Abstract
SP1 is a ubiquitous transcription factor that is involved in the regulation of various house-keeping genes. It is known that it acts by binding to a double-stranded consensus motif. Here, we have discovered that SP1 binds also to a non-canonical DNA structure, a G-quadruplex, with high affinity. In particular, we have studied the SP1 binding site within the promoter region of the c-KIT oncogene and found that this site can fold into an anti-parallel two-tetrad G-quadruplex. SP1 pull-down experiments from cellular extracts, together with biophysical binding assays revealed that SP1 has a comparable binding affinity for this G-quadruplex structure and the canonical SP1 duplex sequence. Using SP1 ChIP-on-chip data sets, we have also found that 87% of SP1 binding sites overlap with G-quadruplex forming sequences. Furthermore, while many of these immuoprecipitated sequences (36%) even lack the minimal SP1 consensus motif, 5′-GGGCGG-3′, we have shown that 77% of them are putative G-quadruplexes. Collectively, these data suggest that SP1 is able to bind both, canonical SP1 duplex DNA as well as G-quadruplex structures in vitro and we hypothesize that both types of interactions may occur in cells.
INTRODUCTION
SP1 is a transcription factor that plays an important role in many cellular activities such as metabolism, cell growth, differentiation, angiogenesis and apoptosis (1). SP1 contains three Cys2His2-type zinc finger motifs and it is generally accepted that its mode of action is mediated mainly by binding to the decanucleotide consensus sequence 5′-(G/T)GGGCGG(G/A)(G/A)(C/T)-3′ in double-stranded DNA (dsDNA) (2). In 2004, a study using chromatin immunoprecipitation (ChIP) and high-density oligonucleotide arrays, identified SP1 binding sites in human chromosomes 21 and 22 (3). SP1 binding sites were not only found within the 5′ promoter regions of coding genes and CpG islands, but also within or 3′ to well-characterized genes. One noteworthy observation was that only a minority of the binding sites contained the consensus binding motif for SP1 using the motif finder MDscan (4). The remaining sites lacked the exact consensus or closely related-sequence variants, raising the question of how SP1 might recognize these other genomic locations (3). While SP1 binding to non-consensus sites may be mediated by alternative mechanisms such as indirect interactions with the DNA via other proteins, or to sequences more distant from the consensus (5), this prompted us to consider the potential recognition by SP1 to other DNA secondary structures. As the SP1 binding site contains consecutive guanine repeats, this raises the possibility of G-quadruplex formation. G-quadruplexes adopt a defined four-stranded core secondary structure by stacking of several consecutive G-quartets. Structural and biophysical experiments have convincingly demonstrated that G-rich sequences, consisting of repeated blocks of guanines, can fold into stable G-quadruplex structures (6). There is growing evidence for the existence and functional consequence of G-quadruplex structure formation in vivo (7–10). In the absence of G-quadruplex stabilizing factors and in the presence of the complementary cytosine-rich strand, the G-quadruplex structure is in competition with its double-stranded counterpart and may therefore be energetically unfavourable. However, during important biological processes, such as transcription or replication, parts of the DNA strands may exist as single strands and in this state the existence of a G-quadruplex-folded structure is more likely (11,12). Additionally, these guanine-rich sequences are significantly enriched around transcription start sites compared to the genome-wide average suggesting a role in gene transcription regulation (13–15). A computational study in 2008 by Todd and Neidle (16) revealed a correlation between putative G-quadruplex sequences (PQS) in cis-upstream regions of the human genome to the SP1-binding consensus sequence. This study showed that many PQS occurring in the immediate upstream region of the transcription start site contain the SP1 consensus sequence. Previous work has also shown that artificially engineered zinc finger proteins bind G-quadruplexes with high affinity (17–19). One such engineered Cys2His2- zinc finger protein, named Gq1, showed high specificity binding in vitro to the intramolecular G-quadruplex formed by the human telomeric sequence 5′-(GGGTTA)5-3′ (19). CNBP, a natural zinc-finger binding protein, has been shown to bind to the c-MYC G-quadruplex (20). More recently, zinc finger transcription factors have been associated with G-quadruplex motifs genome wide in different mammals (21). Collectively, these observations have prompted us to examine whether G-quadruplex structures are recognized by the SP1 transcription factor.
In this study, we have focused on a known SP1 binding site within the human c-KIT promoter. The c-KIT gene encodes a tyrosine kinase receptor and plays a critical role in normal cell growth and stem-cell proliferation and differentiation (22). It also plays a key role in the development of melanocytes, germ cells and hematopoietic cells and dysregulation is linked to the formation of various cancers (23). The regulation of c-KIT expression is complex and relatively little is known about the factors regulating its expression. The c-KIT gene has a minimal core-promoter region of 125 bp proximal to the transcription start site, which is required for maximal activity of c-KIT transcription (23). This region is guanine-rich and lacks a TATA-box. It has been reported that two potential G-quadruplex forming sequences within this core-promoter region, named c-kit1 and c-kit2, with three stacking tetrads can be formed in vitro (Figure 1) (24,25). Extensive biophysical analyses using circular dichroism, thermal difference spectra and UV spectroscopy have described these structures in detail and have revealed all-parallel structures with melting points >60°C under physiological aqueous-buffer conditions. Furthermore, high-resolution NMR spectroscopic studies have provided detailed 3D structures of both G-quadruplexes, c-kit1 (26) and c-kit2 (27,28). C-kit1 and 2 flank a 30 bp long region, which contains several GG-repeats, and previous reporter assays have identified a single SP1 binding site in the proximal promoter region 80–101 bp upstream of the 5′ transcription initiation site (23).
