of ∼1 Å for the Cα of Arg101 of CDR3. Apparently, nanobody A3Nb14 can form a complex with two different proteins while maintaining essentially the same conformation. The interactions of A3Nb14 with A3 and A6 reveals that the target proteins have two rather separate interaction areas with the nanobody (Figure 5). This is reminiscent of the Fab F8.12.18 complexes with the AMA1 domains mentioned above. Interaction Regions 1 of A3OB and A6 provide a similar deep pocket which buries a protruding side chain from CDR3 (Figure 5). In contrast, Interaction Region 2 is rather smooth. Interaction Region 1 of the epitope differs little in structure when comparing A3 and A6 with an r.m.s.d. for Cα atoms of 0.45 Å with 70% sequence identity for 10 amino acids. The differences in Interaction Region 2 are larger with an r.m.s.d. of 2.9 Å with 27% sequence identity for 11 amino acids. It would obviously be interesting to know the affinities of A3Nb14 for A3OB and A6. Unfortunately, determining these affinities is not trivial. First, A3OB by itself is insoluble. Second, while A6 by itself forms soluble homotetramers (54) and nanobody A3Nb14 does bind to A6, gel filtration and SDS–PAGE analysis reveals that the homotetramer dissociates, resulting in formation of (A6-A3Nb14)2 heterotetramers (data not shown). The simultaneous processes of nanobody binding and tetramer dissociation makes determining the affinity of A3Nb14 for A6 a challenge.
The A3OB–A6 heterodimer and tetramer versus the architecture of the editosome
The A3OB–A6 heterodimer structure is the first example of a heterodimer in the widely studied single-strand nucleic acid-binding OB-fold family (72,73,77–80). This unique A3OB–A6 heterodimer appears to be quite similar to that of the A6 homodimer (54), in spite of the fact that the A3OB and A6 chains share only ∼40% sequence identity (Figure 1A). Important for the architecture, and probably also for the biogenesis of the editosome, is that the interface in the A3OB–A6 heterodimer is ∼35% larger than in the A6 homodimer. The main contribution to this enlarged interface stems from extra interactions of the proline-rich L23 loop in A3 with A6. The sequence of L23A3, including the prolines, is highly conserved in trypanosomatids (Supplementary Figure S5) and so are the residues of A6 interacting with L23A3 (Supplementary Figure S6 and Supplementary Table S1). Hence, it is likely that the A3OB–A6 heterodimer is occurring in the actual editosome as well.
Interaction protein A6 is a remarkable editosome protein since it engages with not less than four other editosome proteins and with RNA (35,37,51–53,81). A most intriguing question is how this central interaction protein in the core of the editosome is surrounded by so many other proteins. In the present structure we see for the first time how A6 and the OB-fold of A3OB form extensive interactions. A major remaining question is how the A3OB-A6 heterodimer contacts other OB-folds in the editosome. By a combination of yeast-two hybrid and biochemical studies, the following binary interactions involving A6 have been uncovered previously:
the interaction of A6 with A2OB [Figures 2A and 4A and B in Ref. (51)];
the interaction of A6 with A3OB [Figures 2A and 4C and D in Ref. (51)];
Moreover, a ternary complex of A2OB, A3OB and A6 has been obtained [Figure 7D in Ref. (51)]. In addition, we have performed pull-down experiments which indicate that ternary complexes of A1OB, A3OB and A6 and of A4OB, A3OB and A6 also exist (Supplementary Figure S9).
Interestingly, a suggestion for the mode of association of multiple OB folds in the center of the editosome core comes from the (A3OB-A6)2 heterotetramer observed in the crystals (Figure 3 and Supplementary Figure S7). This heterotetramer is reminiscent of canonical homotetramers of single-strand nucleic acid-binding OB-folds. These homotetramers have D2 symmetry. In the canonical arrangement, the so-called P-dyad of the OB-fold tetramer is shared by the two dimers, i.e. the 2-fold axes of the two dimers coincide (78). But a considerable variation is observed regarding the orientation of the two dimers with respect to each other (74,77,79,80,82,83). As a result, the buried surface area in dimer–dimer interfaces of canonical OB-fold tetramers is highly variable, ranging from 1128 to 3563 Å2 (PDB-IDs: 1EQQ, 1X3E, 1UE1, 2FXQ, and 2VW9). The ∼2000 Å2 buried by the dimers in the (A3OB–A6)2 heterotetramer in our crystals is clearly within the range seen for canonical OB-fold homotetramers.
