Abstract
Patients with mutations in either TGF-β receptor type I (TGFBR1) or TGF-β receptor type II (TGFBR2), such as those with Loeys-Dietz syndrome, have craniofacial defects and signs of elevated TGF-β signaling. Similarly, mutations in TGF-β receptor gene family members cause craniofacial deformities, such as cleft palate, in mice. However, it is unknown whether TGF-β ligands are able to elicit signals in Tgfbr2 mutant mice. Here, we show that loss of Tgfbr2 in mouse cranial neural crest cells results in elevated expression of TGF-β2 and TGF-β receptor type III (TβRIII); activation of a TβRI/TβRIII-mediated, SMAD-independent, TRAF6/TAK1/p38 signaling pathway; and defective cell proliferation in the palatal mesenchyme. Strikingly, Tgfb2, Tgfbr1 (also known as Alk5), or Tak1 haploinsufficiency disrupted TβRI/TβRIII-mediated signaling and rescued craniofacial deformities in Tgfbr2 mutant mice, indicating that activation of this noncanonical TGF-β signaling pathway was responsible for craniofacial malformations in Tgfbr2 mutant mice. Thus, modulation of TGF-β signaling may be beneficial for the prevention of congenital craniofacial birth defects.
Introduction
Craniofacial deformities, such as cleft palate, are among the most common congenital birth defects in humans (1). Altered TGF-β signaling causes syndromic and nonsyndromic cleft palate. For instance, mutations in the genes for TGF-β receptor type I or type II (TGFBR1 [also known as ALK5] or TGFBR2, respectively) are associated with Loeys-Dietz syndrome (previously called Marfan syndrome type II) in humans, which can manifest with craniofacial malformations, including cleft palate, craniosynostosis, hypertelorism, and vascular defects (2, 3). In addition, mutations in FBN1, which encodes an elastic extracellular matrix protein called fibrillin 1, lead to excessive TGF-β signaling and cause Marfan syndrome, which exhibits clinical phenotypes similar to those of Loeys-Dietz syndrome (4–6). Furthermore, individuals with DiGeorge syndrome, which results from a variably sized deletion on chromosome 22 (del22q11), exhibit altered TGF-β signaling (7–9). Thus, TGF-β signaling is crucial in regulating craniofacial development, and altered TGF-β signaling leads to multiple malformations in humans. Interestingly, these studies show that TGF-β signaling is elevated in patients with these syndromes. Nevertheless, we still do not have a clear understanding of the TGF-β signaling mechanism and its functional impact on craniofacial development in patients with mutations in TGFBR genes.
TGF-β signaling mediates a wide range of biological activities in development and disease. TGF-β ligands signal through heterodimeric type I and type II receptors (TGF-β receptor type I [TβRI, also known as ALK5 and TGFBR1] and TβRII) that are members of the serine/threonine kinase family. To date, overwhelming evidence supports the notion that both TβRII and TβRI are indispensable in eliciting the biological response of TGF-β (10). The third member of the TGF-β receptor family, TβRIII (β-glycan), has high binding affinity for TGF-β2 and may be critical for keeping TGF-β at the cell surface for the TβRI/TβRII complex (11). TβRIII may be required for optimal TGF-β2 function during development. Despite the fact that it is missing an intracellular kinase domain, TβRIII plays a crucial role in regulating embryonic development, because loss of Tgfbr3 results in an early embryonic lethal phenotype in mice (12).
Tgfbr1/Alk5, Tgfbr2, and Tgfbr3 are expressed in the developing craniofacial region, and loss of Tgfbr1/Alk5 or Tgfbr2 results in an array of craniofacial deformities, clearly indicating the functional significance of TGF-β signaling in regulating craniofacial development in mice (13–16). In the canonical TGF-β signaling pathway, the TβRII/TβRI complex activates the receptor-regulated SMADs, which form a transcriptional regulatory complex with SMAD4, move into the nucleus, and control downstream target gene expression (11). In parallel, our recent study showed that ectodermal SMAD4 and p38 are functionally redundant in mediating TGF-β signaling and demonstrated the significance of the SMAD-independent pathway in regulating craniofacial development (17).
In order to test the functional significance of TGF-β signaling in regulating the fate of cranial neural crest (CNC) cells, we generated a mutant animal model, in which loss of Tgfbr2 in CNC cells leads to an array of craniofacial deformities, including cleft palate (13). We have shown that CNC cells have a cell autonomous requirement for TβRII to regulate cell proliferation during palatogenesis. In the present study, we investigated the molecular mechanism of TGF-β signaling-mediated palatogenesis by analyzing TGF-β downstream target genes using Tgfbr2fl/fl;Wnt1-Cre mice. Surprisingly, we detected an elevation of TGF-β2 expression that is responsible for the cell proliferation defect and cleft palate in Tgfbr2 mutant mice. Our study demonstrates that elevated TGF-β2 activates an alternative TGF-β signaling pathway through the TβRI/TβRIII receptor complex and induces a SMAD-independent TNF receptor–associated factor 6/TGF-β–activated kinase 1/p38 (TRAF6/TAK1/p38) signaling cascade in the absence of TβRII. Importantly, genetic manipulation of this alternative TGF-β signaling prevents craniofacial deformities in Tgfbr2 mutant mice and offers what we believe to be a unique opportunity for the prevention of craniofacial malformations.
Results
Identification of TGF-β downstream target genes during palatogenesis.
Mice lacking Tgfbr2 in CNC cells exhibit an array of craniofacial deformities, including cleft palate (13). We have shown that CNC cells have a cell autonomous requirement for TβRII to regulate cell proliferation during palatogenesis. To investigate the mechanism of altered TGF-β signaling in Tgfbr2 mutant mice, we performed global gene expression analyses of the palatal tissue of Tgfbr2fl/fl;Wnt1-Cre mice and Tgfbr2fl/fl control mice at E14.5 (during palatal fusion, n = 5 per genotype) and examined the downstream consequences of dysfunctional TGF-β signaling. In this comparison, we uncovered 293 probe sets representing transcripts that were differentially expressed (≥1.5-fold, <5% false discovery rate [FDR]), 149 probe sets that were more abundant in Tgfbr2fl/fl;Wnt1-Cre mice (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI61498DS1), and 144 probe sets that were more abundant in Tgfbr2fl/fl control mice (Supplemental Table 2). The genes identified as altered in Tgfbr2fl/fl;Wnt1-Cre mice were consistent with cell proliferation defects, with significant reductions in the levels of transcripts related to cell cycle control, mitosis, and microtubule-based cell division and with enrichment for transcripts related to the negative regulation of cellular proliferation (Supplemental Table 2).