Figure 1.
Schematic representation of the human c-KIT promoter region and the corresponding G-quadruplex sequences. C-kit1 and c-kit2 refer to previously identified G-quadruplexes, while c-kit* highlights a new predicted two-tetrad G-quadruplex within the SP1 binding site (blue oval). The arrow indicates the transcription start site.
MATERIALS AND METHODS
Sample preparation
DNA oligonucleotides were purchased from Sigma-Aldrich. In general, DNA oligomer solutions were prepared in 50 mM Tris–HCl (pH 7.4) containing 0–100 mM KCl. Samples were heated to 95°C for 5 min and annealed at a cooling rate of 0.2°C/s to room temperature.
Purification of SP1
Nuclear cell extract from HeLa cells was purchased from Cilbiotech, and purified as previously described (29), except 5′-biotinylated dsDNA with sequence 5′-TCG ATG GGC GGA GTT AGG GGC GGG ACT A-3′ and its reverse complement was used for the affinity column. All procedures were performed at 4°C. Briefly, 50 ml crude nuclear extract from 109 HeLa cells was applied to a WGA-agarose resin column (Sigma-Aldrich, 2 ml) pre-equilibrated with a high salt buffer (50 mM Tris, 0.42 M KCl, 20% v/v glycerol, 10% sucrose, 5 mM MgCl2, 0.1 mM EDTA, 1 mM PMSF, 1 mM sodium metabisulfite, 2 mM dithiothreitol). SP1 was eluted with buffer Z [25 mM HEPES pH 7.5, 12.5 mM MgCl2, 20% v/v glycerol, 0.1% v/v Nonidet P-40, 0.01 mM Zn(OAc)2, 1 mM dithiothreitol, 100 mM KCl] containing 0.3 M GlcNAc. The eluate from this purification step was re-applied to a biotinylated dsDNA affinity column. SP1 elution was achieved by washing with buffer Z containing 1 M KCl. Subsequent dialysis against 2× storage buffer [24 mM HEPES pH 7.5, 100 mM KCl, 12 mM MgCl2, 0.01 mM Zn(OAc)2] followed by addition of 50% v/v glycerol yielded 1 ml SP1 at a concentration of 28 nM.
Nuclear magnetic resonance spectroscopy
Nuclear magnetic resonance (NMR) experiments were conducted on a Bruker Avance 500 MHz instrument equipped with a TCI cryo probe. Water suppression was achieved using excitation sculpting. The sample for NMR analysis was dissolved in 500 μl of H2O/D2O (3:1) containing 100 mM KCl, 20 mM PBS (pH 7.4) to a concentration of 100 μM and annealed as above. Variable temperature NMR spectra were recorded at 25°C, 37°C and 44°C. The sample was equilibrated at these temperatures for 20 min prior to data acquisition.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were obtained on a Jasco J-810 spectropolarimeter using software supplied by the manufacturer. Samples measuring 20 μM were prepared containing different concentrations of KCl and annealed as described above. The samples (200 μl) were placed in a quartz cuvette with a path length of 0.1 cm. Measurements were made over a range of 220–320 nm at 25°C. Each curve represents an average of three scans that has been baseline corrected (buffer only).
Thermal denaturation–renaturation spectra were recorded at 290 nm over a temperature range of 15–90°C at a heating/cooling rate of 1°C/min. Melting temperature was estimated from the maximum of the first derivative plot.
Thermal difference spectra
Thermal difference spectra were recorded on a Varian Cary 100 using a 0.1 cm path length. The samples (40 μM) were prepared in 50 mM Tris–HCl (pH 7.4) containing 100 mM KCl. Oligonucleotides were annealed as described above. Absorbance was recorded over a range of 220–320 nm.
Computational analyses
Published microarray ChIP-on-chip data (30) for the genome-wide binding of SP1 were obtained from GEO [accession GSE16078, (31)]. ChIP-enriched regions were identified using the ‘Ringo’ package in the R/bioconductor statistical environment (32,33) and analysed for the presence of the minimal SP1 consensus sequence ‘GGGCGG’ and its reverse complement ‘CCGCCC’. For enriched regions containing the SP1 consensus sequence, a region spanning 20 bp upstream and 20 bp downstream of the SP1 motif was analysed for potential quadruplex forming sequences with the Quadparser algorithm using the search parameters G2+N(1-7)G2+N(1-7)G2+N(1–7)G2+, where ‘G’ was a guanine and ‘N’ any nucleotides and 2+ stands for two Gs or more (34). ChIP-enriched regions either with or without the SP1 minimal consensus motif were analysed for PQS with Quadparser. To test statistical significance of the observed association between SP1-enriched regions and potential quadruplex formation, each of the enriched sequences were permuted to generate random sequences of the same length and base composition. The randomized sequences were analysed by Quadparser with the same parameters. This was repeated 10 000 times and compared with the observed number of quadruplex-associated sequences to generate an empirical P-value.
Fluorescence polarization spectroscopy
Fluorescence polarization (FP) measurements were carried out using a PHERAstar Plus microplate reader at room temperature (20°C) in a 384-well plate (corning, low volume, black flat bottom, polystyrene) and a final volume of 10 μl. 3′-fluorescein-labelled oligonucleotides (2 nM) 5′-GGC GAG GAG GGG CGT GGC CGG C-TTTTT-fluorescein-3′ and the reverse complement 5′-GCC GGC CAC GCC CCT CCT CGC C-3′ or the labelled guanine-rich strand alone, respectively (prior annealed in 50 mM Tris–HCl, pH 7.4 containing 100 mM KCl) were incubated with SP1 in buffer Z for 30 min at room temperature. Measurements were carried out using excitation and emission wavelengths of 480 and 520 nm, respectively. For each sample, xz parallel measurements with an integration time of 2 s were averaged. When comparing dsDNA in the presence and in the absence of SP1, a Z'-factor of 0.86 could be obtained (35). Observed binding-data were fitted to a hyperbola curve using GraphPad Prism Software.