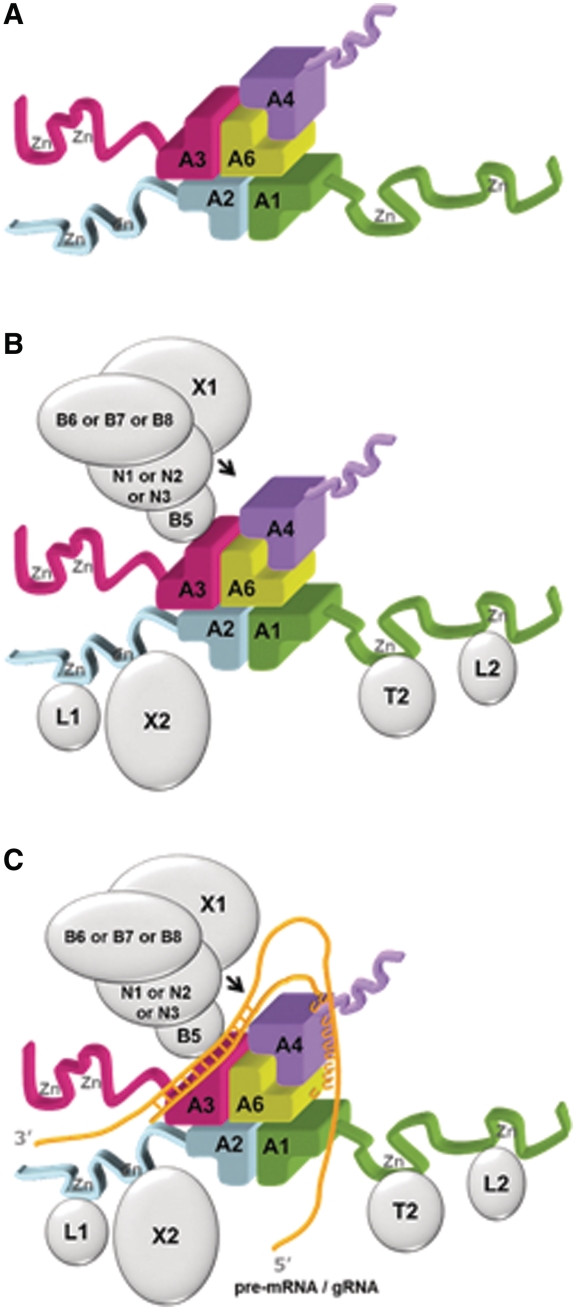
In both the canonical OB-fold tetramer and in the (A3OB–A6)2 heterotetramer, the β-surfaces of the dimers face each other (Figure 3B). However, in contrast to this common feature, there is also a major difference: the 2-fold axes of the two A3OB–A6 heterodimers do not coincide in this heterotetramer, since the two A3OB–A6 dimers are shifted parallel to the β-surface with respect to each other. In a canonical OB-fold homotetramer these 2-fold axes coincide. As a result, one OB-fold in the ‘shifted heterotetramer’ interacts with all three other OB-folds (Figure 3B). This ‘shifted heterotetramer’ serves as a source of inspiration to combine the available solution studies on binary and ternary interactions, listed above, with the new A3OB–A6 structure to arrive at a model for a five OB-fold complex in the editosome core.
In this five OB-fold model, OB-folds from four different editosome proteins are arranged as a shifted heterotetramer (Figure 6A) in such a way that A6 interacts with the OB-folds of three other interaction proteins (35,51), and one additional OB-fold interacts with A6 in a different, yet unknown, manner. The A3OB–A6 heterodimer seen in our current structure is the cornerstone of this model. While the mutual positions of A3OB and A6 are known from our structure, which of the three other OB-folds occupies which position in the five OB-fold model remains to be established. Hence, there are six variants of the model possible by interchanging A1OB, A2OB and A4OB.
Figure 6.