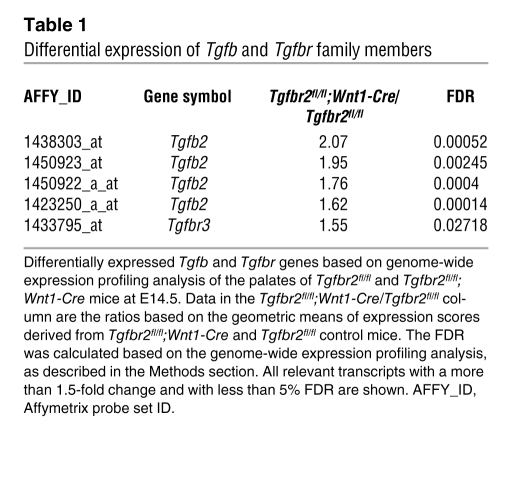
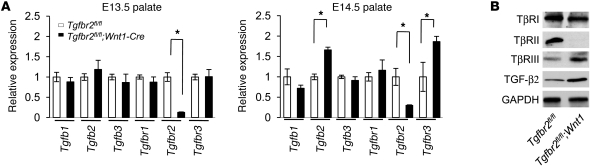
In a focused analysis of Tgfb and Tgfbr gene family member expression, we discovered that Tgfb2 and Tgfbr3 transcript levels were more abundant in the palates of the Tgfbr2fl/fl;Wnt1-Cre mice than in those of the control (Table 1). The microarray data was validated by quantitative RT-PCR analysis of TGF-β ligands and receptors (Figure 1A). As expected, TGF-β2 and TβRIII protein levels were also upregulated in the palatal mesenchyme of Tgfbr2fl/fl;Wnt1-Cre mice at E14.5 (Figure 1B). TGF-β2 has a high affinity for TβRIII, a unique feature that distinguishes TGF-β2 from other TGF-β ligands (18, 19). In in vitro studies, TGF-β isoforms differentially bind to TGF-β receptors and play crucial nonoverlapping roles during craniofacial development (20, 21).
Table 1 .
Differential expression of Tgfb and Tgfbr family members
Figure 1. Altered TGF-β signaling pathway in the palates of Tgfbr2fl/fl;Wnt1-Cre mice.
(A) Quantitative RT-PCR analyses of indicated genes in the palates of Tgfbr2fl/fl (white bars) and Tgfbr2fl/fl;Wnt1-Cre (black bars) mice at E13.5 and E14.5. Three samples were analyzed for each experiment. Error bars represent SD. *P < 0.05 as indicated by a 2-tailed Student’s t test. (B) Immunoblotting analysis of indicated molecules in E14.5 Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre palates.
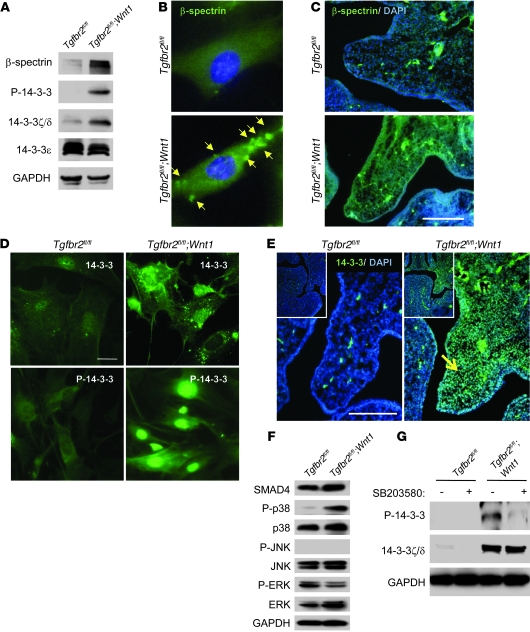
To identify proteins relevant to dysfunctional TGF-β signaling after the loss of Tgfbr2, we analyzed protein extracts from cultured mouse embryonic palatal mesenchymal (MEPM) cells of Tgfbr2fl/fl (control) mice and Tgfbr2fl/fl;Wnt1-Cre mice (Supplemental Figure 1 and Supplemental Table 3). We found that β-spectrin, an adaptor protein of TβRI, was upregulated in Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Figure 2A, Supplemental Figure 1C, and Supplemental Table 3). β-Spectrin is associated with TGF-β receptors, and loss of β-spectrin leads to disruption of TGF-β signaling by SMAD proteins in mice, indicating that β-spectrin is involved in SMAD-dependent TGF-β signaling (22). β-Spectrin knockout mice exhibit a phenotype similar to that of Smad2+/–;Smad3+/– mutant mice, with midgestational death due to gastrointestinal, liver, neural, and heart defects (22). β-Spectrin expression was detectable in primary MEPM cells and palatal mesenchyme of both Tgfbr2fl/fl;Wnt1-Cre and control mice but appears to be increased in Tgfbr2fl/fl;Wnt1-Cre mice (Figure 2, B and C). Consistent with its ability to interact with TGF-β receptors, β-spectrin was mainly localized at the cell membrane in Tgfbr2fl/fl;Wnt1-Cre mice (Figure 2B, arrows).
Figure 2. Identification of molecules with upregulated expression in primary MEPM cells from Tgfbr2fl/fl;Wnt1-Cre mice.
(A) Immunoblotting analysis of indicated molecules in primary MEPM cells of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice. (B) Immunofluorescence analysis of primary MEPM cells of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice using anti–β-spectrin antibody. Arrows indicate expression of β-spectrin. Original magnification, ×400. (C) Immunohistochemical staining of β-spectrin and DAPI staining in sections of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice at E14.0. Scale bar: 50 μm. (D) Immunofluorescence analysis of primary MEPM cells from Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice using anti–14-3-3ζ/δ (14-3-3) or anti–phosphorylated 14-3-3 (P-14-3-3) antibody. Scale bar: 20 μm. (E) Immunohistochemical staining of 14-3-3ζ/δ and DAPI staining in sections of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice at E13.5. 14-3-3ζ/δ expression appears increased in Tgfbr2fl/fl;Wnt1-Cre palate (arrow) compared with that in Tgfbr2fl/fl littermate control. Scale bar: 50 μm. Insets show lower-magnification images (original magnification, ×100). (F) Immunoblotting analysis of indicated molecules in primary MEPM cells from Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice. P-p38, phosphorylated p38; P-JNK, phosphorylated JNK; P-ERK, phosphorylated ERK. (G) Immunoblotting analyses of indicated molecules in primary MEPM cells of Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice treated with (+) or without (–) p38 MAPK inhibitor SB203580. P-14-3-3, phosphorylated 14-3-3.
Similarly, 14-3-3ζ/δ and phosphorylated 14-3-3 were elevated in Tgfbr2fl/fl;Wnt1-Cre MEPM cells compared with those in control cells (Figure 2A). 14-3-3 is a TβRI-interacting protein that is phosphorylated by p38 MAPK α, β, γ, or δ (MAPK14/11/12/13) and plays a key role in protein targeting and protein domain-specific binding (23). 14-3-3 expression appeared to be increased in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, and phosphorylated 14-3-3 was increased in Tgfbr2fl/fl;Wnt1-Cre mice (Figure 2D). Notably, we found elevated 14-3-3 expression in the palatal mesenchyme of Tgfbr2fl/fl;Wnt1-Cre mice compared with that in controls (Figure 2E). Consistent with these results, p38 MAPK was activated only in Tgfbr2 mutant MEPM cells but not in control cells (Figure 2F). To investigate p38 MAPK–mediated 14-3-3 phosphorylation in Tgfbr2fl/fl;Wnt1-Cre mice, we treated MEPM cells with the p38 MAPK inhibitor SB2003580. Phosphorylation of 14-3-3ζ/δ in Tgfbr2fl/fl;Wnt1-Cre MEPM cells was inhibited by SB203580, indicating that 14-3-3 is a downstream substrate of p38 MAPK (Figure 2G).