Enzyme-linked immunosorbent assay
All binding reactions were carried out in buffer Z containing 25 mM HEPES (pH 7.5), 12.5 mM MgCl2, 1 mM dithiothreitol, 20% v/v glycerol, 0.1% v/v nonidet P-40 and 0.1 M KCl. A Highbind Streptaplate (Roche) was hydrated with 1 × PBS buffer for 30 min and blocked with buffer Z containing 3% BSA prior reaction. Subsequently, 200 μl of a 200 nM solution of biotinylated DNA (5′-GGC GAG GAG GGG CGT GGC CGG CTTTTT-biotin-3′) were added per well and allowed to attach for 30 min at 37°C with gentle shaking. Wells were then washed three times with buffer Z containing 3% BSA. SP1 protein was diluted in buffer Z containing 3% BSA and 50 μl added in each well. After incubation for 1 h at room temperature, plates were washed three times with buffer Z. For detection, 50 μl of a rabbit polyclonal SP1 antibody (Millipore) at 1:500 dilution in buffer Z were added per well and incubated for 1 h at room temperature. After washing three times with buffer Z, a HRP-conjugated goat anti-rabbit (Cell Signalling Technology) diluted 1:2000 in buffer Z with 3% BSA was added and incubated for 45 min at room temperature. Wells were washed three times with buffer Z and peroxidase activity detected by adding 100 μl of the BM blue POD substrate (Roche). Reactions were stopped by the addition of 1 M H2SO4. Absorbance at 450 nm was measured using a PowerWave XS Microplate Reader (BioTek Instruments).
SP1 pull-down and western blotting
Streptavidin-coated magnetic beads (Promega) were washed three times with 500 µl of 0.5 × SSC buffer and three times with buffer Z containing 3% of BSA. 200 µl of annealed biotinylated DNA samples (1 µM) were incubated with the beads for 30 min at room temperature. Beads were then washed three times with 500 µl of buffer Z containing 3% of BSA and blocked with the same buffer for 30 min. All subsequent procedures were performed at 4°C. A total of 1 µl (12.8 µg total protein) of a HeLa nuclear-cell extract was incubated in buffer Z containing 3% of BSA in the presence of 0.4 µg/µl salmon sperm DNA for 10 min in a total volume of 500 µl. The reaction mixture was then added to the beads and incubated for another 20 min, followed by 6× 200 µl washing with buffer Z. The beads were resuspended in 20 µl of Laemmli buffer and boiled for 2 min. A total of 18 µl of the bound fraction were then separated on a 8–16% gradient Tris–Glycine polyacrylamide gel (Invitrogen) and transferred to a nitrocellulose membrane. SP1was identified by immunoblotting using the rabbit polyclonal antibody diluted 1:5000. A goat anti-rabbit secondary antibody labelled with IRDye 680 nm (LI-COR) was used for detection with subsequent imaging performed on the Odyssey® Infrared Imaging System (LI-COR).
RESULTS
The SP1 binding site in the c-KIT promoter can fold into a stable and anti-parallel G-quadruplex
The SP1 binding site within the human c-KIT promoter is critical for the maximal transcriptional activity, and is flanked by two well-characterized G-quadruplex motifs, named c-kit1 and c-kit2 (25,36). On further examination of this region, we noted that a third potential G-quadruplex forming sequence was located within the SP1 binding site (named c-kit*, Figure 1). This sequence consists of consecutive GG-repeats with the potential to fold into a non-canonical G-quadruplex structure with two stacked guanine tetrads rather than the three tetrads that have commonly featured in most genomic G-quadruplexes. While some G-quadruplex motif search algorithms have been based on a minimum of three G-tetrads (34,37), biophysical experiments have supported G-quadruplex formation with just two tetrads, as it is the case for the thrombin binding DNA aptamer 5′-GGT TGG TGT GGT TGG-3′ (38) and more recently, a quadruplex motif within the promoter of human thymidine kinase 1 (39).