An OB-fold center in the core of the editosome. (A) Model for a “five OB-fold” center of the editosome. The basis is a shifted heterotetramer (in this example, comprising the OB-folds of A1, A2, A3 and A6) plus an additional A4OB fold interacting with A6. Please note that while the A3OB–A6 heterodimer is experimentally observed, the positions of A1OB, A2OB and A4OB in this figure can be interchanged and still yield a model in agreement with the experimental constraints mentioned in the text. Therefore, the arrangement shown is one of six possible models of these five OB-folds forming the center of the editosome core. (B) The same five OB-fold center as in (A) with the additional domains in the OB-fold interaction proteins shown as ribbons and the proteins interacting with these domains outlined as silver ellipsoids. Note that three different types of editosomes have been reported to exist, containing either N1-B6, or N2-B7, or N3-B8-X1, in addition to the core (52). (C) A global outline of how a pre-mRNA•gRNA duplex might interact with the five OB-fold center shown in (A). The gold wires indicate RNA, with the duplex in the anchor region shown as crossbars, and the poly-U tail of the mature gRNA as a series of U's. The arrow points to an editing site. The surrounding enzymes might display mobility with respect to the five OB-fold center. See also text.
If the assembly of OB-folds in the core of the editosome would be as in our five OB-fold model, then this would have interesting implications for how the editosome might function. After all, the three large interaction proteins A1, A2 and A3 contain extra N-terminal domains which interact, directly or indirectly, with enzymes in the editosome (27,32,35,51,52,81,84,85). These enzymes perform the crucial steps in the RNA editing cascade, i.e. cleaving the pre-mRNA strand at specific sites, removing and inserting U's and sealing the chain again (16,28,84,86–88). The catalytic domains of the RNA ligases L1 and L2, the TUTase T2 and of the 3′→5′exonuclease X2 are linked to OB-folds via flexible linkers between the multiple domains in the full length A1 and A2 proteins. The endonucleases N1, N2 and N3 and the 3′→5′exonuclease X1 are linked to A3 in the OB-fold center via one, or possibly even two, interaction proteins (36,51,52). This arrangement (Figure 6B) then suggests that the five or six [this number is dependent on the editosome type (28)] catalytic domains of the enzymes surrounding the five OB-fold center display a substantial degree of mobility enabling the active sites to reach the editing site of the mRNA.
Results of studies on the RNA-binding properties of OB-folds can be added to this schematic picture. A4 has recently been shown to have very high affinity for poly-U, A3 has a high affinity for dsRNA, and A6 a weak affinity for poly-U (30,38,53). These data lead to a possible association of the pre-mRNA•gRNA duplex with the editosome OB-fold center as sketched in Figure 6C. Establishing whether or not this model captures the global features of the architecture and mechanism of the editosome requires obviously additional experimental studies.
ACCESSION NUMBERS
Protein Data Bank accession number 3STB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–9 and Supplementary References [17,20,25,29,36,51,54,69,89–91].
FUNDING
Funding for open access charge: National Institute of Health (grant RO1 AI34501 to W.G.J.H.); Vrije Universiteit Brussel (VUB GOA65 to J.S.); Vlaams Instituut Biotechnologie (VIB to J.S); the Fund for Scientific Research of Flanders (FWOAL551 to J.S); the Hercules Foundation (HERC2 to J.S) and the Institute for the encouragement of Scientific Research and Innovation of Brussels (Innoviris to J.S & E.P); and the Belgian Government under the framework of the Interuniversity Attraction Poles (I.A.P. P6/19 to E.P).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of BL12.1 beam line at SSRL for invaluable assistance with data collection. We thank Katleen Willibal for the A3-nanobody selections and screening, and Nele Buys for the recloning of the nanobodies. We also thank Stewart Turley and Konstantin Korotkov for scientific discussions and expert technical support.