Alternative TGF-β signaling pathway in the absence of TβRII.
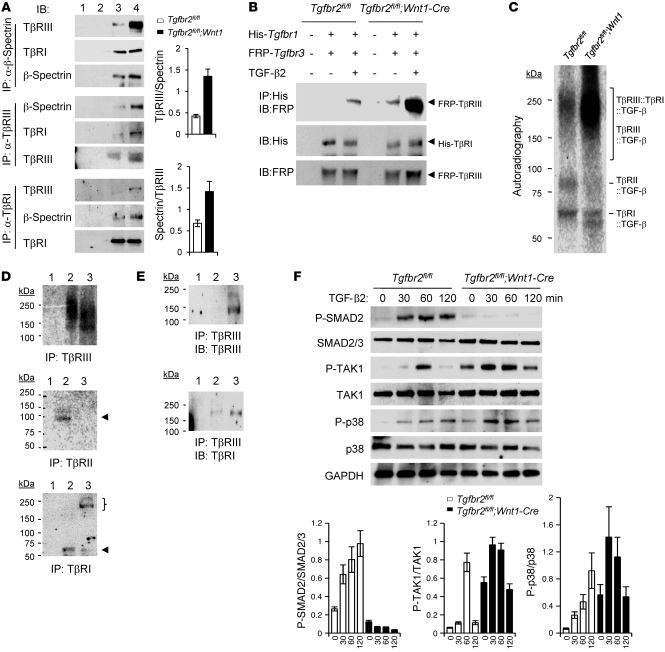
To analyze the mechanism of signaling initiated in the absence of Tgfbr2, we performed coimmunoprecipitation assays using anti-TβRI, TβRIII, and β-spectrin antibodies and extracts of control and Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Figure 3A). β-Spectrin was coimmunoprecipitated with TβRIII and TβRI antibodies in Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Figure 3A). Although the interaction between TβRI and β-spectrin was not altered in Tgfbr2fl/fl;Wnt1-Cre MEPM cells compared with that in control cells, TβRIII was more abundantly coimmunoprecipitated by β-spectrin antibody in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, and vice versa, indicating that β-spectrin stably bound to TβRI and more abundantly bound to TβRIII in the absence of TβRII (Figure 3A). To validate that the interaction between β-spectrin and TβRIII is dependent on TGF-β2, we performed coimmunoprecipitations after treatment with TGF-β2. Although the interaction between TβRI and TβRIII was induced by TGF-β2 in both control and Tgfbr2fl/fl;Wnt1-Cre MEPM cells, the interaction between TβRI and TβRIII was more prominent in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, consistent with upregulated expression of TGF-β2 and TβRIII in Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Supplemental Figure 2). To definitively validate the formation of a TβRI/TβRIII receptor complex in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, we coexpressed His-tagged TβRI and FRP-tagged TβRIII in both control and Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Figure 3B). Notably, we detected colocalization of TβRI and TβRIII in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, whereas we only detected limited colocalization of TβRI and TβRIII in Tgfbr2fl/fl control MEPM cells, supporting our finding that TβRI/TβRIII complex is more abundant in the absence of TβRII (Figure 3B). Furthermore, we performed cross-linking analysis to test whether TGF-β2 binds to TβRI and TβRIII in the absence of Tgfbr2 using radioactive TGF-β2 ligands. In Tgfbr2fl/fl control MEPM cells, radioactive TGF-β2 ligands (12.5 kDa) bind to TβRI (53 kDa), TβRII (70 kDa), and TβRIII (100–200 kDa, highly glycosylated molecule) and form the ligand-receptor complexes of TβRI::TGF-β2 (65.5 kDa), TβRII::TGF-β2 (82.5 kDa), and TβRIII::TGF-β2 (112.5–212.5 kDa) (Figure 3C). In contrast, radioactive TGF-β2 was more abundantly cross-linked with TβRIII and TβRI/TβRIII in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, and complexes of TβRIII::TβRI::TGF-β2 (165.5–265.5 kDa), TβRIII::TGF-β2 (112.5–212.5 kDa), and TβRI::TGF-β2 (65.5 kDa), but not TβRII::TGF-β2 (82.5 kDa), were detectable (Figure 3C). To confirm our findings further, we performed immunoprecipitation–cross-linking analysis (Figure 3D). We found that TGF-β2 can bind to TβRI and TβRIII, but not TβRII, in Tgfbr2fl/fl;Wnt1-Cre MEPM cells and forms a complex with TβRIII and TβRIII/TβRI (Figure 3, D and E). Collectively, our data indicate that a TGF-β2/TβRIII/TβRI complex forms in the absence of Tgfbr2 and likely transduces TGF-β signaling in palatal mesenchymal cells.
Figure 3. Altered ligand/receptor assembly and upregulated TAK1 phosphorylation in the absence of Tgfbr2.
(A) Immunoblotting analysis of immunoprecipitation products derived from IgG control beads with Tgfbr2fl/fl or Tgfbr2fl/fl;Wnt1-Cre MEPM cell extracts (lanes 1 and 2, respectively), Tgfbr2fl/fl MEPM cell extracts (lane 3), and Tgfbr2fl/fl;Wnt1-Cre MEPM cell extracts (lane 4) using the indicated antibodies. The bar graphs show the ratios of TβRIII and β-spectrin after quantitative densitometry analysis of immunoblotting data. Three samples were analyzed for each experiment. Error bars represent SD. (B) Immunoblotting analysis of immunoprecipitation products using anti–His-tag antibody derived from IgG control beads or MEPM cell extracts from Tgfbr2fl/fl or Tgfbr2fl/fl;Wnt1-Cre mice after overexpression of His-tagged Tgfbr1 and FRP-tagged Tgfbr3, with (+) or without (–) TGF-β2 treatment. (C) Cross-linking analysis after treatment with radioactive iodine-125 (125I) TGF-β2 in primary MEPM cells from Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice. (D) Immunoprecipitation products derived from IgG control beads (lane 1), Tgfbr2fl/fl MEPM cell extracts (lane 2), and Tgfbr2fl/fl;Wnt1-Cre MEPM cell extracts (lane 3) after cross-linking with 125I–TGF-β2. The top arrowhead indicates TβRII::TGF-β2 complex after IP with anti-TβRII antibody, and the bottom arrowhead indicates TβRI::TGF-β2 complex after IP with anti-TβRI antibody. The bracket indicates TβRIII::TβRI::TGF-β2 complex. (E) Immunoblotting analysis of immunoprecipitation products derived from IgG control beads (lane 1), Tgfbr2fl/fl MEPM cell extracts (lane 2), and Tgfbr2fl/fl;Wnt1-Cre MEPM cell extracts (lane 3) after cross-linking with 125I–TGF-β2. (F) Immunoblotting analysis of indicated molecules in primary MEPM cells from Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice cultured with TGF-β2 for indicated times. The bar graphs show the ratios of phosphorylated SMAD2 relative to SMAD2/3, phosphorylated TAK1 relative to TAK1, or phosphorylated p38 relative to p38 after quantitative densitometry analysis of immunoblotting data.
