Abstract
The ability of mammals to resist body fat accumulation is linked to their ability to expand the number and activity of “brown adipocytes” within white fat depots. Activation of β-adrenergic receptors (β-ARs) can induce a functional “brown-like” adipocyte phenotype. As cardiac natriuretic peptides (NPs) and β-AR agonists are similarly potent at stimulating lipolysis in human adipocytes, we investigated whether NPs could induce human and mouse adipocytes to acquire brown adipocyte features, including a capacity for thermogenic energy expenditure mediated by uncoupling protein 1 (UCP1). In human adipocytes, atrial NP (ANP) and ventricular NP (BNP) activated PPARγ coactivator-1α (PGC-1α) and UCP1 expression, induced mitochondriogenesis, and increased uncoupled and total respiration. At low concentrations, ANP and β-AR agonists additively enhanced expression of brown fat and mitochondrial markers in a p38 MAPK–dependent manner. Mice exposed to cold temperatures had increased levels of circulating NPs as well as higher expression of NP signaling receptor and lower expression of the NP clearance receptor (Nprc) in brown adipose tissue (BAT) and white adipose tissue (WAT). NPR-C–/– mice had markedly smaller WAT and BAT depots but higher expression of thermogenic genes such as Ucp1. Infusion of BNP into mice robustly increased Ucp1 and Pgc-1α expression in WAT and BAT, with corresponding elevation of respiration and energy expenditure. These results suggest that NPs promote “browning” of white adipocytes to increase energy expenditure, defining the heart as a central regulator of adipose tissue biology.
Introduction
The cardiac natriuretic peptides (NPs), atrial NP (ANP) and its ventricular companion (BNP), are key hormones in fluid and hemodynamic homeostasis. Their actions are mediated by binding to NP receptor A (NPRA), whose intracellular domain possesses guanylyl cyclase activity to generate the second messenger cGMP (1, 2). Another member of the NP receptor family (NPRC, which is referred to as the clearance receptor) also binds ANP and BNP to remove them from circulation (3). Almost 2 decades ago, NP receptors were unexpectedly found to be expressed in adipose tissue of both rats (4) and humans (5), and, interestingly, levels of NPRC in adipose tissue were found to be sharply decreased by fasting in rats (6). Together, these were some of the first results to suggest that perhaps cardiac NPs have a metabolic role in adipocytes, including a putative role for adipose tissue in the clearance of these peptides from the circulation (7).
ANP was subsequently shown to increase lipolysis in human adipocytes, with a potency similar to that of catecholamines (8), which are the well-established physiological pathway controlling lipolysis through activation of the β-adrenergic receptors (β-ARs). Interestingly, the ability of NPs to stimulate lipolysis was reported to be primate specific and apparently absent from rodent adipose tissue (9). To understand this process mechanistically, recall that β-ARs, as the classic stimulator of lipolysis, increase cAMP levels to activate cAMP-dependent protein kinase (PKA). PKA in turn phosphorylates key targets in the fat cell, including hormone-sensitive lipase (HSL) and the abundant lipid droplet–associated perilipins (10). NPs, on the other hand, activate the guanylyl cyclase containing NPRA that generates the second messenger, cGMP, to activate cGMP-dependent protein kinase (PKG) (11–13). PKA and PKG share closely related motifs for substrate phosphorylation (14). Since NPs via PKG have been shown to phosphorylate the same targets in adipocytes as β-agonists through PKA (13), these 2 hormonal systems appear to share a common mechanism to increase lipolysis (reviewed in ref. 15).
A physiological role for NPs in exercise-induced lipolysis in humans has been suggested (16–18). The increased cardiac output with exercise and release of NPs suggests that the heart has a central role in regulating the supply of fatty acids for both cardiac and skeletal muscle under aerobic conditions. Increases in circulating NPs have also been associated with increased postprandial fat oxidation in humans (19, 20) and with weight loss (21, 22). A further connection between the NP system and adipose tissue is highlighted by the fact that obese human subjects with metabolic syndrome often show reduced circulating NPs and biological efficacy (e.g., blood pressure control) (23, 24). Indeed, studies in different populations consistently show an inverse relationship between plasma NPs and BMI (25, 26). As a result, such studies have led to a hypothesis that lower plasma NP levels in the obese are the result of reduced secretion of NPs (due to impaired myocardial release or diminished synthesis). Another possibility is an increased clearance of NPs by NPRC within adipose tissue. These are interesting correlations, but it has been unclear whether changes in body weight and NPs are causally related.
Regulation of body weight or composition by β-AR activation is known from work in animal models as well as human studies (27). We and others have previously shown that the ability of mammals to resist weight gain and body fat accumulation is very closely tied, both genetically and pharmacologically, to the capacity to expand the number and activity of “brown adipocytes” within white fat depots (28–32), and, in most cases, this is also correlated with improved blood glucose levels, insulin sensitivity, and body composition. Because of this link among brown adipocyte activity, thermogenesis, and weight loss, the recent (re)discovery of functional brown adipose tissue (BAT) in adult humans (33–36) has led to reconsideration of its role in body weight regulation, improved cardiovascular status, and other metabolic parameters (37).
Biochemically, the process of adaptive thermogenesis in brown adipocytes is well-known to be mediated by β-AR signaling, in which it coordinates the transcription of key genes, such as mitochondrial uncoupling protein 1 (UCP1) and the transcriptional coregulator, PPARγ coactivator-1α (PGC-1α), the latter contributing to the net increase in mitochondrial capacity required to accommodate the increased metabolic demands for respiratory uncoupling through UCP1. There is also an expansion of brown adipocyte number, activation of lipolysis, and the utilization of those fatty acids as oxidative substrates in situ and as allosteric activators of proton leak by UCP1. Altogether, this results in respiratory uncoupling, net energy expenditure, and heat generation.
Given the parallel actions of β-ARs and cardiac NPs to stimulate lipolysis, the critical questions of the present study were whether NPs function in a parallel manner to the β-ARs to similarly induce a functional “brown-like” adipocyte phenotype and, if so, what is the underlying mechanism. At a molecular level, these events are set in motion by classic β-AR–PKA signaling but, as we previously showed (38), have the additional requirement for the downstream activation of p38α MAPK. Using a combination of approaches that include cultured human adipocytes, mice with targeted deletion of the clearance receptor Npr3 gene (hereafter referred to as the NPR-C gene), and manipulations in mice, including infusions of BNP, we describe what we believe to be a novel pathway, by which cardiac NPs, through NPRA, cGMP, and PKG, can activate p38α MAPK to increase mitochondrial biogenesis and uncoupled respiration. Importantly, we show that ANP and β-AR agonists can act together in an additive manner to more robustly promote brown adipocyte features and functions. Overall our results support the concept of cardiac NPs as cardiometabolic hormones that are able to turn on the machinery characteristic of brown fat thermogenesis in human and mouse adipocytes. Thus, cardiac NPs, together with catecholamine, may modulate energy expenditure to regulate the distribution of body fat and lean mass, with a relevant role in controlling cardiovascular risk and potentially inducing beneficial metabolic effects in the cardiometabolic patients.
Results
ANP and lipolysis.
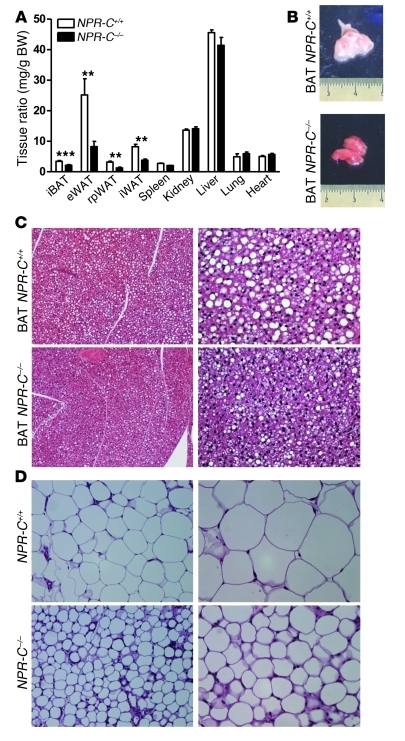
It is well established that lipolysis is stimulated by the sympathetic nervous system (SNS) through the β-AR–cAMP pathway. More recently NPs have been shown to be able to stimulate lipolysis in human adipocytes (ref. 15 and see also Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI59701DS1) but not in other species such as rats and mice (9). This species difference has been suggested to be due to the fact that rodents possess approximately 100-fold greater levels of the NPRC clearance form of the receptor (9, 39), thus greatly reducing signaling through NPRA. To test this idea, we directly compared white adipose tissue (WAT) and BAT from wild-type (NPR-C+/+) mice and NPRC knock-out (NPR-C–/–) mice. First we examined lipolysis. Primary adipocyte cultures from NPR-C+/+ mice showed no lipolytic response to ANP. However, cells from NPR-C–/– mice clearly responded to ANP, with significantly elevated glycerol release (Supplemental Table 1). NPR-C–/– mice also had significantly reduced interscapular BAT (iBAT), inguinal WAT, retroperitoneal WAT, and epididymal WAT fat mass, while body and organ weights were not different from those of wild-type mice (Figure 1A). In comparison with that of NPR-C+/+ mice, the iBAT of NPR-C–/– mice contained no visible superficial WAT and had a deeper reddish-brown tint (Figure 1B), and lipid droplets were visibly reduced (Figure 1C). Similarly, the adipocytes in epididymal WAT of NPR-C–/– mice appeared smaller (Figure 1D). The expression of genes representative of brown adipocytes, including Ucp1, Pgc-1α, and cytochrome c (Cycs), was elevated in BAT (Figure 2, A–C), epididymal WAT (Figure 2, F and G), and inguinal WAT (Figure 2, I–K) from NPR-C–/– mice. The amount of UCP1 detected by immunohistochemistry in epididymal WAT of NPR-C–/– mice was much greater than that in NPR-C+/+ mice (Supplemental Figure 2). Moreover, in the absence of NPRC, the levels of NPRA gene expression were significantly higher in BAT (Figure 2D) and in WAT (Figure 2, H and L). These results suggest that NPR-C–/– mice are more thermogenically active than NPR-C+/+ mice and that, in addition to stimulating lipolysis in human adipocytes, NPs may be able to more generally activate the transcriptional program characteristic of brown adipocyte thermogenesis, depending on the relative ratio of NPRA to NPRC in adipocytes.