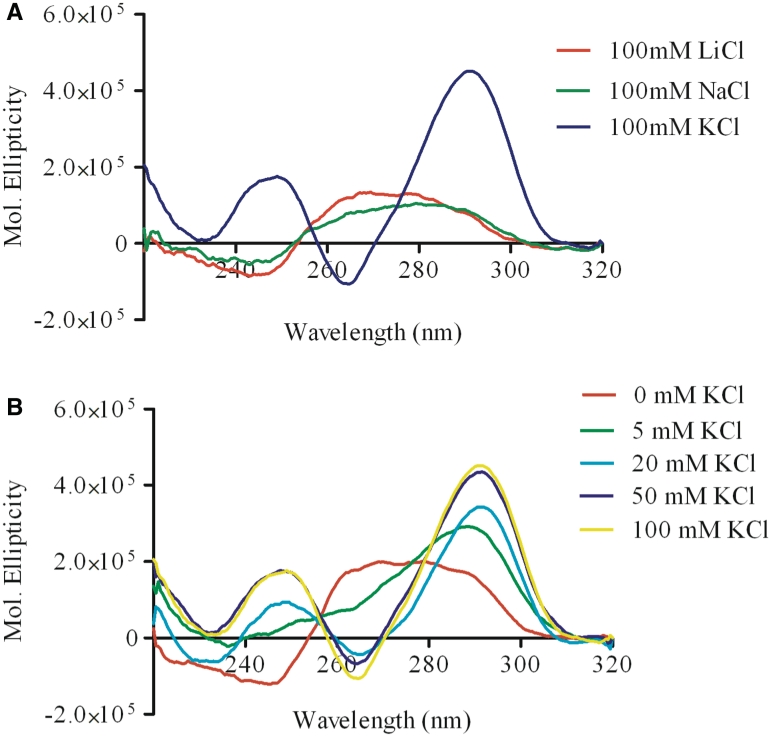
Given the possibility of a potential two-tetrad G-quadruplex in the c-KIT promoter, we employed a range of biophysical in vitro techniques to examine the potential for c-kit* to form a G-quadruplex structure. CD spectra of the annealed oligomer showed a maximum positive signal at 291 and 249 nm and a corresponding negative signal at 263 nm in the presence of 100 mM potassium chloride, but not in the presence of equivalent concentration of sodium or lithium chloride (Figure 2A). These band assignments are consistent with an anti-parallel topology (40), which is to our knowledge, the first example of a non-telomeric human DNA sequence that folds into an anti-parallel two tetrad quadruplex structure. G-quadruplex formation is dependent on the presence of physiological concentrations of potassium (41), and the c-kit* structure showed typical strong potassium dependency typical for a G-quadruplex structure (42) (Figure 2B).
Figure 2.
(A) CD analysis showing the folding of the c-kit* oligonucleotide [(20 μM, (5′-GGC GAG GAG GGG CGT GGC CGG C-3′)] in different salt conditions. No formation of the G-quadruplex could be observed in the presence of lithium chloride (red) and sodium chloride (green). When annealed in the presence of potassium chloride (blue), the spectra showed characteristic bands centred at 290 and 250 nm indicative of the formation of an anti-parallel G-quadruplex. (B) CD analysis showing the dependency on KCl for G-quadruplex formation of the c-kit* oligonucleotide (20 μM). Typical G-quadruplex structure formation is promoted by increasing concentration of KCl. 0 mM KCl (red), 5 mM KCl (green), 20 mM KCl (light bue), 50 mM KCl (dark blue) and 100 mM KCl (yellow).
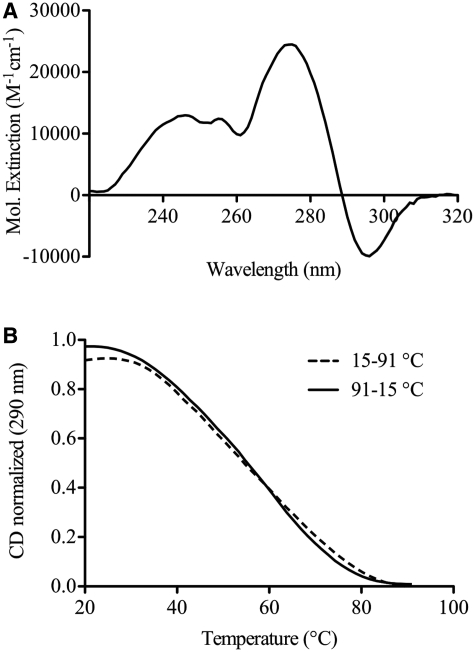
Thermal difference spectral (TDS) analysis supports that c-kit* forms a G-quadruplex structure. TDS provides structural insights into nucleic acid structures, as each spectral shape is unique to particular structures (43). For c-kit*, the spectrum obtained by recording the UV absorbance in a temperature range above and below the melting point showed major positive bands centred at 246 and 275 nm and a negative band centred at 296 nm, a profile characteristic of a G-quadruplex motif (Figure 3A). In agreement with our CD spectra, no G-quadruplex profile can be observed in the presence of 100 mM sodium chloride (Supplementary Figure S1). We also performed CD-melting experiments with repeated heating and cooling of the sample that resulted in the determination of a melting temperature of 55°C. The heating and cooling traces superimposed and the lack of hysteresis was indicative of reversible and intramolecular G-quadruplex formation with fast folding kinetics (Figure 3B). CD melting experiments with various oligomer concentrations (2, 10 and 40 μM) resulted in superimposable melting curves indicating the formation of an intramolecular G-quadruplex (see Supplementary Figure S2).
Figure 3.
(A) The TDS spectrum of the c-kit* oligonucleotide (5′-GGC GAG GAG GGG CGT GGC CGG C-3′) was obtained from the subtraction of the 15°C spectrum from the 91°C spectrum. This indicates the formation of an anti-parallel G-quadruplex due to positive band centred at 246 and 275 nm and a negative band at 296 nm. (B) CD thermal denaturation–renaturation studies show fast-folding kinetics with little hysteresis. The broken line represents recorded data while heating from 15°C to 91°C and the solid line while cooling form 91°C to 15°C (20 μM DNA).
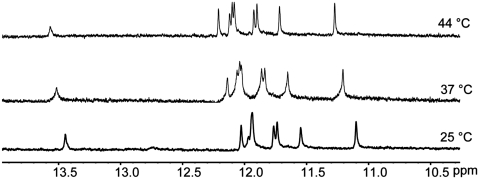
One-dimensional 1H-NMR spectroscopy on c-kit* folded in potassium showed distinct signals for the exchangeable iminoprotons resonating between 11–12.5 ppm characteristics of the formation of a G-quadruplex (44). Upon heating to 44°C, the imino signals sharpened up to afford eight clearly resolvable singlets. This is consistent with the formation of two stacked guanine tetrads with each guanine in a unique environment (Figure 4) and suggests a homogeneous folded structure. The slight downfield shift of the imino signals (∼0.2 ppm over a 19°C temperature range) at these temperatures might indicate an increase in the dynamics of the bases near the melting temperature (55°C). Beside the imino signals, another singlet was also observed around 13.5 ppm, indicating a hydrogen bond, which we suggest, may be from a loop base.