TGF-β signals through a receptor complex of both TβRII and TβRI, causing phosphorylation of SMAD proteins (11, 24). We found that SMAD-dependent TGF-β signaling was compromised in Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Figure 3F). In parallel, patients with TGFBR2 mutations show elevated TGF-β signaling (3, 25). We hypothesized that elevated TGF-β2 levels in Tgfbr2 mutant cells initiate an intracellular signal in the absence of Tgfbr2, most likely via a SMAD-independent p38 MAPK pathway, using a hitherto unidentified mechanism.
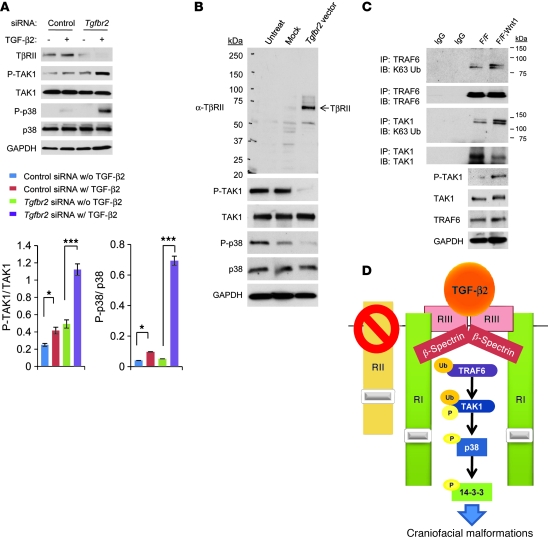
MAPK kinase kinase 7 (MAP3K7), also known as TAK1, was originally identified as an activator of TGF-β–induced p38 activation (26). SMAD-dependent TGF-β signaling is compromised in the palates of Tgfbr2fl/fl;Wnt1-Cre mice (Figure 3F). Based on our observation that p38 MAPK was activated in the palates of Tgfbr2fl/fl;Wnt1-Cre mice, TGF-β2 may activate SMAD-independent TGF-β signaling through a TAK1/p38 MAPK cascade. We found that TAK1 activation was upregulated in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, which correlated with p38 MAPK activation (Figure 3F). In contrast, although TAK1 is also involved in BMP signaling, BMP signaling was not altered in Tgfbr2 mutant cells (Supplemental Figure 3), indicating that BMP signaling was not responsible for TAK1/p38 MAPK activation. To test our model that loss of TβRII leads to the activation of a TAK1/p38 pathway, we performed siRNA knockdown of Tgfbr2 in wild-type MEPM cells. Indeed, reduction of Tgfbr2 in wild-type MEPM cells activated a TAK1/p38 MAPK cascade (Figure 4A). In addition, reintroduction of Tgfbr2 in Tgfbr2 mutant cells reversed ectopic TAK1/p38 MAPK activation (Figure 4B), suggesting that compromised TβRII expression is crucial for the TAK1/p38 MAPK activation. TAK1 can bind to the TAK-associated binding protein 1 (TAB1), TAB2, and TAB3 (27). The activity of this complex is dependent on the amino-terminal RING domain in the E3 TRAF6 (28). Therefore, we speculated that the activity of the TAK1/TAB complex might affect p38 MAPK activation in MEPM cells. To test this hypothesis, we performed siRNA knockdown of Tak1 or Tab1 (Supplemental Figure 4). siRNA knockdown for Tak1, but not Tab1, blocked p38 MAPK activation in Tgfbr2fl/fl;Wnt1-Cre MEPM cells, indicating that TAK1 is crucial for p38 MAPK activation.
Figure 4. TβRII is crucial in regulating TAK1/p38 MAPK activity.
(A) Immunoblotting analysis of indicated molecules in primary MEPM cells from Tgfbr2fl/fl control mice treated with control or Tgfbr2 siRNA and cultured with (+; w/) or without (–; w/o) TGF-β2 (10 ng/ml) for 30 minutes. The bar graphs show the ratios of phosphorylated TAK1 relative to TAK1 and phosphorylated p38 relative to p38 after quantitative densitometry analysis of immunoblotting data. Three samples were analyzed for each experiment. Error bars represent SD. *P < 0.05; ***P < 0.001. (B) Immunoblotting analysis with indicated antibodies of untreated Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Untreat) or empty vector treated Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Mock) or cells after reintroduction of Tgfbr2 (Tgfbr2 vector). (C) Immunoblotting analysis with anti-K63 ubiquitin (K63 Ub) antibody after immunoprecipitation by anti-TRAF6 or anti-TAK1 antibody of extracts from MEPM cells from Tgfbr2fl/fl (F/F) and Tgfbr2fl/fl;Wnt1-Cre (F/F;Wnt1) mice. (D) Schematic diagram depicts our model of the mechanism of p38 MAPK activation in the Tgfbr2fl/fl;Wnt1-Cre palate, leading to craniofacial malformations. TGF-β2 is upregulated and binds to TβRIII (RIII), followed by assembly with TβRI (RI) and β-spectrin. TRAF6 activates TAK1 ubiquitination (Ub) and phosphorylation (P) after TGF-β2 binding. Finally, p38 and 14-3-3 proteins are phosphorylated, leading to downstream signaling and cell proliferation defect. RII, TβRII.
We next addressed the possibility that disruption of Tgfbr2-mediated signaling might lead to signaling through an alternative type II receptor. However, we detected no change in the expression of other type II receptors in the TGF-β family (Bmpr2, Acvr2a, Acvr2b) in Tgfbr2 mutant mice (Supplemental Tables 1 and 2), and siRNA knockdown of Bmpr2, Acvr2a, and Acvr2b had no effect on the TAK1 activation (Supplemental Figure 5). To explore the downstream target of the TβRIII/TβRI signaling complex in the absence of TβRII, we performed siRNA knockdown for Tgfbr3 and Tgfbr1. We found that siRNA knockdown for Tgfbr3 and Tgfbr1 resulted in a reduction of the ectopic TAK1 activation in Tgfbr2 mutant cells, indicating that lack of TβRII activates the TAK1 cascade through the TβRI/TβRIII complex (Supplemental Figure 6).
Previous studies indicated that MAPK pathways (p38 and c-Jun NH2-terminal kinase [JNK1, JNK2, and JNK3, also known as MAPK8, MAPK9, and MAPK10, respectively]) are activated by SMAD-independent pathways via lys63 ubiquitination (K63 ubiquitination) and phosphorylation of TAK1 after binding of TGF-β ligands (29, 30). TβRI induces ubiquitination of TRAF6 on K63 after the binding of TGF-β. TRAF6-induced polyubiquitination of TAK1 leads to TAK1 activation, resulting in “juxtaposition-induced autophosphorylation” (29). Therefore, we investigated K63 ubiquitination of TRAF6 and TAK1 in Tgfbr2 mutant cells. We found that K63 ubiquitination of both TRAF6 and TAK1 was upregulated in Tgfbr2fl/fl;Wnt1-Cre MEPM cells (Figure 4C). Thus, our results show that, in the absence of Tgfbr2, TGF-β2 forms a complex with TβRIII/TβRI/β-spectrin and induces a TRAF6/TAK1/p38 MAPK signaling cascade in the palatal mesenchyme (Figure 4D).