Figure 1. Mice lacking NPRC have reduced fat mass.
(A) Ratio between weight of isolated tissues and total body weight of NPR-C+/+ mice compared with that of NPR-C–/– mice. eWAT, epididymal WAT; rpWAT, retroperitoneal WAT; iWAT, inguinal WAT. **P < 0.01, ***P < 0.001 versus control. (B) Representative images of BAT from NPR-C+/+ mice compared with those from NPR-C–/– mice. (C and D) Representative hematoxylin-eosin staining of iBAT and epididymal WAT sections, respectively, taken from NPR-C+/+ mice compared with those from NPR-C–/– mice. Original magnification, ×20 (left column); ×40 (right column).
Figure 2. Adipose tissues from NPR-C–/– mice express higher levels of brown adipocyte marker genes as well as NPRA.
Gene expression analysis of Ucp1, Pgc-1α, Cycs, and NPRA in (A–D) BAT, (E–H) epididymal WAT, and (I–L) inguinal WAT from NPR-C+/+ and NPR-C–/– mice. Typical cycle threshold values for NPR-C+/+ samples are presented above each bar. *P < 0.05 versus NPR-C+/+; **P < 0.01 versus NPR-C+/+.
ANP triggers the acquisition of a brown fat cell phenotype.
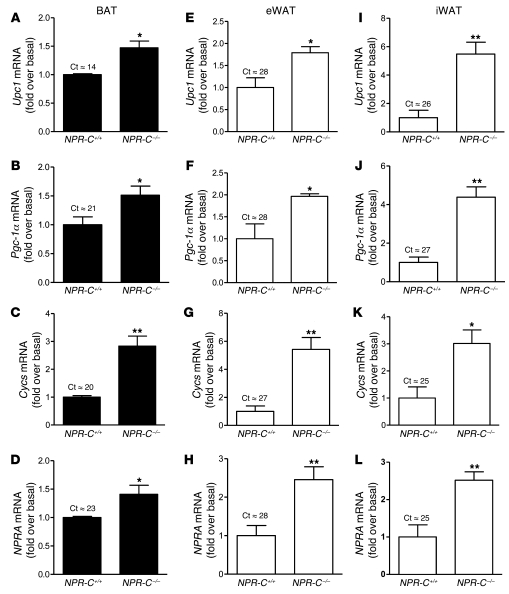
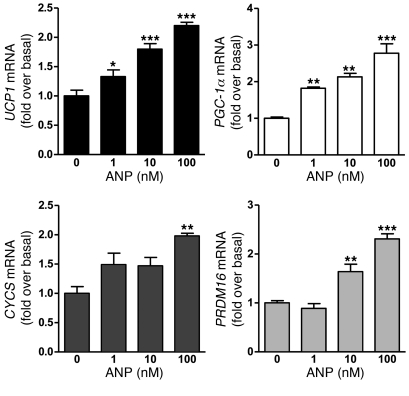
Since human adipocytes have a relatively low level of NPRC compared with NPRA (9), we examined whether ANP could increase expression of UCP1, PGC-1α, cytochrome c. In these experiments, we used differentiated human multipotent adipose-derived stem (hMADS) cells (40). Treatment with ANP over a range of concentrations from 1–100 nM, as shown in Figure 3, was able to significantly increase the transcripts for these genes as well as PRDM16; the latter is noted to be a significant factor in brown adipogenesis (41). Western blotting (Figure 4A) showed that the ability of ANP to increase expression was similar to the effect of the nonselective β-AR agonist isoproterenol (Iso) or the β3-AR agonist L755,507 (L755). BNP, which also activates NPRA, elicited these same responses (Figure 4B). Since NPRA activation results in cGMP production, we assessed the role of cGMP to induce these brown adipocyte markers. As shown in Figure 4C, pretreatment with the synthetic cGMP analog Rp-8-pCPT-cGMPS, which is a PKG inhibitor (hereafter referred to as PKGi), completely blocked the effect of ANP. PKGi had a powerful impact on CYCS expression, suggesting that basal PKG activity is important for its expression. In primary cultures of human subcutaneous adipocytes, qualitatively similar results were obtained as those in hMADS adipocytes, although the levels of UCP1, PGC-1α, and cytochrome c expression were lower (Figure 4D). Each of these genes could be induced by ANP and prevented by PKGi (Figure 4E).
Figure 3. ANP increases brown adipocyte marker genes in hMADS cells.
Cells were treated with ANP (1–100 nM) for 6 hours and levels of UCP1, PGC-1α, CYCS, and PRDM16 gene expression were measured. *P < 0.05, **P < 0.01, ***P < 0.001 versus untreated. Typical cycle threshold values in untreated cells are as follows: UCP1, 27; PGC-1α, 23; cytochrome c, 20; and PRDM16, 29.
Figure 4. NPs induce UCP1, PGC-1α, and cytochrome c levels in human adipocytes through PKG.
(A) hMADS cells were treated or not with ANP, Iso, or the β3-AR agonist L755 (100 nM each), and protein levels were measured by Western blot with GAPDH as the internal standard, as described in Methods. CYTO C, cytochrome c; C, control. (B) As in A after treatment or not with 100 nM BNP. (C) hMADS cells were pretreated or not for 30 minutes with 500 μM PKGi followed by ANP (100 nM, 6 hours), and mRNA levels for indicated genes were measured. Typical cycle threshold values in untreated cells are as follows: UCP1, 27; PGC-1α, 23; CYCS, 20. (D) Human subcutaneous adipocytes were treated as in A and C, and samples were analyzed for protein levels by Western blotting or (E) mRNA levels of indicated genes. Typical cycle threshold values in untreated cells are as follows: UCP1, 29; PGC-1, 24; and cytochrome c, 23. Results are mean ± SD of 3 to 5 independent experiments. **P < 0.01, ***P < 0.001 ANP versus untreated controls; ††P < 0.01 PKGi versus untreated controls.
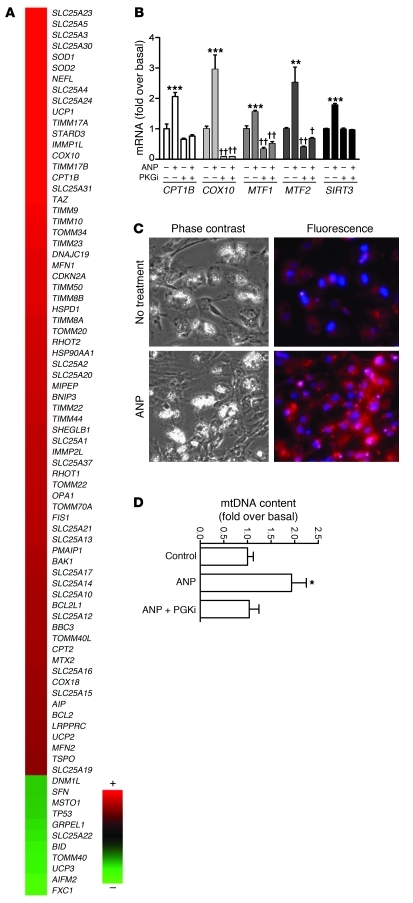
The results obtained from NPR-C–/– mice and human adipocytes suggest that ANP can turn on a brown-like adaptive thermogenic response in a manner quite similar to activation of the β-ARs via the SNS. Therefore, we examined whether ANP could increase a broader set of parameters indicative of mitochondrial biogenesis and respiration, including gene expression, mitochondrial mass, and respiratory uncoupling. Approximately 80% of the genes in the Human Mitochondria RTβ Profiler PCR Array (SABiosciences) were increased after exposure to ANP for 6 hours (Figure 5A). Validation of some of these genes by qRT-PCR is shown in Figure 5B, and, for each, their response to ANP was PKG mediated. Again, the expression of certain genes, such as COX10 and the mitofusins, was suppressed below basal levels by PKGi. To determine whether the effect of ANP on expression of genes for mitochondrial components translates into greater mitochondrial content, a fluorescent mitochondrion-selective dye (MitoTracker; see Methods) was used, and mitochondrial DNA (mtDNA) content was also measured. MitoTracker staining was markedly greater in hMADS cells treated with ANP compared with that in control cells, consistent with increased mitochondrial capacity (Figure 5C). mtDNA measurements also revealed that ANP treatment increased the mtDNA content, and PKGi was able to block the ANP effect (Figure 5D). This provided additional evidence supporting a role for ANP to promote mitochondrial biogenesis in adipocytes and is consistent with observations that cGMP produced in response to nitric oxide can increase mitochondrial content in some cell types (42, 43). All together, these results support the conclusion that ANP can increase UCP1 expression as well as activate the machinery to increase overall mitochondrial content.
Figure 5. ANP promotes mitochondrial biogenesis in human adipocytes.
(A) Heat map of mitochondrial PCR array results from hMADS adipocytes that were treated or not with 100 nM ANP for 6 hours. The range of fold change versus basal was 3.1 to –1.4. (B) Validation of response to ANP and effect of PKGi on select genes from PCR array data. SIRT3 was also measured. **P < 0.01, ***P < 0.001 for ANP versus untreated cells; †P < 0.05, ††P < 0.01 PKGi versus untreated control. (C) Phase-contrast and MitoTracker fluorescence images of hMADS cells that were treated or not with 100 nM ANP, as described in Methods. Mitotracker is identified by the orange-red color. DAPI-stained nuclei are blue (original magnification, ×20). (D) mtDNA content was quantified by qPCR, as described in Methods. Bars represent mean ± SEM. *P < 0.05 compared with control.