Figure 4.
The imino region of the 1H NMR spectra of the c-kit* sequence (5′-GGC GAG GAG GGG CGT GGC CGG C-3′) recorded at 25°C (bottom spectrum), 37°C (middle spectrum) and 44°C (top spectrum) showing eight defined peaks. Conditions: 100 μM DNA, 100 mM KCl and 20 mM PBS (pH 7.4).
SP1 displays high-affinity binding to a novel G-quadruplex in the c-KIT promoter
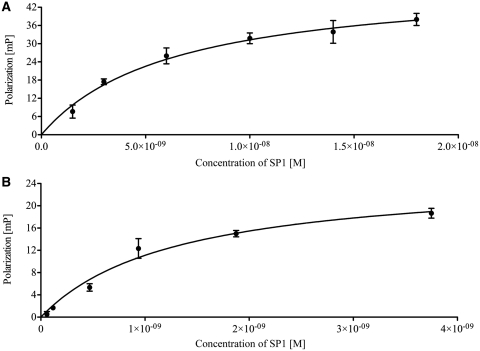
As the c-kit* G-quadruplex motif falls within the SP1 binding site in the c-KIT promoter, we next examined whether SP1, purified from HeLa cells (29) could bind to the corresponding G-quadruplex structure. We established FP and enzyme-linked immunosorbant assay (ELISA) as independent methods to evaluate the binding of SP1 to the c-kit* quadruplex and the canonical double-stranded binding site (Supplementary Figure S3). For FP, a 3′-fluorescein labelled oligonucleotide spanning the SP1 site, either double stranded (ds c-kit*) or the respective guanine-rich strand alone (ss c-kit*) that was annealed to form a G-quadruplex, were incubated with SP1 protein for 30 min at 20°C. The equilibrium dissociation constants (Kd) for the SP1–DNA interaction were determined by non-linear regression by fitting to a hyperbolic binding curve. We found that, for SP1 binding to ds c-kit* and ss c-kit*, the Kd-values were 6.3 ± 1.7 nM and 1.3 ± 0.3 nM, respectively (Figure 5). We then confirmed these observations by an ELISA approach. Biotinylated derivatives of ds c-kit* or ss c-kit* oligonucleotides were immobilized on streptavidin-coated microwells and SP1 protein added to the wells at different concentrations to evaluate binding (see Supplementary Figure S4). The Kd values determined by ELISA for the ds c-kit* and ss c-kit* G-quadruplex were 6.2 ± 2.1 and 8.5 ± 4.7 nM, respectively, close to the values determined by FP. Our obtained data are in agreement with previously reported Kd values (low nanomolar range) (45). Furthermore, we performed single point ELISA experiments with other G-quadruplex forming sequences found in the promoter region from DNMT1 (mouse), c-KIT (human), and a repeat sequence from the Simian virus 40 (see Supplementary Figure S5), demonstrating that SP1 is generally recognizing G-quadruplex structures with high affinity.
Figure 5.
SP1 protein binding to the c-KIT double-stranded SP1 binding site or to the single stranded c-kit* G-quadruplex by fluorescence polarization. (A) Binding curve for SP1 binding to double-stranded c-kit* sequence, 5′-GGC GAG GAG GGG CGT GGC CGG C-TTTTT-fluorescein-3′ annealed to its reverse complement 5′-GCC GGC CAC GCC CCT CCT CGC C-3′ (Kd 6.3 ± 1.7 nM). (B) Binding curve for SP1 binding to single-stranded c-kit* G-quadruplex, 5′-GGC GAG GAG GGG CGT GGC CGG C-TTTTT-fluorescein-3′ (Kd 1.3 ± 0.3 nM).
SP1 binds the c-kit* G-quadruplex from a HeLa nuclear extract
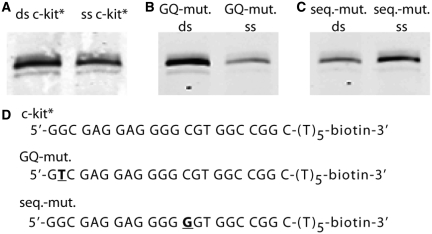
We next aimed to confirm that native SP1 present in human cells showed affinity for the c-kit* G-quadruplex. To achieve this, we assessed the ability of SP1 protein in nuclear extracts prepared from HeLa cells to be captured by immobilized 3′-biotinylated c-kit* oligonucleotides. First, the pull down experiment was established with a positive control using ds c-kit* and a negative control where beads not coupled to any DNA were used (see Supplementary Figure S6). As seen in the Western blots shown in Figure 6A, SP1 was pulled out of the nuclear cell extract with both, the double-stranded sequence as well as the folded c-kit* oligonucleotide.
Figure 6.