Rescue of cleft palate by manipulation of the alternative TGF-β signaling pathway.
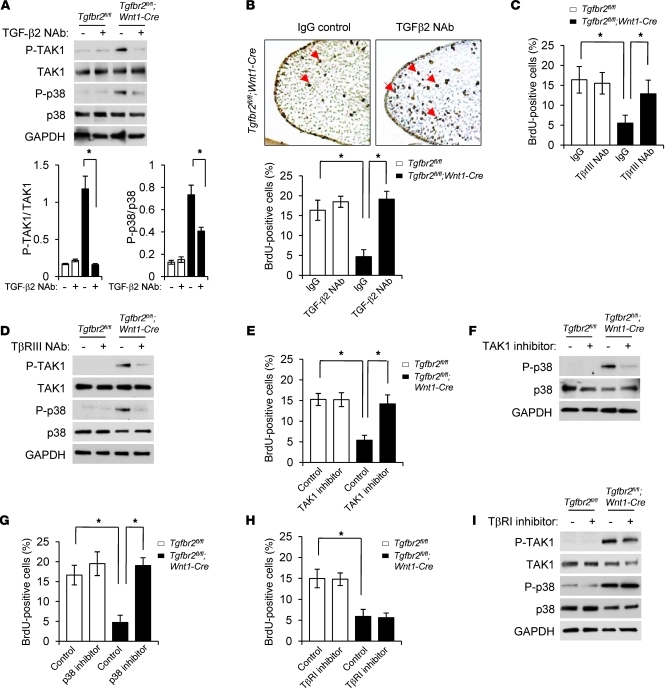
Our previous study showed that loss of Tgfbr2 results in cleft palate with reduced CNC cell proliferation in the palatal mesenchyme (13). In this current study, we show that elevated TGF-β2 triggers a SMAD-independent TGF-β signaling cascade via a TβRI/TβRIII complex in the absence of Tgfbr2 and may adversely affect CNC cell proliferation during palate formation. Therefore, reduction of TGF-β2–induced TAK1/p38 MAPK activation may increase CNC cell proliferation in Tgfbr2fl/fl;Wnt1-Cre palates. To test this hypothesis, we took an in vitro approach and treated palatal explants with TGF-β2–neutralizing antibody (TGF-β2 NAb). Inhibition of altered TGF-β signaling with TGF-β2 NAb blocked phosphorylation of both TAK1 and p38 in Tgfbr2fl/fl;Wnt1-Cre MEPM cells in a cell-autonomous manner (Figure 5A). Furthermore, TGF-β2 NAb treatment restored cell proliferation in the palatal mesenchymal cells in Tgfbr2fl/fl;Wnt1-Cre palates in the ex vivo organ culture system (Figure 5B). In order to test the hypothesis that activation of alternative TGF-β signaling in Tgfbr2fl/fl;Wnt1-Cre mice depends on the noncanonical TGF-β signaling mediated by TβRI/TβRIII, we cultured palates with a NAb for TβRIII (Figure 5, C and D). The cell proliferation defect was rescued by treatment with the NAb for TβRIII in the ex vivo organ culture system, whereas cell proliferation was not affected in control samples treated with a TβRIII NAb (Figure 5C). To examine the requirements of TAK1 and p38 MAPK activation for the cell proliferation defect in Tgfbr2fl/fl;Wnt1-Cre palates, we cultured palates with a TAK1 or p38 MAPK inhibitor, (5Z)-7-Oxozeaenol or SB203580, respectively, and found that the cell proliferation defect was also rescued (Figure 5, E–G).
Figure 5. Inhibition of TGF-β2/TβRI/TβRIII–mediated TAK1/p38 MAPK activation rescues a cell proliferation defect in Tgfbr2fl/fl;Wnt1-Cre palates.
(A) Immunoblotting analysis of indicated molecules in MEPM cells from Tgfbr2fl/fl and Tgfbr2fl/fl;Wnt1-Cre mice cultured with (+) or without (–) TGF-β2 NAb. The bar graphs show the ratios of indicated molecules after quantitative densitometry analysis of immunoblotting data. *P < 0.05. (B) BrdU incorporation in the palate of Tgfbr2fl/fl;Wnt1-Cre mice after IgG or TGF-β2 NAb treatment. Arrows indicate BrdU-positive cells. Original magnification, ×200. The bar graph shows the percentage of BrdU-labeled nuclei in the indicated genotyping palate treated with TGF-β2 NAb or IgG. *P < 0.05. (C) Quantitation of the percentage of BrdU-labeled nuclei in the indicated genotyping palates treated with TβRIII NAb or IgG. *P < 0.05. (D) Immunoblotting analysis of indicated molecules in indicated genotyping MEPM cells cultured with (+) or without (–) TβRIII NAb. (E) Quantitation of the percentage of BrdU-labeled nuclei in the indicated genotyping palates treated with TAK1 inhibitor or vehicle. *P < 0.05. (F) Immunoblotting analysis of indicated molecules in indicated genotyping MEPM cells cultured with (+) or without (–) TAK1 inhibitor. (G) Quantitation of the percentage of BrdU-labeled nuclei in the indicated genotyping palates treated with p38 MAPK inhibitor or vehicle. *P < 0.05. (H) Quantitation of the percentage of BrdU-labeled nuclei in the indicated genotyping palates treated with TβRI kinase inhibitor or vehicle. *P < 0.05. (I) Immunoblotting analysis of indicated molecules in primary indicated genotyping MEPM cells cultured with (+) or without (–) TβRI kinase inhibitor or vehicle.
Previous reports suggest that ubiquitination of TRAF6 by TGF-β is independent of TβRI kinase activity (29). Therefore, we tested whether TβRI kinase activity is necessary for the activation of TAK1/p38 MAPK and inhibition of TβRI kinase activity can rescue a defect in cell proliferation in Tgfbr2fl/fl;Wnt1-Cre palates in the ex vivo organ culture system. An inhibitor of TβRI kinase, A83-01 or SB431542, failed to rescue the cell proliferation defect in Tgfbr2fl/fl;Wnt1-Cre palates, consistent with the ectopic noncanonical TGF-β signaling being independent of TβRI kinase activity (Figure 5, H and I, and data not shown). Thus, elevated TGF-β2 triggers a SMAD-independent TAK1/p38 MAPK signaling cascade via a TβRI/TβRIII complex in the absence of TβRII and adversely affects CNC cell proliferation during palate formation.