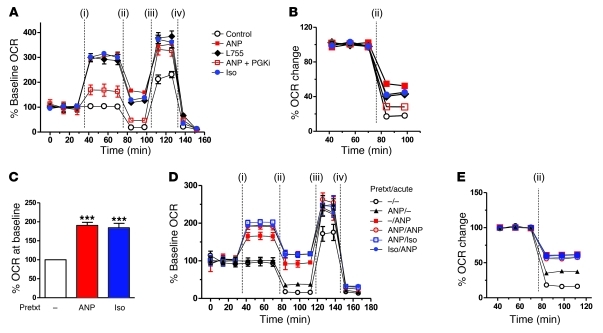
To assess the potential for respiratory uncoupling in response to ANP, we measured cellular respiration using the XF24 Extracellular Flux Analyzer (Seahorse Bioscience), in which continuous measurements of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were collected over time (Figure 6 and Supplemental Figure 3). First, we examined the acute effect of ANP on the respiratory parameters of the cells after their differentiation. As shown in Figure 6A, after an initial period of steady basal OCR measurements, addition of Iso, L755, or ANP increased OCR by more than 250%, which was relatively stable over the 40-minute interval. This increase in OCR by ANP was largely blunted by the PKGi. A significant fraction of the increased OCR in response to β-agonists and ANP could be attributed to mitochondrial uncoupling. This is illustrated in Figure 6B. Under basal conditions, oligomycin inhibited OCR by 80%. However, in Iso-, L755-, or ANP-treated cells, OCR was less sensitive to oligomycin. Note that Iso and ANP tended to enhance uncoupling more than the β3-AR agonist. In addition, much of the oligomycin-insensitive OCR from ANP treatment was prevented by inhibition of PKG. This is congruent with our previous findings that Iso-induced increases in OCR correspondingly depend on PKA (44). We also measured maximal respiration by the addition of the global uncoupler FCCP, which was higher in the agonist-treated samples. Addition of the complex I inhibitor rotenone reduced the OCR to between 9% and 13% of total OCR. This experiment tested the acute effects of ANP on respiration and uncoupling. However, since a longer exposure to β-agonists or ANP is able to substantially increase the expression of UCP1, PGC-1α, and overall mitochondrial content, we next determined whether the acute response to Iso or ANP would be even greater if preceded by pretreatment with these agonists. As shown in Figure 6C, pretreatment with either ANP or Iso increased basal OCR, and they responded to the subsequent acute challenge with a greater rise in OCR than that in those samples that received the acute injection alone (Figure 6D). This was true whether the pretreatment was with Iso or ANP. In addition, the degree of oligomycin insensitive respiration, shown in Figure 6E, was greater in the pretreated cells, suggesting that the increased mitochondria and UCP1 allowed for higher respiratory uncoupling. Altogether these results support the conclusion that the entire machinery for adaptive thermogenesis can be induced by ANP as well as β-adrenergic agonists.
Figure 6. Respiration and uncoupling are increased by β-agonists and ANP.
(A) Representative measurements of the percentage increase in OCRs and relative baseline rates in hMADS cells in response to ANP, Iso, or L755, and effects of respiratory chain modulators. OCR measurements before drug injections (i) were set as 100%. At the time points indicated in the figures, the following were injected sequentially: (ii) oligomycin (ATP synthase inhibitor), (iii) FCCP, and (iv) rotenone (complex I inhibitor). Each data point is a mean of 9 to 10 wells. Supplemental Figure 1 shows histograms summarizing the average maximal percentage increase of OCRs or ECARs over their baseline rates. (B) Representative measurements of the percentage change in OCRs in hMADS cells after injection of oligomycin, as an index of uncoupled respiration. The OCR before oligomycin injection was set as 100%. Each data point is the mean of 9 to 10 wells. (C) Basal levels of percentage OCR in hMADS cells before treatment overnight with 100 nM ANP or ISO. The OCR measured in control cells (no pretreatment) was set as 100%. ***P < 0.001 versus control. (D) Representative measurements of OCRs in hMADS cells that had been pretreated (Prtxt) or not treated (–) overnight with ANP or Iso (100 nM) and analyzed as in A. At the times indicated, acute injections of drugs (i) and of the respiratory chain modulators (ii–iv) were provided. (E) The percentage of oligomycin-insensitive OCRs in pretreated and acutely treated hMADS cells as in B.
β-ARs and ANP use a parallel signaling cascade from PKA/PKG to p38 MAPK.
Although β-ARs and NPRA are structurally different classes of receptors, they produce closely related chemical messengers that result in the activation of functionally homologous protein kinases: PKA and PKG (45). We previously showed that p38α MAPK is activated in white and brown adipocytes in response to stimulation of β-ARs and cAMP/PKA (46), and p38α MAPK is necessary for transcription of the genes for Ucp1 and Pgc-1α in mice (38). Because PKG can recognize and phosphorylate the same generic target motif (e.g., RRXS/T) as PKA (45), we examined whether ANP might use a pathway from PKG to p38 MAPK in human adipocytes.
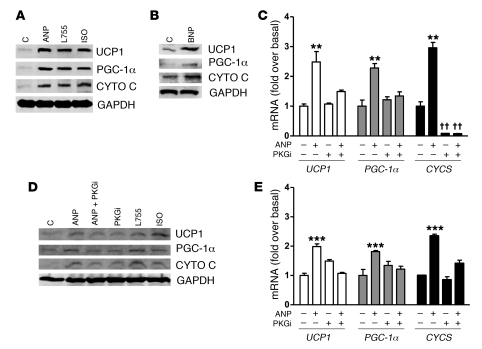
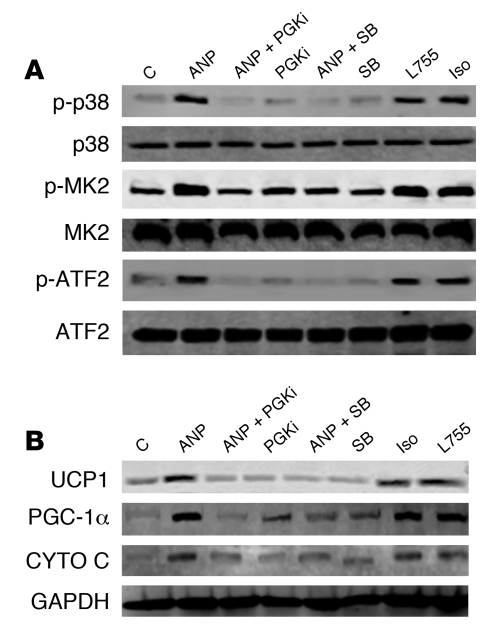
As shown in Figure 7A, ANP could indeed increase p38 MAPK activity in hMADS adipocytes, as indicated by the phosphorylation of p38 MAPK and its downstream targets MAPKAPK-2 (MK2) and the transcription factor ATF2. This was suppressed by PKGi and the p38 MAPK catalytic inhibitor SB203850 (SB). Importantly, as shown in Figure 7B, blockade of p38 MAPK with SB completely suppressed the ability of ANP to increase UCP1, PGC-1α, and cytochrome c and was as effective as inhibition of PKG itself. Similar results were obtained when examining mRNA levels for these proteins in white and brown adipocytes from NPR-C–/– mice (Supplemental Figure 4), indicating that the dependence on p38 MAPK is preserved between rodent and human. In a separate approach, immunofluorescent staining of UCP1 in hMADS adipocytes was also examined in response to ANP (Supplemental Figure 5). Under basal conditions (Supplemental Figure 5, A and B), there was a low but detectable amount of UCP1. The intensity of the UCP1 immunofluorescence was markedly increased by ANP (Supplemental Figure 5, C and D), and this was prevented in the presence of either PKGi (Supplemental Figure 5, E and F) or SB (Supplemental Figure 5, G and H). Background staining with secondary antibody alone was low (Supplemental Figure 5, I and J). All together these results demonstrate that, in human hMADS as well as in NPR-C–/– mouse adipocytes, ANP increases the expression of UCP1 and mitochondrial content through a pathway from PKG to p38 MAPK. Thus ANP, through NPRA and PKG, uses the same signaling mechanism as the catecholamines, through β-AR and PKA.
Figure 7. Activation of brown adipocyte marker genes by ANP and PKG requires p38 MAPK.
(A) hMADS adipocytes were pretreated or not for 30 minutes with p38 inhibitor SB (10 μM) or PKGi (500 μM) as indicated, followed by 100 nM ANP for 1 hour. Cell lysates were processed for Western blotting, as described in Methods, to detected phosphorylated and total p38 MAPK, MK2, and ATF2, with GAPDH as the internal standard. (B) hMADS adipocytes were treated as in A for 6 hours. Cell lysates were prepared, and UCP1, PGC-1α, cytochrome c, and GAPDH were measured by immunoblotting using specific antisera, as indicated in Methods.
Transcription factor recruitment to the human Ucp1 promoter by ANP.
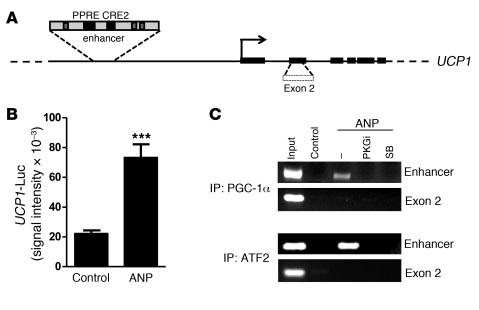
The transcriptional activation of the UCP1 gene is regulated through a critical enhancer region (47–49). This enhancer confers brown adipocyte–specific expression as well as the response to cAMP and is well conserved among species (50, 51). It contains several transcription factor binding sites, including two key elements that we previously showed to be critically dependent on the β-AR/PKA/p38α MAPK cascade: a peroxisome proliferator receptor response element (PPRE) and a sequence originally termed a cAMP-response element (CRE2) (ref. 50 and see Figure 8A). In earlier work, we showed that p38α MAPK phosphorylates PGC-1α to interact with PPARγ at the PPRE, p38α MAPK also phosphorylates ATF2 to bind CRE2, and interference with either of these 2 elements is detrimental to transcription of the UCP1 gene (50). Here, we show that, in these human adipocytes, ANP was also able to significantly increase the activity of luciferase reporter constructs containing the UCP1 enhancer (Figure 8B) or the CRE-containing proximal promoter of the PGC-1α gene (data not shown). Since ANP can increase expression of UCP1 in a PKG- and p38 MAPK–dependent manner (Figures 4 and 7), we examined by ChIP assays whether ANP could promote binding of PGC-1α and ATF2 to the corresponding elements in the human UCP1 enhancer. As shown in Figure 8C, after treatment of human adipocytes with ANP, immunoprecipitation of PGC-1α or ATF2 was able to recover the UCP1 enhancer, and these interactions were not observed if PKG or p38 MAPK activity was inhibited. Altogether, these results strongly support a mechanism by which NP activation of human UCP1 gene transcription proceeds from PKG to p38α MAPK; a process which is analogous to the pathway from PKA to p38α MAPK that we previously described for β-ARs in mouse adipocytes (38, 46).