SP1 pull-down from Hela cell nuclear extracts by c-kit* G-quadruplex structures. Nuclear extracts were incubated with biotinylated oligonucleotides bound to streptavdin beads in buffer Z containing 100 mM KCl and the resulting proteins analysed by western blotting using an SP1 antibody. The detected SP1 protein appears as a multimer around 115 and 95 kDa, consistent with the known profile of SP1 in Hela cells (29). (A) SP1 is pulled down by the double-stranded consensus motif (ds c-kit*) and the ss c-kit* G-quadruplex. (B) Less SP1 is pulled down by a single stranded G-quadruplex mutation (GQ-mut.) but still binds to the SP1 double-stranded oligonucleotide with the same mutation (C) Conversely, more SP1 is pulled-down by a G-quadruplex structure (seq-mut ss) carrying a mutation known to disrupt SP1 binding to the double-strand SP1 motif (seq.-mut. ds). (D) DNA sequences of the immobilized oligomers (either single- or double-stranded).
Next, we performed pull-down experiments using two mutated c-kit* oligonucleotide to assess whether a mutation that disrupts the G-quadruplex or the double-stranded SP1 binding site would prohibit SP1 protein binding. The first mutation (GQ-mut) does not alter the SP1 binding site in the duplex, but disrupts the G-quadruplex formation in the single-stranded DNA, as confirmed by measurement of the CD and TDS spectrum in the presence of 100 mM KCl (see Supplementary Figure S8). Incubating this oligonucleotide with nuclear extracts only gave a faint SP1 band compared with dsDNA from the same sequence (Figure 6B, Supplementary Figure S7 for relative band-intensity quantification). The second mutation (seq-mut) is based on the observations of Letovsky and Dynan showing the importance of the central C within the Sp1 recognition site 5′-GGGGCGGGGC-3′ for DNA binding. Mutation of this C to A, G or T resulted in reduced binding (45). Following this observation, in the c-kit* context, a central C-G mutation should reduce SP1 binding but would still allow G-quadruplex formation (see Supplementary Figure S8). Indeed, the amount of SP1 pulled-down with a mutated double-stranded oligonucleotide was reduced, while binding to the mutated single stranded c-kit* G-quadruplex was maintained (Figure 6C, Supplementary Figure S7 for band quantification). Overall, these results support interaction of SP1 protein with G-quadruplex structures, in addition to the canonical double-stranded consensus sequence, in the context of a nuclear extract.
Genome-wide SP1-binding sites correlate with G-quadruplex structure forming sequences
After showing that SP1 binds to a G-quadruplex in the c-kit sequence context, we next investigated whether SP1 binding sites throughout the genome have potential for G-quadruplex formation. To complement the approach of previous reports (13,16,21), who asked computationally whether predicted G-quadruplex sequences overlapped with predicted SP1 consensus, we analysed genome-wide SP1 ChIP-on-chip data (30) for the correlation of SP1 binding sites to potential G-quadruplexes using a sequence search algorithm (34).
Genomic regions with a log2 fold binding ratio of ≥1.5 were identified as significantly enriched, resulting in 5810 genomic fragments. In order to avoid eliminating any potential SP1 consensus binding sites, we employed a minimal consensus sequence ‘GGGCGG’ and its reverse complement ‘CCGCCC’, and identified 7204 and 6917 motifs, respectively. To investigate whether these SP1 sites correlate with a G-quadruplex forming sequence, we defined a 46 bp window, 20 bp upstream and downstream of the minimal consensus, to search for potential quadruplex forming sequences (PQS) using Quadparser software (34). This algorithm considered GG-repeats of the type G2+N(1−7)G2+N(1−7)G2+N(1−7)G2+, where ‘G’ was a guanine, ‘N’ any of the four nucleotides and ‘2+’ stands for two Gs or more. It should be noted that this analysis included predicted quadruplexes with at least two tetrads, in the light of our findings with the c-kit* G-quadruplex. The analysis revealed that 87% of the enriched SP1 motif sequences overlapped with a PQS, indicating that the majority of experimentally determined SP1 protein binding sites correlate with G-quadruplex structure forming sequences.
Given that the majority of the SP1 ChIP-enriched sequences were reported to lack the consensus SP1 binding motif in a limited analysis of chromosomes 21 and 22 (3), we analysed genome-wide ChIP-enriched sequences (30). In this case, 2083 sequences (36% of all enriched sequences) were found to lack the minimal SP1 consensus binding motif. Despite this, the majority of these fragments (77.4%) were found to contain at least one PQS (Table 1). Of the remaining 3727 enriched sequences (64%) containing one or more SP1 consensus motifs with the sequence ‘GGGCGG’ or its reverse complement ‘CCGCCC’, 99.7% contained at least one PQS. Permutation analyses of respective SP1-enriched regions, showed that none of the permutations had the same or a higher number of PQS sequences, giving empirical P < 0.0001. These data demonstrate a strong correlation between experimentally determined SP1 binding sites and G-quadruplex structure forming sequences regardless of the presence or absence of the SP1 consensus motif.
Table 1.
PQS in SP1 ChIP-on-chip enriched sequences (30) with and without the SP1 consensus motif sequence (total number of enriched sequences = 5810)
| Sequence characteristics | Number of sequences | Number of sequences with ≥ 1 PQS | Percentage of all 5810 sequences |
|---|---|---|---|
| ≥1 consensus binding motif (‘GGGCGG’) and reverse complement (‘CCGCCC’) | 3727 | 3716**** | 99.7 |
| number of sequences without the binding motif | 2083 | 1612**** | 77.4 |
****P < 0.0001
DISCUSSION
We have identified a previously unknown G-quadruplex folding structure motif situated 80–101 bp upstream of the transcription initiation site of c-KIT that also completely overlaps with a known SP1 binding site that is known to be essential for the maximum promoter activity (23). This G-quadruplex structure displayed a stable intramolecular all-anti-parallel topology with eight clearly resolved guanine imino protons in the NMR spectrum, consistent with a two-tetrad system. The thermal stability was determined by UV-melting (Figure 3) and its melting transition point of 55°C is in good agreement with the Quadpredict software estimating 59°C ± 10°C (46). This suggests that the two-tetrad structure from this genomic sequence could, in principle, exist in physiological conditions. There has been relatively little attention paid to PQS with only two-tetrads in the literature, with most studies generally considering G-quadruplexes with three or more tetrads. Our finding suggests that motifs capable of forming two-tetrads should not be excluded. It is noteworthy that this particular quadruplex is unusual in being the first all-anti-parallel G-quadruplex except from human non-telomeric DNA. Indeed, most DNA G-quadruplexes studied have tended to adopt a parallel or a mixed parallel/anti-parallel topology.