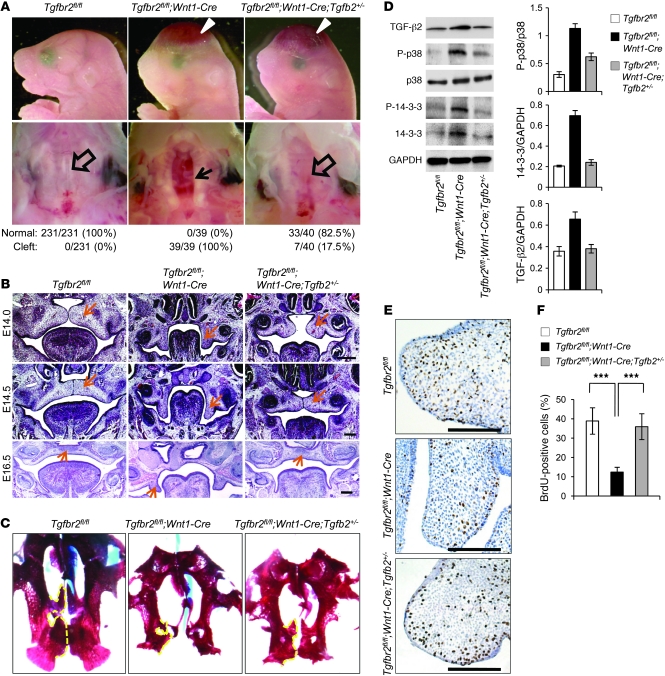
Increased TGF-β2 seems to be a trigger for alternative TGF-β signaling activation. Therefore, reduction of TGF-β2 expression may lead to a rescue of cleft palate in Tgfbr2fl/fl;Wnt1-Cre mice. To test this hypothesis further, we took an in vivo approach and generated Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice in order to reduce TGF-β2 ligand expression level. Indeed, Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice formed normal palates, with 82.5% phenotype penetrance (33 out of 40 mice had normal palates), although they still exhibited a calvaria defect and small mandible (Figure 6A and Supplemental Figure 7A). Histological analysis showed that the growth and elevation of the palatal shelf in Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice was similar to that in the control at E14.5 (Figure 6B). At E16.5, palatal shelves fused normally in Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice (Figure 6B). At birth, the palatal processes of the maxilla and the palatine bones were partially rescued in Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice compared with those in the control, whereas there was little palatine bone formation in Tgfbr2fl/fl;Wnt1-Cre mice (Figure 6C).
Figure 6. Rescue of cleft palate in Tgfbr2fl/fl;Wnt1-Cre mice via reduction of TGF-β2.
(A) Morphologies of newborn Tgfbr2fl/fl control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice. The bottom row shows the macroscopic appearance of palates at the newborn stage. Arrowheads show calvaria defects. The arrow shows cleft palate, and open arrows show normal palates. Palates were scored as normal or cleft at birth. (B) Hematoxylin and eosin staining of histological sections of Tgfbr2fl/fl, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice at E14.0, E14.5, and E16.5. Arrows indicate palate. Scale bar: 50 μm. (C) Whole-mount Alcian blue–Alizarin red skeletal staining of Tgfbr2fl/fl, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– newborn mice. Dotted lines indicate the palatal process of maxilla and palatine bones. (D) Immunoblotting analysis of E14.5 Tgfbr2fl/fl, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– palates. The bar graphs show the ratios of phosphorylated p38 relative to p38, 14-3-3 relative to GAPDH, or TGF-β2 relative to GAPDH after quantitative densitometry analysis of immunoblotting data in Tgfbr2fl/fl, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– palates. Three samples were analyzed for each experiment. Error bars represent SD. (E) BrdU staining of Tgfbr2fl/fl, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice at E14.0. Scale bar: 50 μm. (F) Quantitation of the number of BrdU-labeled nuclei in the palate of Tgfbr2fl/fl, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice at E14.0. Three samples were analyzed for each experiment. Error bars represent SD. ***P < 0.001.
At the molecular level, the upregulated phosphorylated p38 MAPK and phosphorylated 14-3-3 expression in Tgfbr2fl/fl;Wnt1-Cre palates was restored to control levels in E14.5 Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– palates (Figure 6D). Cell proliferation activity in the CNC-derived palatal mesenchyme was restored to control levels in Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– palates (Figure 6, E and F). Thus, reducing the TGF-β2 level reversed the activation of p38 MAPK signaling and restored cell proliferation in the palatal mesenchyme of Tgfbr2fl/fl;Wnt1-Cre mice.
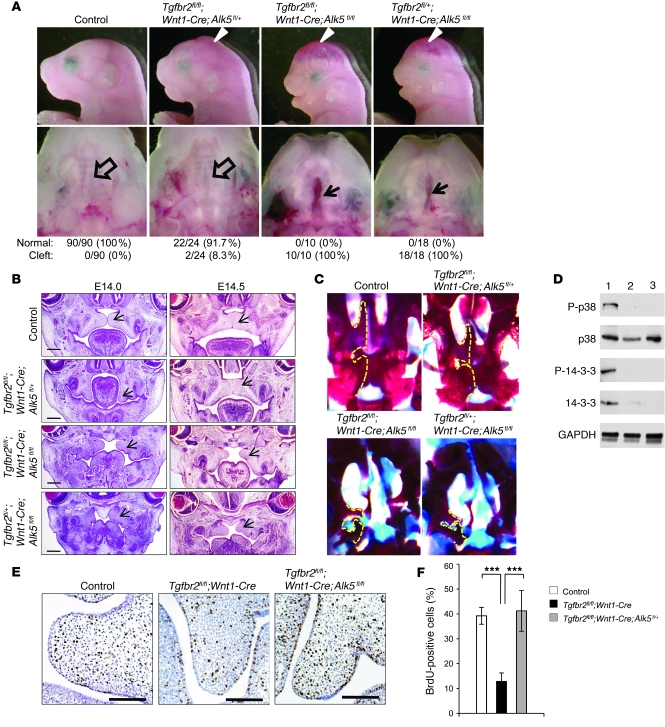
In our model, TGF-β2–mediated TAK1/p38 MAPK is activated through TβRI. In order to test that activation of alternative TGF-β signaling in Tgfbr2fl/fl;Wnt1-Cre mice depends on the presence of TβRI, we generated Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Figure 7 and Supplemental Figure 7B). In fact, Tgfbr1/Alk5 haploinsufficiency resulted in the rescue of palatal fusion in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice, with 91.7% penetrance (22 out of 24 mice had normal palates), whereas Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/fl mice still exhibited cleft palate (Figure 7A). The other craniofacial deformities, such as calvaria defect and small mandible, were mostly rescued in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice as well (Figure 7A), suggesting that alternative TGF-β signaling activation in the absence of TβRII is a mechanism conserved in other craniofacial regions. Based on histological analysis, the growth and elevation of the palatal shelf in most Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice were similar to those in the control at E14.5 (Figure 7B). At birth, the palatal processes of the maxilla and the palatine bones were mostly rescued in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice compared with those in the control, whereas there was little palatine bone formation in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/fl and Tgfbr2fl/+;Wnt1-Cre;Alk5fl/fl mice (Figure 7C). Protein expression analysis showed that elevated phosphorylated p38 and phosphorylated 14-3-3 levels detectable in Tgfbr2fl/fl;Wnt1-Cre mice (Figure 7D, lane 1) were restored to the control level (Figure 7D, lane 2) in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Figure 7D, lane 3). Moreover, haploinsufficiency of Alk5 restored cell proliferation activity in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice (Figure 7, E and F). Finally, haploinsufficiency of Tak1 in Tgfbr2 mutant mice (Tgfbr2fl/fl;Wnt1-Cre;Tak1fl/+ mice) also resulted in a rescue of cleft palate via a reduction of alternative p38 MAPK activation (Supplemental Figure 8). Thus, our findings indicate that SMAD-independent, TRAF6/TAK1/p38 MAPK activation mediated through TβRI/TβRIII is responsible for the CNC cell proliferation defect and failure of palatal fusion in Tgfbr2fl/fl;Wnt1-Cre mice.