Figure 8. ANP signaling increases transcriptional activity and transcription factor recruitment to the UCP1 enhancer.
(A) Schematic representation of the human UCP1 gene with PPRE and CRE2 indicated. (B) UCP1 enhancer reporter gene activity in transfected hMADS cells treated or not with ANP (100 nM), as described in Methods. Results are mean ± SEM. ***P < 0.001 versus untreated cells (n = 6). (C) ChIP assay. hMADS adipocytes were pretreated or not with PKGi or SB, followed by ANP, as described in Methods. Chromatin was prepared and immunoprecipitated with antisera against PGC-1α or ATF2. The final DNA extraction was amplified by PCR using a primer encompassing the UCP1 enhancer region or exon 2 as a control region (ENSG00000109424).
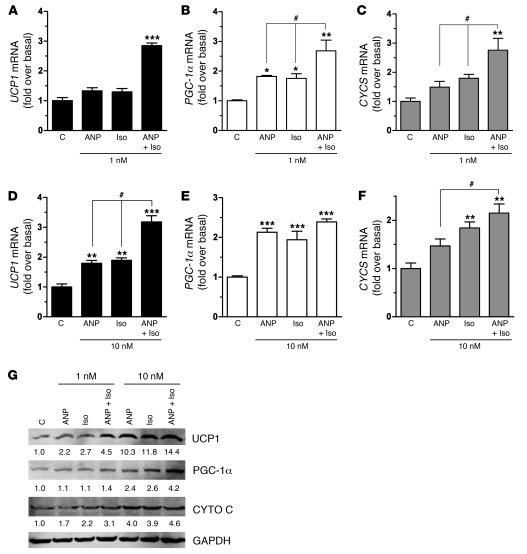
Additive response of NPs and b-ARs increases a brown adipocyte-like pattern of gene expression.
Since ANP can activate transcription of the UCP1 and PGC-1a genes and both pathways converge on p38 MAPK as the mediator, to increase mitochondrial biogenesis and other features of brown adipocytes in the same manner as b-adrenergic agonists, we assessed whether these 2 signaling pathways might act in an additive manner. hMADS adipocytes were treated with low concentrations of ANP or Iso, either alone or in combination, and transcripts for UCP1, PGC-1a, and cytochrome c were measured. As shown in Figure 9, A–F, the response to the combination of Iso and ANP was always greater than that to either agent individually. This is particularly evident at the lower concentrations, in which gene expression was increased in an additive manner (e.g., PGC-1a and CYCS) or even synergistically (e.g., UCP1). UCP1, PGC-1a, and cytochrome c protein levels were also higher when hMADS cells were treated with ANP and Iso together (Figure 9G). These results, together with those in Figure 3, show that ANP can induce these genes at the low concentrations commonly found in patients (19, 20).
Figure 9. Combining β-agonist and ANP treatment augments their individual capacities to induce brown adipocyte markers.
hMADS adipocytes were treated with (A–C) 1 nM or (D–F) 10 nM of ANP or Iso, both alone and together (ANP+Iso), for 6 hours, and mRNA levels were measured for (A and D) UCP1, (B and E) PGC-1α, and (C and F) CYCS. *P < 0.05, **P < 0.01, ***P < 0.001 versus untreated cells; #P < 0.01 for ANP plus Iso versus ANP or Iso treatment alone. (G) In a sample experiment, similarly treated hMADS adipocytes were harvested, cell lysates were prepared, and Western blotting using specific antisera was performed, as indicated in Methods, and normalized for GAPDH (values are reported under each band.
NPs increase brown adipocyte characteristics and energy expenditure in vivo.
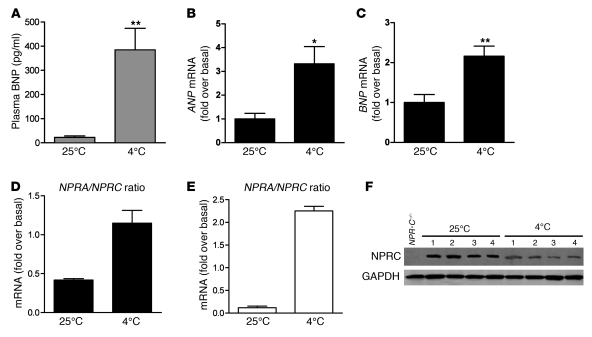
Increases in SNS activity are well-established outcomes of cold-temperature challenge, which then sets in motion the activation of brown fat mechanisms of thermogenesis. Given the parallel nature of β-AR and NP signaling in adipocytes and their cooperative responses on gene expression, we investigated whether changes in the NP system were stimulated by cold exposure. Wild-type mice were maintained at room temperature (25°C) or exposed to 4°C for 6 hours. As shown in Figure 10, compared with mice maintained at room temperature, mice at 4°C had significantly increased levels of plasma BNP (Figure 10A) and increased levels of ANP (Figure 10B) and BNP (Figure 10C) transcripts in the heart. Interestingly, in both iBAT and epididymal WAT, the ratio of gene expression of NPRA to NPR-C in mice placed at 4°C was also increased (Figure 10, D and E), because NPRA mRNA levels were elevated with a contemporaneous decrease in NPR-C levels (Supplemental Figure 6). This reduction in NPRC expression after cold exposure was confirmed by Western blot (Figure 10F). (Western blotting for NPRA could not be performed due to lack of sufficient quality antisera to NPRA.) Although only a correlation, these changes in circulating NPs, coupled with a shift in the ratio of receptor expression in adipose tissue toward NPRA, would seem to favor activation of lipolysis and thermogenesis under these conditions. Therefore, the NP system may function together with the SNS in the activation of cold-induced thermogenesis and, in humans, might partly explain the reported inability of β-blockers to inhibit cold-induced thermogenesis (52).
Figure 10. Physiological modulation of the NP system in C57BL/6J mice after 6 hours of cold exposure (4°C; n = 9) compared with control mice maintained at room temperature (25°C; n = 8).
(A) BNP circulating plasma levels. (B and C) Gene expression levels of ANP and BNP in the heart. *P < 0.05, **P < 0.01 versus 25°C. (D and E) NPRA/NPR-C gene expression ratio in iBAT and epididymal WAT, respectively. (F) Representative NPRC protein levels in WAT of C57BL/6 mice maintained at 25°C or at 4°C for 6 hours. GAPDH was used as internal control. Samples from NPR-C–/– mice were used as controls for NPRC antibody specificity.
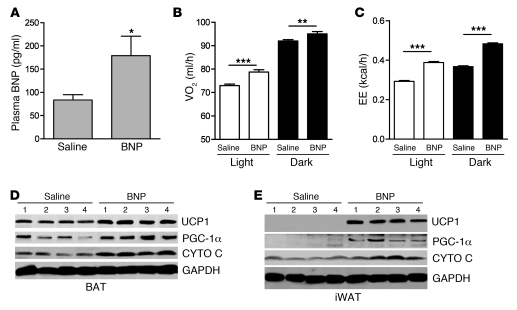
Finally, we assessed the metabolic consequences of infusing BNP. BNP is more commonly measured than ANP in clinical settings (53, 54), and it is administered in the clinic. Mini-pumps containing saline or BNP were implanted into 10-week-old mice, as detailed in Methods. As shown in Figure 11A, plasma levels of BNP were significantly higher in the mice receiving BNP, while within relevant physiological range (55). Although there was a trend for mice receiving BNP to consume more food, there were no significant differences in food intake or physical activity between the 2 groups (data not shown). Mice treated with BNP exhibited greater oxygen consumption (Figure 11B) and energy expenditure (Figure 11C) during both the light and dark periods than the saline control group. Respiratory exchange ratio (RER) was modestly decreased only during the light period (saline, 0.845 ± 0.004 kcal/l; BNP, 0.805 ± 0.006 kcal/l; P < 0.01). Whether treatment for longer duration would have had a greater impact on RER is unknown. BNP treatment had a powerful effect on the expression of brown adipocyte markers in adipose tissue (Figure 11, D and E, and Supplemental Figure 7). This result was particularly striking in the inguinal WAT, in which the levels of UCP1 and PGC-1α were undetectable in the saline-treated group but quite robust in the BNP-treated group. Together, these results suggest that mice treated with BNP expend more energy than control mice through the activation of BAT and the browning of WAT.
Figure 11. BNP infusion increased oxygen consumption, energy expenditure, and expression of brown adipocyte markers.
C57BL/6 mice were treated for 7 days with saline (0.9% NaCl) or with BNP (2 ng/kg/ml) via Mini-pump. Food intake, physical activity, and respiration were measured by the Oxymax Comprehensive Lab Animal Monitoring System. (A) Plasma BNP levels measured on day 7. (B) Oxygen consumption and (C) energy expenditure (EE) during the light and dark periods. *P < 0.05; **P < 0.01; ***P < 0.001. Western blot in (D) iBAT (BAT) and (E) inguinal WAT for UCP1, PGC-1α, and cytochrome c protein levels. GAPDH was used as internal standard.