Previous reports have shown that zinc finger proteins have the potential to bind non-B DNA structures. Studies on an artificial, engineered zinc finger protein have shown that a relatively modest adaptation of a classical three zinc finger transcription factor can lead to a protein that recognizes the G-quadruplex fold of DNA (17,18). Xodo and co-workers demonstrated that the murine Myc-associated zinc finger protein, MAZ, recognizes the duplex and G-quadruplex conformations of the GA-element in the KRAS promoter (47). Our data show that the zinc finger protein, SP1, can bind to the G-quadruplex DNA structure and double-stranded structure within the target site of the c-KIT promoter with low nanomolar affinity. We performed SP1 pull-down experiments from HeLa cell nuclear extracts to determine the binding of SP1 in the presence of numerous other naturally occurring proteins known to interact with G-quadruplex DNA that include Poly(ADP-Ribose)polymerase-1 (48), Nucleolin (49), FANCJ (50) among others. We intentionally compare only pull-downs within the same sequence context either double- or single-stranded considering that mutations alter the sequence context in a way that SP1 might compete with different and additional proteins. The pull-down assay showed that both, ds c-kit* DNA and ss c-kit* G-quadruplex, isolated SP1 protein, thus demonstrating binding to a G-quadruplex in the presence of other cellular proteins. This observation is consistent with our FP and ELISA binding data using purified SP1; however, at this point we cannot rule out indirect effects such as protein–protein interactions. The latter effect might also be the reason why the pull-down using ss c-kit* G-quadruplex showed to be less effective than the ds c-kit*.
A selective mutation in the duplex binding site for SP1 led to reduced efficiency in the isolation of SP1 from nuclear extract, as anticipated based on previous reports in a different sequence context (45). A mutation in c-kit* that destabilizes G-quadruplex formation also led to a reduction in the efficiency of SP1 isolation (Figure 6). These findings support our hypothesis that SP1 recognizes the G-quadruplex DNA structure as an alternative-binding motif.
A computational study (16) has previously suggested that there might be a link between SP1 binding sites and putative G-quadruplex forming sequences. That particular study was established using the minimal consensus sequences GGGCGG rather than experimental SP1 binding data. Our analysis used genome wide SP1 ChIP-on-chip data and showed that there was a significant overlap (P < 0.0001) between G-quadruplexes and SP1 binding sites. However, one has to be cautious interpreting the P-values because potential guanine quadruplex sequences are non-randomly distributed in the genome as suggested in previous analyses (34,37) and artificial randomization might not reflect real genomes. Our binding data obtained by FP, ELISA and pull-down, together with the fact that the majority of the enriched sequences with no consensus could possibly fold into a G-quadruplex, gives new insight into previously unidentified binding targets for SP1 other than the generic double stranded consensus sequence.
These data raise a number of questions regarding how the recognition of a transcription factor such as SP1 to G-quadruplex structures might have relevance for the cellular function. In relation to a potential transcription-related role, we have noted that SP1 elements protect CpG islands from de novo methylation (51). In addition, a genome-wide study has shown that G-quadruplex structures restricts methylation of CpG dinucleotides (52). Thus, one possibility is that SP1 might bind to G-quadruplex regions and prevent subsequent methylation of the sequence. Many promoter regions of housekeeping genes, which are regulated by SP1, contain methylation free islands suggesting an epigenetic role for SP1 (53,54). We conclude that our results show a hitherto unknown recognition property of SP1 that may prove to be important for the regulation of gene expression.
ADDITIONAL NOTES
We noted in interest that a paper recently appeared, whilst this paper was under review, that suggests the potential SP1 binding to other three-tetrad G-quadruplexes in the promoter region of HRAS as shown by EMSA (55).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures S1–S8.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC); programme funding from Cancer Research UK; a Postdoctoral fellowship from the Deutscher Akademischer Austausch Dienst (DAAD) (to R.K.). Funding for open access charge: BBSRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor S.P. Jackson for helpful advice on the purification of SP1. We thank Dr D. Tannahill for critical reading of this article.