Figure 7. Rescue of cleft palate in Tgfbr2fl/fl;Wnt1-Cre mice via reduction of TGF-β receptor type I.
(A) Morphologies of newborn Tgfbr2fl/fl;Alk5fl/fl (control), Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+, Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/fl, and Tgfbr2fl/+;Wnt1-Cre;Alk5fl/fl mice. The bottom row shows the macroscopic appearance of palates at the newborn stage. Arrowheads show calvaria defects. Arrows show cleft palate, and open arrows show normal palates. Palates were scored as normal or cleft at birth. (B) Hematoxylin and eosin staining of sections of Tgfbr2fl/fl;Alk5fl/fl (control), Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+, Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/fl, and Tgfbr2fl/+;Wnt1-Cre;Alk5fl/fl palates at E14.0 and E14.5. Arrows indicate palate. Scale bar: 50 μm. (C) Whole-mount Alcian blue–Alizarin red skeletal staining of Tgfbr2fl/fl;Alk5fl/fl (control), Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+, Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/fl, and Tgfbr2fl/+;Wnt1-Cre;Alk5fl/fl newborn mice. Dotted lines indicate the palatal process of maxilla and palatine bones. (D) Immunoblotting analysis of E14.5 Tgfbr2fl/fl;Wnt1-Cre (lane 1), Tgfbr2fl/fl control (lane 2), and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ (lane 3) palates. (E) BrdU staining of Tgfbr2fl/fl control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice at E14.0. Scale bar: 50 μm. (F) Quantification of the number of BrdU-labeled nuclei in the palate of Tgfbr2fl/fl control, Tgfbr2fl/fl;Wnt1-Cre, and Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice at E14.0. Three samples were analyzed for each experiment. Error bars represent SD. ***P < 0.001.
Discussion
TGFBR1 and TGFBR2 mutations have been reported in humans with craniofacial malformations and/or vascular abnormalities (2, 3, 31–34). The majority are missense substitutions or nonsense mutations in exon 4 of TGFBR1 and exons 4–7 of TGFBR2 that are predicted to disrupt the kinase domain (34, 35). Our findings may be relevant in human disease, given that altered TGF-β signaling is implicated in multiple congenital malformations and syndromes in humans. Our observation that TGF-β2 and TβRIII expression is elevated in the palatal mesenchyme of Tgfbr2fl/fl;Wnt1-Cre mice led to our discovery of an alternative TGF-β signaling mechanism in the absence of TβRII that is detrimental to craniofacial morphogenesis. Specifically, our study shows that elevated TGF-β2 signals through a TβRI/TβRIII complex to induce a SMAD-independent TRAF6/TAK1/p38 signaling cascade and results in cleft palate in Tgfbr2 mutant mice. This is an important advancement in our understanding of the mechanism of TGF-β signaling. Previous studies suggested that TβRIII appears to be dispensable for TGF-β–mediated signal transduction, because it lacks an intracellular domain and most cells that lack functional TβRIII still respond to TGF-β (12, 36). However, these studies did not consider how TGF-β signals in the absence of TβRII. Furthermore, TβRIII is required for murine somatic development, because loss of Tgfbr3 results in an early embryonic lethal phenotype (12). In this study, we show that alternative TGF-β signaling can be activated through the TβRI/TβRIII complex, which in turn activates the TRAF6/TAK1/p38 MAPK signaling cascade to inhibit CNC-derived palatal mesenchymal cell proliferation and cause cleft palate.
Although Tgfb2 haploinsufficiency rescued the CNC cell proliferation defect and cleft palate in Tgfbr2fl/fl;Wnt1-Cre mice, calvarial and mandibular defects were not rescued. In contrast, Tgfbr1/Alk5 haploinsufficiency completely rescued the CNC cell proliferation defect as well as palatal fusion and mostly rescued calvarial, maxillary, and mandibular defects in Tgfbr2fl/fl;Wnt1-Cre mice. One possible explanation is that TGF-β ligands other than TGF-β2 can signal through the TβRI/TβRIII complex to cause adverse effects on craniofacial development (Supplemental Figure 9). Therefore, a reduction of TGF-β2 signaling alone does not prevent these developmental defects, but a reduction of Tgfbr1/Alk5 does. This reasoning is supported by the finding that almost all craniofacial defects, such as cleft palate, microglossia, small mandible, and calvaria defects, are rescued in Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice. It is important to note that alternative TGF-β signaling involving TβRI/TβRIII is a widely used mechanism and can affect multiple craniofacial organogeneses during development.
The lack of phenotype rescue other than cleft palate in Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice could also be explained as the result of the CNC-derived palatal mesenchyme being more sensitive than other tissues to the level of noncanonical TGF-β signaling. In support of this theory, progenitor cell differentiation into hepatocytes and biliary cells is dependent on a gradient of TGF-β signaling (37). It is conceivable that TGF-β levels, in conjunction with Tgfbr activity, differentially modulate distinct signaling pathways that control unique sets of downstream target genes.
The fact that Alk5fl/fl;Wnt1-Cre mice exhibit more severe developmental defects than Tgfbr2fl/fl;Wnt1-Cre mice suggests that the TβRI may mediate signals from additional members of the TGF-β family (other than TGF-β1, TGF-β2, and TGF-β3) and regulate downstream target genes during craniofacial development. The gene expression pattern of TGF-β ligands and receptors may also contribute more widespread craniofacial malformations in Tgfbr1/Alk5 mutant mice. We found that there is increased p38 signaling in Tgfbr2fl/fl;Wnt1-Cre mice and are differentially affected downstream signaling mediators in Alk5fl/fl;Wnt1-Cre mice (J. Iwata et al., unpublished observations), suggesting that noncanonical TGF-β signaling may be controlled via multiple molecular mechanisms during craniofacial development. In parallel, we found that there is no activation of p38 signaling in Alk5fl/fl;Wnt1-Cre mice. Further studies are necessary to explore the TGF-β signaling mechanism in the absence of Alk5 during embryonic development.
Recent studies revealed that noncanonical TGF-β signaling is also activated in some physiological (17, 38, 39) and pathological conditions, including Marfan and Loeys-Dietz syndromes (25, 40). Previous reports have shown that the p38 MAPK activation mediated by TGF-β is able to signal through a kinase-dead TβRI or constitutively active TβRI in the presence of TβRII in cell lines (29, 30). In contrast, this current study shows that noncanonical TGF-β signaling in the palatal mesenchyme is activated even in the absence of TβRII and results in cleft palate. We found that a low level of activation of noncanonical TGF-β signaling is induced in wild-type MEPM cells, suggesting that some TβRI/TβRII complex may also control p38 MAPK activation in the palatal mesenchyme during normal palate formation.