Discussion
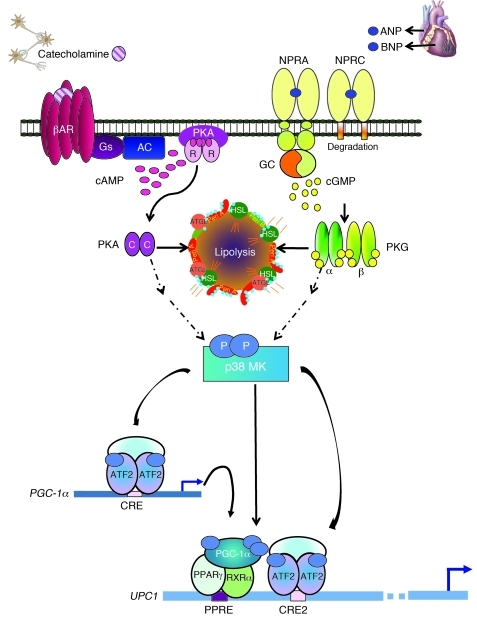
Our findings in human adipocytes and mouse models emphasize a role of the heart as a central regulator of adipose tissue biology through its NPs ANP and BNP. We have used a combination of in vitro and in vivo approaches to show that NPs, via cGMP and PKG, turn on the machinery typical of brown fat thermogenesis in adipocytes as effectively as catecholamines through cAMP and PKA. It is well established that the cAMP-dependent transcription of the Ucp1 gene involves a key enhancer region that is highly conserved among species (reviewed in refs. 56 and 57), 2 key elements of which are a PPRE and a CRE. Previous studies in mouse adipocytes and animal models showed that β-AR stimulation triggers a kinase cascade from PKA to p38 MAPK (46, 50), which phosphorylates PGC-1α and ATF2, as illustrated in Figure 12. Thus activated, the PGC-1α–PPARγ complex and ATF2 are recruited to the PPRE and CRE, respectively. These events contribute to the orchestrated response to increase mitochondriogenesis and the overall thermogenic capacity of brown adipocytes. Here we show that in both human and mouse adipocytes, ANP (and BNP) treatment, at concentrations similar to those found in vivo (55), leads to the same increase in PGC-1α and UCP1 expression, mitochondrial content, and uncoupled respiration as occurs in response to β-agonists, all in a p38 MAPK–dependent manner (Figure 12). As observed for the mouse Ucp1 gene, this includes the recruitment of PGC-1α and ATF2 to the equivalent enhancer region of the human UCP1 gene, indicating conservation of this mechanism between mouse and human adipocytes. An important finding from these studies is that ANP and β-agonists can work together through p38 MAPK to produce additive or synergistic effects.
Figure 12. Model for parallel β-AR and NPRA activation of p38 MAPK to trigger expression of the brown fat thermogenic gene program.
Catecholamines bind the heptahelical β-ARs on adipocytes to activate the G protein Gs and increase cAMP (pink ovals). cAMP binds the regulatory (R) subunits of PKA. The released catalytic (C) subunits (purple ovals) can then phosphorylate targets, including HSL and perilipin (Peri A), to allow lipolysis of stored triglycerides. Lipolysis can also be activated by NPs. ANP and BNP bind to guanylyl cyclase (GC) receptor NPRA to increase cGMP (yellow circles). Adipocytes also express NPRC that mainly removes NPs from circulation. The cGMP produced by NPRA activates PKG (α and β subunits are represented as thin green and yellow ovals, respectively), whose substrate specificity for phosphorylation closely overlaps that of PKA. Thus, PKG can phosphorylate the same targets as PKA to elicit lipolysis. β-ARs and PKA can also activate a protein kinase cascade, culminating in the activation of p38α MAPK (p38 MK). Now, we can add NPs and PKG as parallel activators of p38α MAPK. Thus, in response to β-agonist or NPs, p38α MAPK phosphorylates (light blue ovals) the transcriptional regulators ATF2 and PGC-1α. PGC-1α interacts with PPARγ and RXRα. These phosphorylated and activated factors are recruited to specific motifs within the UCP1 enhancer (PPRE, CRE2) to increase its gene expression (blue arrow). ATF2 also binds the CRE in the PGC-1α promoter to increase transcription (blue arrow) and increase the amount of PGC-1α.
In earlier work, the ability of NPs to stimulate lipolysis was noted to be specific to primate adipocytes (9). Our studies confirm that, irrespective of species, these metabolic effects of NPs largely depend on the ratio in cells of the signaling receptor NPRA to the clearance receptor NPRC (3) (see Figure 12). As shown here using adipose tissues and cultured cells from mice with a targeted deletion of the NPR-C gene, the lean phenotype of NPR-C–/– mice is likely a direct consequence of the absence of NPRC from the adipose tissue, allowing circulating NPs to induce a browning of the adipocytes, such that they are more metabolically active than those in NPR-C+/+ mice. For example, NPR-C–null mice had significantly reduced adipose tissue mass, and the typical brown adipocyte markers were elevated in both BAT and WAT. ANP treatment of cultured adipocytes from NPR-C–/– mice produced a lipolytic response and increased expression of UCP1, PGC-1α, and other brown adipocyte markers that were not detected in NPR-C+/+ cells. Interestingly, NPRA–/– mice tended to show the opposite phenotype from that of NPR-C–/– mice. NPRA–/– animals not only had higher heart weight associated with cardiac hypertrophy (58) but significantly higher adipose tissue weights compared with those of NPRA+/+ mice (Supplemental Figure 8). Also of note, NPR-C–/– mice had higher NPRA expression in their adipose tissue (Figure 2).
Results from other mouse models, in which the PKG axis has been manipulated in mice, have some connections to our findings. For example, PKG type I–deficient mice had reduced mitochondrial content and impaired adipogenesis in vitro (59), while mice with forced whole body overexpression of NP/PKG pathway components (BNP, 100 fold normal plasma levels; catalytic subunit of PKG, 10-fold higher than normal tissue levels) (60) had increased skeletal muscle mitochondrial content and function; less lipid accumulation in adipose, liver, and skeletal muscle; and were protected from obesity and insulin resistance. Furthermore, in a clinical study, carriers of a genetic variant in the promoter of the BNP gene had approximately 20% higher BNP levels than noncarriers and were associated with a 15% reduction in risk for type 2 diabetes (61, 62).
The results we have obtained with human and mouse cell models have allowed us to define molecular mechanisms of how the NPs can promote the acquisition of brown adipocyte features and functions. Better physiological support for these results was obtained by examining the effects of cold challenge on the NP system and the response to infusing mice with physiological levels of BNP. The effect of cold exposure to increase NP levels and to modulate the expression of NPRA (increased; Supplemental Figure 6, A and C) and NPRC (decreased; Supplemental Figure 6, B and D) in mouse WAT and BAT is interesting, as this shift in the ratio of receptor expression in adipose tissue toward NPRA would seem to favor activation of lipolysis and thermogenesis under these conditions. Although not proof, this observation is consistent with the idea that the NP system is actively integrated in vivo in the thermogenic response together with the SNS via catecholamines. (It would also seem that a much better understanding of the molecular basis for changes in NPRA and NPRC expression is needed, since their regulation now has implications for impacting not only blood pressure control but body weight and composition.) More definitive physiological evidence of NP action to affect the white-to-brown adipocyte signature was obtained when BNP was infused into mice in vivo. BNP was a potent stimulator of the appearance of brown-like adipocytes expressing UCP1, PGC-1α, and mitochondrial biogenesis (implied by the increase in cytochrome c), which was associated with increased oxygen consumption and energy expenditure. Given the “rediscovery” of brown adipocytes in adult humans (33–36), and the intense interest surrounding their biology and therapeutic implications, our results imply that the NPs add another dimension to how this tissue may be regulated.
The clinical implications of our findings suggest that the ability of NPs together with catecholamines to modulate uncoupled respiration and control white fat mass may serve as a therapeutic target for the management of obesity and the metabolic complications that accompany it. However, another aspect to consider is the development of weight loss in heart failure, known as pathological cardiac cachexia. High levels of NPs that are characteristic of heart failure are used as diagnostic markers of disease severity (63, 64). Cardiac cachexia is a commonly encountered clinical problem (65), which has marked neurohormonal and metabolic abnormalities (66), and is associated with a poor prognosis (67). Interestingly, cachexia — associated with advanced chronic obstructive airway disease (68) and with various malignancies (69) — is also characterized by elevated plasma levels of NPs. An attractive hypothesis is that the pathological weight loss in heart failure associated with high circulating NPs and/or catecholamine levels may involve increased brown fat and energy expenditure.
In conclusion, to our knowledge our results demonstrate a novel role for cardiac NPs as potent cardiometabolic hormones, revealing a new aspect in the metabolic relationship between heart and adipose tissue. With a deeper molecular and clinical understanding of how NPs control lipid metabolism and energy balance, we may find new ways to manage the patient with vascular and metabolic disease.
Methods
Reagents and antibodies
Iso, insulin, dexamethasone, isobutylmethylxanthine, triiodothyronine, and the small molecule kinase inhibitors H89 (catalog no. B-1427) and SB (catalog no. S-7067) were obtained from Sigma-Aldrich. The human β3-AR–selective agonist L755 was a gift from Merck Research Laboratories. The PKGi Rp-8-pCPT-cGMPS was from BIOLOG. It was aliquoted and stored in liquid nitrogen, due to its sensitivity to moisture and light, to avoid desulfurization yielding a PKG activator. ANP and BNP were obtained from Anaspec. Other reagents included a Proteinase Inhibitor Cocktail (cOmplete Mini) and a Phosphatase Inhibitor Cocktail (PhosSTOP) (Roche Diagnostics). Antisera against p38α MAPK, MK2, and ATF2 as well as phosphorylated forms of MK2 and ATF2 were all obtained from Cell Signaling. Other antisera used include anti–phospho-p38 MAPK (Promega), anti-UCP1 (Abcam), anti–PGC-1α (EMD), and anti–cytochrome c (Santa Cruz Biotechnology Inc.). Antisera against NPRC was a gift from William Gower at the James A. Haley VA Medical Center, Tampa, Florida, USA. Secondary antibodies anti-rabbit (catalog no. A3687) and anti-mouse (catalog no. A3562) conjugated with alkaline phosphatase were from Sigma-Aldrich.
Animal studies
Male NPR-C+/+ and NPR-C–/– mice (8 weeks old; backcrossed to C57BL/6J at least 8 times) were a gift from Nobuyo Maeda, Department of Pathology and Laboratory Medicine, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA. Total body weight and weight of individual organs (spleen, kidney, liver, lung, heart, and several adipose depots) were measured after careful dissection. Tissues isolated for gene expression analyses were rapidly collected, frozen in liquid nitrogen, and stored at –80°C. In some experiments preadipocytes were isolated from epididymal WAT and iBAT, cultured, and differentiated.
Cold challenge experiment.
Wild-type C57BL/6J male mice (8 weeks old) were obtained from The Jackson Laboratory and individually housed at either the control (25°C) temperature (n = 8 mice) or low (4°C) temperature (n = 9 mice) for 6 hours.
BNP infusion study.