REFERENCES
- 1.Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 2006;19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fairlamb AH. Chemotherapy of human African trypanosomiasis: current and future prospects. Trends. Parasitol. 2003;19:488–494. doi: 10.1016/j.pt.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Sachs SE, Sachs JD, Savioli L. Control of neglected tropical diseases. N. Engl. J. Med. 2007;357:1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- 4.Tarleton RL, Reithinger R, Urbina JA, Kitron U, Gurtler RE. The challenges of Chagas Disease–grim outlook or glimmer of hope. PLoS Med. 2007;4:e332. doi: 10.1371/journal.pmed.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moyersoen J, Choe J, Fan E, Hol WG, Michels PA. Biogenesis of peroxisomes and glycosomes: trypanosomatid glycosome assembly is a promising new drug target. FEMS Microbiol. Rev. 2004;28:603–643. doi: 10.1016/j.femsre.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Hammarton TC. Cell cycle regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153:1–8. doi: 10.1016/j.molbiopara.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Souza W. From the cell biology to the development of new chemotherapeutic approaches against trypanosomatids: dreams and reality. Kinetoplastid Biol. Dis. 2002;1:3. doi: 10.1186/1475-9292-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 9.Simpson L, Wang SH, Thiemann OH, Alfonzo JD, Maslov DA, Avila HA. U-insertion/deletion Edited Sequence Database. Nucleic Acids Res. 1998;26:170–176. doi: 10.1093/nar/26.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinz S, Goringer HU. The guide RNA database (3.0) Nucleic Acids Res. 1999;27:168. doi: 10.1093/nar/27.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart K, Allen TE, Heidmann S, Seiwert SD. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart K, Panigrahi AK. RNA editing: complexity and complications. Mol. Microbiol. 2002;45:591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- 13.Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madison-Antenucci S, Grams J, Hajduk SL. Editing machines: the complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- 15.Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl Acad. Sci. USA. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnaufer A, Panigrahi AK, Panicucci B, Igo RP, Jr, Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Ernst NL, Palazzo SS, Panigrahi AK, Salavati R, Stuart K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot. Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Li F, Ge P, Hui WH, Atanasov I, Rogers K, Guo Q, Osato D, Falick AM, Zhou ZH, Simpson L. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc. Natl Acad. Sci. USA. 2009;106:12306–12310. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golas MM, Bohm C, Sander B, Effenberger K, Brecht M, Stark H, Goringer HU. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 2009;28:766–778. doi: 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panigrahi AK, Gygi SP, Ernst NL, Igo RP, Jr, Palazzo SS, Schnaufer A, Weston DS, Carmean N, Salavati R, Aebersold R, et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panigrahi AK, Schnaufer A, Carmean N, Igo RP, Jr, Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng J, Schnaufer A, Salavati R, Stuart KD, Hol WG. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 30.Brecht M, Niemann M, Schluter E, Muller UF, Stuart K, Goringer HU. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Drozdz M, Palazzo SS, Salavati R, O'Rear J, Clayton C, Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang X, Falick AM, Nelson RE, Gao G, Rogers K, Aphasizhev R, Simpson L. Disruption of the zinc finger motifs in the Leishmania tarentolae LC-4 (=TbMP63) L-complex editing protein affects the stability of the L-complex. J. Biol. Chem. 2004;279:3893–3899. doi: 10.1074/jbc.M310185200. [DOI] [PubMed] [Google Scholar]
- 33.Law JA, O'Hearn S, Sollner-Webb B. In Trypanosoma brucei RNA editing, TbMP18 (band VII) is critical for editosome integrity and for both insertional and deletional cleavages. Mol. Cell Biol. 2007;27:777–787. doi: 10.1128/MCB.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salavati R, Ernst NL, O'Rear J, Gilliam T, Tarun S, Jr, Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- 36.Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo X, Ernst NL, Stuart KD. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei. Mol. Cell Biol. 2008;28:6939–6953. doi: 10.1128/MCB.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kala S, Salavati R. OB-fold domain of KREPA4 mediates high-affinity interaction with guide RNA and possesses annealing activity. RNA. 2010;16:1951–1967. doi: 10.1261/rna.2124610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang CE, O'Hearn SF, Sollner-Webb B. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Hearn SF, Huang CE, Hemann M, Zhelonkina A, Sollner-Webb B. Trypanosoma brucei RNA editing complex: band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law JA, O'Hearn SF, Sollner-Webb B. Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion. RNA. 2008;14:1187–1200. doi: 10.1261/rna.899508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson DG, Kowalczykowski SC. SSB protein controls RecBCD enzyme nuclease activity during unwinding: a new role for looped intermediates. J. Mol. Biol. 1998;282:275–285. doi: 10.1006/jmbi.1998.2013. [DOI] [PubMed] [Google Scholar]
- 43.Eggington JM, Haruta N, Wood EA, Cox MM. The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 2004;4:2. doi: 10.1186/1471-2180-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 45.Lohman TM, Bujalowski W, Overman LB. E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends Biochem. Sci. 1988;13:250–255. [PubMed] [Google Scholar]