REFERENCES
- 1.Deniaud E, Baguet J, Chalard R, Blanquier B, Brinza L, Meunier J, Michallet MC, Laugraud A, Ah-Soon C, Wierinckx A, et al. Overexpression of transcription factor Sp1 leads to gene expression perturbations and cell cycle inhibition. PLoS One. 2009;4:e7035. doi: 10.1371/journal.pone.0007035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song J, Ugai H, Ogawa K, Wang Y, Sarai A, Obata Y, Kanazawa I, Sun K, Itakura K, Yokoyama KK. Two consecutive zinc fingers in Sp1 and in MAZ are essential for interactions with cis-elements. J. Biol. Chem. 2001;276:30429–30434. doi: 10.1074/jbc.M103968200. [DOI] [PubMed] [Google Scholar]
- 3.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 4.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 5.Reid JE, Evans KJ, Dyer N, Wernisch L, Ott S. Variable structure motifs for transcription factor binding sites. BMC Genomics. 2010;11:30. doi: 10.1186/1471-2164-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Paeschke K, Capra JA, Zakian VA. DNA Replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol. Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pontier DB, Kruisselbrink E, Guryev V, Tijsterman M. Isolation of deletion alleles by G4 DNA-induced mutagenesis. Nat Methods. 2009;6:655–657. doi: 10.1038/nmeth.1362. [DOI] [PubMed] [Google Scholar]
- 13.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todd AK, Neidle S. The relationship of potential G-quadruplex sequences in cis-upstream regions of the human genome to SP1-binding elements. Nucleic Acids Res. 2008;36:2700–2704. doi: 10.1093/nar/gkn078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isalan M, Patel SD, Balasubramanian S, Choo Y. Selection of zinc fingers that bind single-stranded telomeric DNA in the G-quadruplex conformation. Biochemistry. 2001;40:830–836. doi: 10.1021/bi001728v. [DOI] [PubMed] [Google Scholar]
- 18.Ladame S, Schouten JA, Roldan J, Redman JE, Neidle S, Balasubramanian S. Exploring the recognition of quadruplex DNA by an engineered Cys2-His2 zinc finger protein. Biochemistry. 2006;45:1393–1399. doi: 10.1021/bi050229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel SD, Isalan M, Gavory G, Ladame S, Choo Y, Balasubramanian S. Inhibition of human telomerase activity by an engineered zinc finger protein that binds G-quadruplexes. Biochemistry. 2004;43:13452–13458. doi: 10.1021/bi048892t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borgognone M, Armas P, Calcaterra NB. Cellular nucleic-acid-binding protein, a transcriptional enhancer of c-Myc, promotes the formation of parallel G-quadruplexes. Biochem. J. 428:491–498. doi: 10.1042/BJ20100038. [DOI] [PubMed] [Google Scholar]
- 21.Kumar P, Yadav VK, Baral A, Saha D, Chowdhury S. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Res. 2011;39:8005–8016. doi: 10.1093/nar/gkr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai M, Takeishi T, Thompson H, Langley KE, Zsebo KM, Metcalfe DD, Geissler EN, Galli SJ. Induction of mast cell proliferation, maturation, and heparin synthesis by the rat c-kit ligand, stem cell factor. Proc. Natl Acad. Sci. USA. 1991;88:6382–6386. doi: 10.1073/pnas.88.14.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park GH, Plummer HK, III, Krystal GW. Selective Sp1 binding is critical for maximal activity of the human c-kit promoter. Blood. 1998;92:4138–4149. [PubMed] [Google Scholar]
- 24.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu ST, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuryavyi V, Patel DJ. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 18:73–82. doi: 10.1016/j.str.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson SP, Tjian R. Purification and analysis of RNA polymerase II transcription factors by using wheat germ agglutinin affinity chromatography. Proc. Natl Acad. Sci. USA. 1989;86:1781–1785. doi: 10.1073/pnas.86.6.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M, Dietmaier W, Thiede C, Holler E, Andreesen R, et al. General transcription factor binding at CpG islands in normal cells correlates with resistance to de novo DNA methylation in cancer cells. Cancer Res. 2010;70:1398–1407. doi: 10.1158/0008-5472.CAN-09-3406. [DOI] [PubMed] [Google Scholar]
- 31.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Team RDC. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 33.Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo–an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 36.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 39.Basundra R, Kumar A, Amrane S, Verma A, Phan AT, Chowdhury S. A novel G-quadruplex motif modulates promoter activity of human thymidine kinase 1. FEBS J. 2010;277:4254–4264. doi: 10.1111/j.1742-4658.2010.07814.x. [DOI] [PubMed] [Google Scholar]
- 40.Joachimi A, Benz A, Hartig JS. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 41.Cameron IL, Sparks RL, Seelig LL., Jr Concentration of calcium and other elements at a subcellular level in the lactating epithelium of rat. Cytobios. 1980;27:89–96. [PubMed] [Google Scholar]
- 42.Hud NV, Smith FW, Anet FA, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 43.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Searle MS, Williams HE, Gallagher CT, Grant RJ, Stevens MF. Structure and K+ ion-dependent stability of a parallel-stranded DNA quadruplex containing a core A-tetrad. Org Biomol Chem. 2004;2:810–812. doi: 10.1039/b314559j. [DOI] [PubMed] [Google Scholar]
- 45.Letovsky J, Dynan WS. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989;17:2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong HM, Stegle O, Rodgers S, Huppert JL. A toolbox for predicting g-quadruplex formation and stability. J. Nucleic Acids. 2010 doi: 10.4061/2010/564946. June 8 (doi:10.4061/2010/564946; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cogoi S, Paramasivam M, Membrino A, Yokoyama KK, Xodo LE. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soldatenkov VA, Vetcher AA, Duka T, Ladame S. First evidence of a functional interaction between DNA quadruplexes and poly(ADP-ribose) polymerase-1. ACS Chem. Biol. 2008;3:214–219. doi: 10.1021/cb700234f. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 52.Halder R, Halder K, Sharma P, Garg G, Sengupta S, Chowdhury S. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 2010;6:2439–2447. doi: 10.1039/c0mb00009d. [DOI] [PubMed] [Google Scholar]
- 53.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 54.Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 55.Membrino A, Cogoi S, Pedersen EB, Xodo LE. G4-DNA Formation in the HRAS Promoter and Rational Design of Decoy Oligonucleotides for Cancer Therapy. PLoS One. 2011;6:e24421. doi: 10.1371/journal.pone.0024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.