Two groups of 5 male C57BL/6J mice (10 weeks of age) each were used. BNP was dissolved in sterile water, and 50 μl was added to sterile saline (0.9% NaCL) for a final BNP concentration of 100 ng/ml. Mini-osmotic pumps (model 2001; Alzet Corporation) were filled with 200 μl of this BNP solution to release 2 ng/h/g body weight over 7 days. The control group received pumps containing saline only. All mice were individually housed. During days 5–7, mice were transferred to Oxymax chambers (Columbus Instruments) maintained at 24°C ± 1°C and given free access to chow and water to measure oxygen consumption (VO2), carbon dioxide production (VCO2), physical activity, and food and water intake. All measurements were made at 15-minute intervals for 3 days, with the first day of acclimation discarded from the analysis.
At the end of all experiments, blood was collected into tubes containing EDTA (5 mM final concentration) and protease inhibitors, and tissues (iBAT, epididymal and inguinal WAT, heart) were rapidly isolated, frozen in liquid nitrogen, and stored at –80°C. Plasma was prepared from blood by centrifugation at 2,000 g for 10 minutes at 4°C. Plasma levels of BNP were measured using the RayBio BNP Enzyme Immunoassay Kit (RayBiotech).
Cell culture
Primary adipocyte cultures from NPR-C+/+ and NPR-C–/– mice.
iBAT and epididymal WAT were collected for primary cell cultures essentially as described previously (70). When reaching confluency, the cells were differentiated in 1:1 DMEM/F12 media containing 10% FBS with 0.5 mM isobutylmethylxanthine, 0.4 μg/ml dexamethasone, 5 μg/ml insulin, and 1 μM rosiglitazone. After 2 days, cells received fresh DMEM plus 10% FBS medium without additives, and cells were replenished with the same media every 3 days for 8 to 9 days.
Human subcutaneous adipocytes.
Human subcutaneous adipocytes were obtained from Zen-Bio as previously described (44, 71, 72). These were pooled samples from healthy, nondiabetic women (n = 7; average BMI, 26.8 kg/m2; range, 25.00–29.57; average age, 49 years; range, 40–68 years) and were differentiated into adipocytes according to the supplier’s protocol.
hMADS cells.
hMADS cells (40, 73), which express not only white adipocyte genes, such as leptin and adiponectin, but also exhibit features of functional brown adipocytes, were maintained as a stock in DMEM (low glucose), 10% fetal bovine serum, 2 mM l-glutamine, 10 mM HEPES buffer, 50 units/ml penicillin, and 50 μg/ml streptomycin, supplemented with 2.5 ng/ml human fibroblast growth factor-2. Cell growth and differentiation were performed as follows. Briefly, when reaching confluency, the cells were differentiated in DMEM/F12 (50:50) media supplemented with 10 μg transferrin, 0.85 μM insulin, and 1 μM rosiglitazone. After day 2, fresh media was added without dexamethasone and isobutyl methylxanthine. On day 6, rosiglitazone was removed. All experiments were performed between days 10 and 12, when at least 80% of hMADS cells were well differentiated. Oil Red O staining for lipid droplets was used to evaluate adipocyte differentiation.
Lipolysis and kinase assays
Human and mouse adipocytes were treated or not with 100 nM ANP or 100 nM Iso for a time course of lipolysis. Glycerol released in the cultured medium was measured by the Free Glycerol Determination Kit (Sigma-Aldrich), and concentrations were determined by comparison with a standard curve. Intraassay and interassay CV were less than 6% and 9.1%, respectively. For measurement of intracellular cyclic nucleotide levels, human adipocytes were treated at 37°C for 0, 20, 40 minutes with ANP or Iso (100 nM for each). The medium was then rapidly removed, 0.5 ml of 0.1 M HCl was added, and an aliquot of the lysate was used to measure cAMP and cGMP concentrations using an enzyme immunoassay kit from Cayman Chemical.
RNA isolation and analysis
Total RNA was extracted from human or mouse adipocytes or mouse tissues using TRIzol (Invitrogen). Reverse transcription (RT) of 2 μg RNA was performed with the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems). Each single gene expression experiment was performed in triplicate. Differences in total RNA or different efficiency of cDNA synthesis among samples were normalized using human GAPDH expression for hMADS and mouse 36B4 for mouse samples. The primer sequences are noted in Supplemental Table 2.
Human mitochondria PCR array
RNA was extracted from ANP-treated cells and control cells, incubated with RNase-free DNase, and purified using the RNeasy Mini Kit (Qiagen). The RT reaction was performed with 400 μg total RNA using the RT2 First-Strand Kit (SABiosciences). Two hundred nanograms of random-primed cDNAs were processed for quantitative real-time reverse-transcriptase PCR of 84 genes involved in biogenesis and function of mitochondria and 12 housekeeping genes, including internal controls, by using an RT2 Profiler PCR Array Kit (RT2 Profiler PCR Array Human Mitochondria, PAHS-087A, SABioscience) and an Eppendorf RealPlex2 Mastercycler PCR system. PCR products were quantified by measuring SYBR Green (Invitrogen) fluorescent dye incorporation with ROX dye reference. Using an integrated web-based software package for the PCR Array System, the ΔΔCt-based fold-change calculations from the uploaded raw threshold cycle data were obtained.
mtDNA content
Genomic and mtDNA was isolated from hMADS cells as previously described (74). In this assay, NADH dehydrogenase (ND1) primers are used to evaluate mitochondria DNA, and lipoprotein lipase (LPL) primers are used to measure nuclear DNA. ND1 expression level is normalized to that of LPL. The primer sequences are noted in Supplemental Table 2.
Western blotting
Cells or tissues were lysed and sonicated in buffer containing 25 mM HEPES (pH 7.4), 150 mM NaCL, 5 mM EDTA, 5 mM EGTA, 5 mM glycerophosphate, 0.9% Triton X-100, 0.1% IGEPAL, 5 mM sodium pyrophosphate, 10% glycerol, plus 1 tablet each of complete protease inhibitor cocktail (Roche) and PhoSTOP phosphatase inhibitors (Roche) per 10 ml of lysis buffer. For Western blot analysis, 50 μg total protein (15 μg for BAT lysates) was resolved in 4%–20% Tris-glycine gels (Invitrogen), transferred to nitrocellulose membranes, and probed overnight at 4°C with specific primary antibodies. Secondary antisera against rabbit or mouse IgG conjugated with alkaline phosphatase was used for specific protein detection. Image acquisition was performed on a Typhoon FLA9000 variable mode imager and analyzed using ImageQuant TL software. In some cases, membranes were “stripped” by incubation in a buffer (0.76 g Tris, 2 g SDS, 700 μl β-mercaptoethanol in 100 ml) at 37°C for 45 minutes, in order to be subsequently probed with additional antibodies.
Cell respiration measurements
Cells were seeded into 0.2% gelatin-covered 24-well plates (XF24) (20,000 cells/well) or 96-well plates (XF96) (10,000 cells/well) (both from Seahorse Bioscience) for OCRs and ECARs. On the day of experiments, the cells were washed with 1 ml XF-DMEM (no. D5030, Sigma-Aldrich) containing 1 mM sodium pyruvate, 2 mM GlutaMAX-1, 17.5 mM glucose, 1.85 g/l NaCl, and 15 mg/l phenol red (pH 7.4), and 500 or 100 μl of this media were added per well of cells in XF24 or XF96 plates, respectively. Cellular OCRs and ECARs were measured as described previously (44, 75). As indicated for specific experiments, bioenergetics of hMADS cells were tested in response to 100 nM Iso, ANP, or L755; oxygen consumption was blocked by oligomycin (1 μg/ml), an ATP synthase inhibitor; the ionophore FCCP (carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; 0.6 μmol/l) assayed maximal respiratory capacity of mitochondria, whereas rotenone (3 μmol/l), a mitochondrial inhibitor, was used to block mitochondrial respiration. For some experiments, cells were pretreated with Rp-8-CPT-cGMPS (500 μM) or prestimulated overnight with 100 nM ANP or Iso. Optimal drug concentrations were established in preliminary experiments.
Transient transfection and reporter gene assays
Three μg of pGL4-UCP1 enhancer-TATA-Luc (subcloned from a previous vector; ref. 46) or pGL3-PGC-1α-Luc (gift from Daniel P. Kelly, Diabetes and Obesity Research Center, Sanford-Burnham Medical Research Institute at Lake Nona, Orlando, Florida, USA) were transfected into differentiated hMADS cells, together with 1 μg of pCMV-β-galactosidase by calcium phosphate precipitation as previously described (76). Media was changed after 6 hours, and cells were treated or not with ANP (100 nM). Twenty-four hours later, cells were washed with PBS, and the lysates were prepared for Luciferase Assay (Promega) and β-galactosidase activity.
ChIP
These assays were performed essentially as described previously (77). Briefly, hMADS cells were grown and differentiated in 10-cm dishes, pretreated for 30 minutes with the p38 inhibitor SB (10 μM) or PKGi (500 μM), followed by ANP (100 nM) for 2 hours, and then processed for ChIP assay. Polyclonal antibody against PGC-1α or ATF2 (see Reagents and antibodies) was used for immunoprecipitation, and PCR was performed using primers for the enhancer region of the human Ucp1 gene, which generates a 402-bp fragment. The primer sequences are noted in Supplemental Table 2.
Histology and microscopy
Cultured cells.
For UCP1 staining, hMADS cells were differentiated on chamber slides and were treated or not with PKGi or SB, followed by overnight incubation with ANP (100 nM). Cells were washed in PBS and fixed with 3% paraformaldehyde and 0.2% Triton-X 100 in PBS for 20 minutes at 4°C. Fixed cells were washed 3 times in blocking solution, 10% bovine serum albumin in PBS, incubated overnight with UCP1 antibody, washed, and incubated with secondary antibody conjugated with FITC. Cells were washed 3 times in PBS and mounted in SlowFade Gold Antifade Reagent with DAPI for nuclear staining (Invitrogen). Fluorescence analysis was performed using a IX81 Olympus Microscope and image acquired with cooled CCD monochrome CCD camera QuantEM. Two wells of cells treated with ANP were incubated only with secondary antibody and used as a IgG negative control. To image mitochondria, 250 nM MitoTracker (M7513, Invitrogen) was added directly into the media of untreated and treated adipocytes (ANP 100 nM) and incubated for 45 minutes at 37°C. MitoTracker was oxidized, sequestered, and conjugated in mitochondria. After incubation, the cells were washed with 1:1 DMEM/F12 and fixed with 3.7% paraformaldehyde for 1 hour at room temperature. Fixed cells were processed and imaged as above.