- 46.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol. Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bogden CE, Fass D, Bergman N, Nichols MD, Berger JM. The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell. 1999;3:487–493. doi: 10.1016/s1097-2765(00)80476-1. [DOI] [PubMed] [Google Scholar]
- 48.Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- 49.Murzin AG. OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnaufer A, Wu M, Park YJ, Nakai T, Deng J, Proff R, Hol WG, Stuart KD. A protein-protein interaction map of trypanosome ∼20S editosomes. J. Biol. Chem. 2010;285:5282–5295. doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carnes J, Soares CZ, Wickham C, Stuart K. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J. Biol. Chem. 2011;286:19320–19330. doi: 10.1074/jbc.M111.228965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarun SZ, Jr, Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu M, Park YJ, Pardon E, Turley S, Hayhurst A, Deng J, Steyaert J, Hol WG. Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies. J. Struct. Biol. 2011;174:124–136. doi: 10.1016/j.jsb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 56.Lam AY, Pardon E, Korotkov KV, Hol WG, Steyaert J. Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus. J. Struct. Biol. 2009;166:8–15. doi: 10.1016/j.jsb.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korotkov KV, Pardon E, Steyaert J, Hol WGJ. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure. 2009;17:255–265. doi: 10.1016/j.str.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 59.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 60.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 61.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 62.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 65.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 66.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 67.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 70.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 71.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang CY, Hsu CH, Sun YJ, Wu HN, Hsiao CD. Complexed crystal structure of replication restart primosome protein PriB reveals a novel single-stranded DNA-binding mode. Nucleic Acids Res. 2006;34:3878–3886. doi: 10.1093/nar/gkl536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 74.Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J. Mol. Biol. 2009;388:508–519. doi: 10.1016/j.jmb.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 75.Lescar J, Pellegrini M, Souchon H, Tello D, Poljak RJ, Peterson N, Greene M, Alzari PM. Crystal structure of a cross-reaction complex between Fab F9.13.7 and guinea fowl lysozyme. J. Biol. Chem. 1995;270:18067–18076. doi: 10.1074/jbc.270.30.18067. [DOI] [PubMed] [Google Scholar]
- 76.Igonet S, Vulliez-Le Normand B, Faure G, Riottot MM, Kocken CH, Thomas AW, Bentley GA. Cross-reactivity studies of an anti-Plasmodium vivax apical membrane antigen 1 monoclonal antibody: binding and structural characterisation. J. Mol. Biol. 2007;366:1523–1537. doi: 10.1016/j.jmb.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 77.Jedrzejczak R, Dauter M, Dauter Z, Olszewski M, Dlugolecka A, Kur J. Structure of the single-stranded DNA-binding protein SSB from Thermus aquaticus. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1407–1412. doi: 10.1107/S0907444906036031. [DOI] [PubMed] [Google Scholar]
- 78.Saikrishnan K, Manjunath GP, Singh P, Jeyakanthan J, Dauter Z, Sekar K, Muniyappa K, Vijayan M. Structure of Mycobacterium smegmatis single-stranded DNA-binding protein and a comparative study involving homologus SSBs: biological implications of structural plasticity and variability in quaternary association. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1140–1148. doi: 10.1107/S0907444905016896. [DOI] [PubMed] [Google Scholar]
- 79.Saikrishnan K, Jeyakanthan J, Venkatesh J, Acharya N, Sekar K, Varshney U, Vijayan M. Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications. J. Mol. Biol. 2003;331:385–393. doi: 10.1016/s0022-2836(03)00729-0. [DOI] [PubMed] [Google Scholar]
- 80.Yang C, Curth U, Urbanke C, Kang C. Crystal structure of human mitochondrial single-stranded DNA binding protein at 2.4 A resolution. Nat. Struct. Biol. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 81.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DiDonato M, Krishna SS, Schwarzenbacher R, McMullan D, Jaroszewski L, Miller MD, Abdubek P, Agarwalla S, Ambing E, Axelrod H, et al. Crystal structure of a single-stranded DNA-binding protein (TM0604) from Thermotoga maritima at 2.60 A resolution. Proteins. 2006;63:256–260. doi: 10.1002/prot.20841. [DOI] [PubMed] [Google Scholar]
- 83.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl Acad. Sci. USA. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ernst NL, Panicucci B, Igo RP, Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3' terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 85.Gao G, Simpson AM. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- 86.Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 87.Kang X, Rogers K, Gao G, Falick AM, Zhou S, Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl Acad. Sci. USA. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cruz-Reyes J, Zhelonkina AG, Huang CE, Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ernst NL, Panicucci B, Carnes J, Stuart K. Differential functions of two editosome exoUases in Trypanosoma brucei. RNA. 2009;15:947–957. doi: 10.1261/rna.1373009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang CE, Cruz-Reyes J, Zhelonkina AG, O'Hearn S, Wirtz E, Sollner-Webb B. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J. 2001;20:4694–4703. doi: 10.1093/emboj/20.17.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.