Isolated tissues.
iBAT and epididymal WAT isolated from NPR-C+/+ and NPR-C–/– mice were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned (5-μm thickness). Sections were stained VWR hematoxylin (catalog no. 95057-844) and VWR eosin (catalog no. 95057-848) and examined under bright-field microscopy with a Nikon 80i. Epididymal WAT from NPR-C+/+ and NPR-C–/– mice was also embedded in OCT medium (Tissue-Tek, Sakura Finetek Europe), frozen, and stored at –80°C for UCP1 immunohistochemistry as previously described (78).
Statistics
Results are presented as mean ± SEM unless otherwise indicated. Data were analyzed using 2-tailed Student’s t test or 1-way ANOVA, followed by post-hoc Newman-Keuls tests when F was significant. Food and water intake, physical activity, and respiration data were analyzed by repeated measures ANOVA, and post-hoc comparisons were made using Bonferroni test. Differences were considered significant at P < 0.05. Prism 5.0 software was used for statistical analyses; P values for significance are indicated for each data set.
Study approval
All animal experiments were approved by the Institutional Animal Care and Use Committees of the Sanford-Burnham Medical Research Institute at Lake Nona and University of North Carolina at Chapel Hill in accordance with NIH Guides for the Care and Use of Laboratory Animals.
Supplementary Material
Acknowledgments
We thank Nobuyo Maeda and Oliver Smithies for the gift of tissues from NPR-C+/+ and NPR-C–/– mice, Zen-Bio Inc. for human subcutaneous adipocytes, the Sanford-Burnham Histology Core, Jian-Liang Li for assistance with PCR array representation, William Wetsel for statistical advice and consultation, Phil Wood for comments on the manuscript, Julio Ayala and SBMRI Cardiometabolic Phenotyping Core, and our colleagues for helpful suggestions. This work was supported by an International Fellowship from The Italian Society of Hypertension (to M. Bordicchia), NIH R01DK53092 (to S. Collins), and the Sanford-Burnham Medical Research Institute.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):1022–1036. doi:10.1172/JCI59701.
See the related Commentary beginning on page 804.
References
- 1.Chinkers M, et al. A membrane form of guanylate cyclase is an atrial natriuretic peptide receptor. Nature. 1989;338(6210):78–83. doi: 10.1038/338078a0. [DOI] [PubMed] [Google Scholar]
- 2.Waldman SA, Rapoport RM, Murad F. Atrial natriuretic factor selectively activates particulate guanylate cyclase and elevates cyclic GMP in rat tissues. . J Biol Chem. 1984;259(23):14332–14334. [PubMed] [Google Scholar]
- 3.Maack T, et al. Physiological role of silent receptors of atrial natriuretic factor. Science. 1987;238(4827):675–678. doi: 10.1126/science.2823385. [DOI] [PubMed] [Google Scholar]
- 4.Sarzani R, Paci VM, Dessi-Fulgheri P, Espinosa E, Rappelli A. Comparative analysis of atrial natriuretic peptide receptor expression in rat tissues. . J Hypertens Suppl. 1993;11(5):S214–215. [PubMed] [Google Scholar]
- 5.Sarzani R, Dessi-Fulgheri P, Paci VM, Espinosa E, Rappelli A. Expression of natriuretic peptide receptors in human adipose and other tissues. J Endocrinol Invest. 1996;19(9):581–585. doi: 10.1007/BF03349021. [DOI] [PubMed] [Google Scholar]
- 6.Sarzani R, et al. Fasting inhibits natriuretic peptides clearance receptor expression in rat adipose tissue. J Hypertens. 1995;13(11):1241–1246. doi: 10.1097/00004872-199511000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Dessi-Fulgheri P, et al. Plasma atrial natriuretic peptide and natriuretic peptide receptor gene expression in adipose tissue of normotensive and hypertensive obese patients. J Hypertens. 1997;15(12 pt 2):1695–1699. doi: 10.1097/00004872-199715120-00074. [DOI] [PubMed] [Google Scholar]
- 8.Sengenes C, Berlan M, De Glisezinski, I, Lafontan M, Galitzky J. Natriuretic peptides: a new lipolytic pathway in human adipocytes. FASEB J. 2000;14(10):1345–1351. doi: 10.1096/fj.14.10.1345. [DOI] [PubMed] [Google Scholar]
- 9.Sengenes C, et al. Natriuretic peptide-dependent lipolysis in fat cells is a primate specificity. Am J Physiol Regul Integr Comp Physiol. 2002;283(1):R257–R265. doi: 10.1152/ajpregu.00453.2001. [DOI] [PubMed] [Google Scholar]
- 10.Carmen GY, Victor SM. Signalling mechanisms regulating lipolysis. Cell Signal. 2006;18(4):401–408. doi: 10.1016/j.cellsig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Potter LR, Hunter T. Guanylyl cyclase-linked natriuretic peptide receptors: structure and regulation. J Biol Chem. 2001;276(9):6057–6060. doi: 10.1074/jbc.R000033200. [DOI] [PubMed] [Google Scholar]
- 12.Garbers DL, Lowe DG. Guanylyl cyclase receptors. J Biol Chem. 1994;269(49):30741–30744. [PubMed] [Google Scholar]
- 13.Sengenes C, et al. Involvement of a cGMP-dependent pathway in the natriuretic peptide-mediated hormone-sensitive lipase phosphorylation in human adipocytes. J Biol Chem. 2003;278(49):48617–48626. doi: 10.1074/jbc.M303713200. [DOI] [PubMed] [Google Scholar]
- 14.Wood JS, Yan X, Mendelow M, Corbin JD, Francis SH, Lawrence DS. Precision substrate targeting of protein kinases. The cGMP- and cAMP-dependent protein kinases. J Biol Chem. 1996;271(1):174–179. doi: 10.1074/jbc.271.1.174. [DOI] [PubMed] [Google Scholar]
- 15.Lafontan M, Moro C, Berlan M, Crampes F, Sengenes C, Galitzky J. Control of lipolysis by natriuretic peptides and cyclic GMP. Trends Endocrinol Metab. 2008;19(4):130–137. doi: 10.1016/j.tem.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Moro C, et al. Atrial natriuretic peptide contributes to physiological control of lipid mobilization in humans. FASEB J. 2004;18(7):908–910. doi: 10.1096/fj.03-1086fje. [DOI] [PubMed] [Google Scholar]
- 17.Moro C, et al. Atrial natriuretic peptide stimulates lipid mobilization during repeated bouts of endurance exercise. Am J Physiol Endocrinol Metab. 2006;290(5):E864–E869. doi: 10.1152/ajpendo.00348.2005. [DOI] [PubMed] [Google Scholar]
- 18.Moro C, et al. Exercise-induced lipid mobilization in subcutaneous adipose tissue is mainly related to natriuretic peptides in overweight men. Am J Physiol Endocrinol Metab. 2008;295(2):E505–E513. doi: 10.1152/ajpendo.90227.2008. [DOI] [PubMed] [Google Scholar]
- 19.Birkenfeld AL, et al. Lipid mobilization with physiological atrial natriuretic peptide concentrations in humans. J Clin Endocrinol Metab. 2005;90(6):3622–3628. doi: 10.1210/jc.2004-1953. [DOI] [PubMed] [Google Scholar]
- 20.Birkenfeld AL, et al. Atrial natriuretic peptide induces postprandial lipid oxidation in humans. Diabetes. 2008;57(12):3199–3204. doi: 10.2337/db08-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chainani-Wu N, et al. Relation of B-type natriuretic peptide levels to body mass index after comprehensive lifestyle changes. Am J Cardiol. 2010;105(11):1570–1576. doi: 10.1016/j.amjcard.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Chen-Tournoux A, et al. Effect of weight loss after weight loss surgery on plasma N-terminal pro-B-type natriuretic peptide levels. Am J Cardiol. 2010;106(10):1450–1455. doi: 10.1016/j.amjcard.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TJ, et al. Impact of obesity on plasma natriuretic peptide levels. Circulation. 2004;109(5):594–600. doi: 10.1161/01.CIR.0000112582.16683.EA. [DOI] [PubMed] [Google Scholar]
- 24.Khan AM, et al. Cardiac natriuretic peptides, obesity, and insulin resistance: evidence from two community-based studies. J Clin Endocrinol Metab. 2011;96(10):3242–3249. doi: 10.1210/jc.2011-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Chu WS, Lu T, Hasstedt SJ, Kern PA, Elbein SC. Uncoupling protein-2 polymorphisms in type 2 diabetes, obesity, and insulin secretion. Am J Physiol Endocrinol Metab. 2004;286(1):E1–E7. doi: 10.1152/ajpendo.00231.2003. [DOI] [PubMed] [Google Scholar]
- 26.Sugisawa T, Kishimoto I, Kokubo Y, Makino H, Miyamoto Y, Yoshimasa Y. Association of plasma B-type natriuretic peptide levels with obesity in a general urban Japanese population: the Suita Study. Endocr J. 2010;57(8):727–733. doi: 10.1507/endocrj.K10E-067. [DOI] [PubMed] [Google Scholar]
- 27.Cinti S. The role of brown adipose tissue in human obesity. Nutr Metab Cardiovasc Dis. 2006;16(8):569–574. doi: 10.1016/j.numecd.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Himms-Hagen J, et al. Effect of CL-316,243, a thermogenic β3-agonist, on energy balance and brown and white adipose tissues in rats. . Am J Physiol. 1994;266(4 pt 2):R1371–R1382. doi: 10.1152/ajpregu.1994.266.4.R1371. [DOI] [PubMed] [Google Scholar]
- 29.Surwit RS, et al. Differential effects of fat and sucrose on the development of obesity and diabetes in C57BL/6J and A/J mice. Metabolism. 1995;44(5):645–651. doi: 10.1016/0026-0495(95)90123-X. [DOI] [PubMed] [Google Scholar]
- 30.Collins S, Daniel KW, Petro AE, Surwit RS. Strain-specific response to β3-adrenergic receptor agonist treatment of diet-induced obesity in mice. . Endocrinology. 1997;138(1):405–413. doi: 10.1210/en.138.1.405. [DOI] [PubMed] [Google Scholar]
- 31.Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. J Clin Invest. 1998;102(2):412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almind K, Kahn CR. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53(12):3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- 33.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293(2):E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 34.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 36.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 37.van Marken Lichtenbelt WD, Schrauwen P. Implications of non-shivering thermogenesis for energy balance regulation in humans. Am J Physiol Regul Integr Comp Physiol. 2011;301(2):R285–R296. doi: 10.1152/ajpregu.00652.2010. [DOI] [PubMed] [Google Scholar]
- 38.Cao W, et al. p38 MAP kinase is the central regulator of cAMP-dependent transcription of the brown fat uncoupling protein-1 gene. Mol Cell Biol. 2004;24(7):3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsukawa N, et al. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci U S A. 1999;96(13):7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elabd C, et al. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells. 2009;27(11):2753–2760. doi: 10.1002/stem.200. [DOI] [PubMed] [Google Scholar]
- 41.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121(1):96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nisoli E, et al. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299(5608):896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 43.Nisoli E, Carruba MO. Nitric oxide and mitochondrial biogenesis. J Cell Sci. 2006;119(pt 14):2855–2862. doi: 10.1242/jcs.03062. [DOI] [PubMed] [Google Scholar]
- 44.Yehuda-Shnaidman E, Buehrer B, Pi J, Kumar N, Collins S. Acute stimulation of white adipocyte respiration by PKA-induced lipolysis. Diabetes. 2010;59(10):2474–2483. doi: 10.2337/db10-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lincoln TM, Corbin JD. Adenosine 3′:5′-cyclic monophosphate- and guanosine 3′:5′-cyclic monophosphate-dependent protein kinases: possible homologous proteins. Proc Natl Acad Sci U S A. 1977;74(8):3239–3243. doi: 10.1073/pnas.74.8.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao W, Medvedev AV, Daniel KW, Collins S. β-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein-1 (UCP1) gene requires p38 MAP kinase. J Biol Chem. 2001;276(29):27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 47.Kozak UC, Kopecky J, Teisinger J, Enerback S, Boyer B, Kozak LP. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol Cell Biol. 1994;14(1):59–67. doi: 10.1128/mcb.14.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.del Mar Gonzalez-Barroso M, et al. Transcriptional activation of the human ucp1 gene in a rodent cell line. Synergism of retinoids, isoproterenol, and thiazolidinedione is mediated by a multipartite response element. J Biol Chem. 2000;275(41):31722–31732. doi: 10.1074/jbc.M001678200. [DOI] [PubMed] [Google Scholar]
- 49.Cassard-Doulcier AM, Gelly C, Bouillaud F, Ricquier D. A 211-bp enhancer of the rat uncoupling protein-1 (UCP-1) gene controls specific and regulated expression in brown adipose tissue. Biochem J. 1998;333(pt 2):243–246. doi: 10.1042/bj3330243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao W, et al. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. 2004;24(7):3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, et al. Liver X receptor is a transcriptional repressor of the uncoupling protein-1 gene and brown adipocyte phenotype. Mol Cell Biol. 2008;28(7):2187–2200. doi: 10.1128/MCB.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wijers SL, Schrauwen P, van Baak MA, Saris WH, van Marken Lichtenbelt WD. Beta-adrenergic receptor blockade does not inhibit cold-induced thermogenesis in humans: possible involvement of brown adipose tissue. J Clin Endocrinol Metab. 2011;96(4):E598–E605. doi: 10.1210/jc.2010-1957. [DOI] [PubMed] [Google Scholar]
- 53.Davidson NC, Naas AA, Hanson JK, Kennedy NS, Coutie WJ, Struthers AD. Comparison of atrial natriuretic peptide B-type natriuretic peptide, and N-terminal proatrial natriuretic peptide as indicators of left ventricular systolic dysfunction. Am J Cardiol. 1996;77(10):828–831. doi: 10.1016/S0002-9149(97)89176-X. [DOI] [PubMed] [Google Scholar]
- 54.Maisel AS, et al. Rapid measurement of B-type natriuretic peptide in the emergency diagnosis of heart failure. N Engl J Med. 2002;347(3):161–167. doi: 10.1056/NEJMoa020233. [DOI] [PubMed] [Google Scholar]
- 55.Kohno T, et al. Role of high-mobility group box 1 protein in post-infarction healing process and left ventricular remodelling. Cardiovasc Res. 2009;81(3):565–573. doi: 10.1093/cvr/cvn291. [DOI] [PubMed] [Google Scholar]
- 56.Collins S, Cao W, Robidoux J. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol Endocrinol. 2004;18(9):2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Rogers PM, Su C, Varga G, Stayrook KR, Burris TP. Regulation of cholesterologenesis by the oxysterol receptor, LXRalpha. J Biol Chem. 2008;283(39):26332–26339. doi: 10.1074/jbc.M804808200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliver PM, et al. Hypertension, cardiac hypertrophy, and sudden death in mice lacking natriuretic peptide receptor A. Proc Natl Acad Sci U S A. 1997;94(26):14730–14735. doi: 10.1073/pnas.94.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haas B, et al. Protein kinase G controls brown fat cell differentiation and mitochondrial biogenesis. Sci Signal. 2009;2(99):ra78. doi: 10.1126/scisignal.2000511. [DOI] [PubMed] [Google Scholar]
- 60.Miyashita K, et al. Natriuretic peptides/cGMP/cGMP-dependent protein kinase cascades promote muscle mitochondrial biogenesis and prevent obesity. Diabetes. 2009;58(12):2880–2892. doi: 10.2337/db09-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meirhaeghe A, et al. Association between the T-381C polymorphism of the brain natriuretic peptide gene and risk of type 2 diabetes in human populations. Hum Mol Genet. 2007;16(11):1343–1350. doi: 10.1093/hmg/ddm084. [DOI] [PubMed] [Google Scholar]
- 62.Choquet H, et al. The T-381C SNP in BNP gene may be modestly associated with type 2 diabetes: an updated meta-analysis in 49 279 subjects. Hum Mol Genet. 2009;18(13):2495–2501. doi: 10.1093/hmg/ddp169. [DOI] [PubMed] [Google Scholar]
- 63.Maisel A, et al. State of the art: using natriuretic peptide levels in clinical practice. Eur J Heart Fail. 2008;10(9):824–839. doi: 10.1016/j.ejheart.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 64.Noveanu M, Potocki M, Mueller C. Natriuretic peptides and their evolving clinical applications. Future Cardiol. 2008;4(6):593–598. doi: 10.2217/14796678.4.6.593. [DOI] [PubMed] [Google Scholar]
- 65.Sharma R, Anker SD. Cardiac cachexia is a world-wide problem. Int J Cardiol. 1999;71(2):113–114. doi: 10.1016/S0167-5273(99)00130-8. [DOI] [PubMed] [Google Scholar]
- 66.Anker SD, et al. Hormonal changes and catabolic/anabolic imbalance in chronic heart failure and their importance for cardiac cachexia. Circulation. 1997;96(2):526–534. doi: 10.1161/01.cir.96.2.526. [DOI] [PubMed] [Google Scholar]
- 67.Anker SD, et al. Wasting as independent risk factor for mortality in chronic heart failure. Lancet. 1997;349(9058):1050–1053. doi: 10.1016/S0140-6736(96)07015-8. [DOI] [PubMed] [Google Scholar]
- 68.Bando M, Ishii Y, Sugiyama Y, Kitamura S. Elevated plasma brain natriuretic peptide levels in chronic respiratory failure with cor pulmonale. Respir Med. 1999;93(7):507–514. doi: 10.1016/S0954-6111(99)90094-X. [DOI] [PubMed] [Google Scholar]
- 69.Fiorica JV, Graham L, Rao PS, Hoffman MS, Roberts WS, Cavanagh D. Atrial natriuretic factor in gynecologic malignancy. Obstet Gynecol. 1995;85(5 pt 1):740–744. doi: 10.1016/0029-7844(95)00036-q. [DOI] [PubMed] [Google Scholar]
- 70.Rehnmark S, et al. Brown adipocytes differentiated in vitro can express the gene for the uncoupling protein thermogenin: effects of hypothyroidism and norepinephrine. Exp Cell Res. 1989;182(1):75–83. doi: 10.1016/0014-4827(89)90280-2. [DOI] [PubMed] [Google Scholar]
- 71.Halvorsen YD, et al. Thiazolidinediones and glucocorticoids synergistically induce differentiation of human adipose tissue stromal cells: biochemical, cellular, and molecular analysis. Metabolism. 2001;50(4):407–413. doi: 10.1053/meta.2001.21690. [DOI] [PubMed] [Google Scholar]
- 72.Sen A, et al. Adipogenic potential of human adipose derived stromal cells from multiple donors is heterogeneous. J Cell Biochem. 2001;81(2):312–319. doi: 10.1002/1097-4644(20010501)81:2<312::AID-JCB1046>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez AM, et al. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem Biophys Res Commun. 2004;315(2):255–263. doi: 10.1016/j.bbrc.2004.01.053. [DOI] [PubMed] [Google Scholar]
- 74.Lai L, et al. Transcriptional coactivators PGC-1alpha and PGC-1beta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22(14):1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol. 2007;292(1):C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 76.Collins S, Bouvier M, Bolanowski MA, Caron MG, Lefkowitz RJ. cAMP stimulates transcription of the β2-adrenergic receptor gene in response to short-term agonist exposure. . Proc Natl Acad Sci U S A. 1989;86(13):4853–4857. doi: 10.1073/pnas.86.13.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nielsen R, Grontved L, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Mol Cell Biol. 2006;26(15):5698–5714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cinti S, Zingaretti MC, Cancello R, Ceresi E, Ferrara P. Morphologic techniques for the study of brown adipose tissue and white adipose tissue. In: Ailhaud G, ed.Methods in Molecular Biology . Totowa, New Jersey, USA: Humana Press; 2001:21–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.