Previous studies indicate that TGF-β can signal only through TβRI/TβRII complex. In response to TGF-β binding, the cytoplasmic domains of the type I receptors are phosphorylated by the constitutively active type II receptor kinase (41, 42). TβRII kinase activity is crucial for the activation of TβRI. In this study, we have demonstrated that TβRI/TβRIII complex without TβRII mediates elevated TGF-β2 signaling without requiring the kinase activity of TβRI, consistent with the previous study indicating that TAK1/p38 MAPK activation is independent of TβRI kinase activity (29).
A haploinsufficiency of Tak1 in Tgfbr2 mutant mice (Tgfbr2fl/fl;Wnt1-Cre;Tak1fl/+ mice) results in reduced p38 MAPK activation and rescues the craniofacial abnormalities, including cleft palate, consistent with ectopic TAK1/p38 MAPK activation being responsible for these developmental defects in Tgfbr2 mutant mice. In addition, siRNA knockdown for Tak1 inhibited p38 MAPK activation. The TAK1 kinase inhibitor (5Z)-7-Oxozeaenol also blocked p38 MAPK activation and rescued the cell proliferation defect in Tgfbr2fl/fl;Wnt1-Cre palates in the ex vivo organ culture system. Thus, TAK1 is crucial for the activation of alternative TGF-β signaling in the absence of TβRII. Regarding the p38 MAPK activity, SB203580 is a selective inhibitor of p38 MAPK, and it inhibits the activation of downstream targets such as 14-3-3. SB203580 inhibits p38 catalytic activity by binding to the ATP-binding pocket but does not inhibit phosphorylation of p38 by upstream kinases. The upregulated p38 phosphorylation in Tgfbr2 mutant cells is partially reversed after treatment with SB203580 (Supplemental Figure 10), suggesting that p38 activation may be attributed in part to p38 autophosphorylation caused by a feedback mechanism or other stimuli (43), but mainly results from the TAK1 cascade. Collectively, our data suggest that the TRAF6/TAK1 pathway contributes to 14-3-3 phosphorylation through the activation of p38 MAPK.
14-3-3 proteins comprise a large family of highly conserved acidic polypeptides (44, 45). Multiple isoforms of 14-3-3 proteins play an essential role in regulating cell proliferation and differentiation and can also interact with a wide variety of proteins related to signal transduction, cell cycle control, vesicular transport, and more (46). 14-3-3 interacts with TβRI and positively regulates TGF-β signaling through TβRI (47). 14-3-3 may thus modulate TGF-β signaling via protein interactions in both SMAD-dependent and -independent pathways and regulate gene expression related to cell proliferation during palatogenesis. These functions will be addressed in future studies.
Our findings that elevated TGF-β2 and ectopic p38 MAPK activation is the molecular mechanism responsible for adversely affecting cell proliferation in the CNC-derived palatal mesenchyme and that reduction of Tgfb2, Tgfbr1/Alk5, or Tak1 rescues cleft palate in Tgfbr2fl/fl;Wnt1-Cre mice have the potential for longer-term clinical applications. We propose that elevated TGF-β2 and ectopic TAK1/p38 MAPK activation could serve as biomarkers for identifying individuals with an increased risk of having a child with congenital craniofacial malformations. In addition, our molecular mechanisms could be useful for interpreting the results of large-scale population-based genetic studies aimed at identifying inherited risk factors for craniofacial malformations (48, 49). Finally, in the longer term, our results could prove useful in efforts to identify safe and effective maternal nutritional and therapeutic interventions that reduce the risk of offspring developing these congenital malformations.
Methods
Animals.
Wnt1-Cre mice were obtained from The Jackson Laboratory and crossed to Tgfbr2fl/fl mice (a gift from Harold L. Moses, Vanderbilt University, Nashville, Tennessee, USA). To generate Tgfbr2fl/fl;Wnt1-Cre;Tgfb2+/– mice, we mated Tgfbr2fl/+;Wnt1-Cre;Tgfb2+/– mice with Tgfbr2fl/fl mice. Tgfb2+/– mice were gifted by Thomas Doetschman (University of Cincinnati, Cincinnati, Ohio, USA). To generate Tgfbr2fl/fl;Wnt1-Cre;Alk5fl/+ mice, we mated Tgfbr2fl/+;Wnt1-Cre;Alk5fl/+ mice with Tgfbr2fl/fl;Alk5fl/fl mice. Alk5fl/fl mice were provided by Stefan Karlsson (Lund University Hospital, Lund, Sweden). To generate Tgfbr2fl/fl;Wnt1-Cre;Tak1fl/+ mice, we mated Tgfbr2fl/+;Wnt1-Cre;Tak1fl/+ mice with Tgfbr2fl/fl mice. Tak1fl/fl mice were provided by Michael D. Schneider (Baylor College of Medicine, Houston, Texas, USA). Genotyping was performed using PCR primers, as described previously (13, 50, 51).
Histological examination.
Hematoxylin and eosin staining and BrdU staining were performed as described previously (13, 52). Immunohistochemical staining was performed as described previously (53). Antibodies used for immunohistochemistry were anti–β-spectrin rabbit polyclonal (Abcam) and anti–14-3-3ζ/δ rabbit polyclonal antibodies (Abcam). Fluorescence images were obtained using a fluorescence microscope (model IX71, Olympus).
Primary MEPM cells and transfection.
Primary MEPM cells were obtained from E13.5 embryos. Briefly, palatal shelves were dissected at E13.5 and trypsinized for 30 minutes at 37°C in a CO2 incubator. After pipetting thoroughly, cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum supplemented with penicillin, streptomycin, l-glutamate, sodium pyruvate, and nonessential amino acids. MEPM cells were treated with or without p38 MAPK inhibitor SB203580 (1 μM), TAK1 inhibitor (5Z)-7-Oxozeaenol (8 nM), or TβRI kinase inhibitor A83-01 (12 nM) for 24 hours. MEPM cells were treated with TGF-β2 (10 ng/ml) for the indicated time. MEPM cells were cultured with NAb for TGF-β2 (2 μg/ml) or TβRIII (2 μg/ml) for 24 hours. pCMV5B-Tgfbr2, His-Tgfbr1, or mFRP-Tgfbr3 (addgene) was transiently introduced into MEPM cells using Lipofectamine LTX Plus (Invitrogen) according to standard procedures. The cells were lysed 48 hours later, and then the lysates were analyzed by immunoblotting, as described previously (53). For 14-3-3ζ/δ, phosphorylated 14-3-3, and β-spectrin staining, MEPM cells were fixed and stained with anti–14-3-3ζ/δ (Abcam), phosphorylated 14-3-3 (Affinity BioReagents), and β-spectrin (Abcam) antibodies as described previously (53). All fluorescence images were obtained using a fluorescence microscope (model IX71, Olympus). Pictures were taken using MicroSuite Analytical Suite software (Olympus).
Immunological analysis.
Immunoblots were performed as described previously (54, 55). Antibodies used for immunoblotting were as follows: rabbit polyclonal antibodies against TβRII, p38, JNK, phosphorylated JNK, phosphorylated SMAD1/5/8, phosphorylated SMAD1/5, TAK1, phosphorylated TAK1 (Ser412), and TAB1 (all from Cell Signaling Technology); SMAD1/5/8, TRAF6, 14-3-3ζ/δ, TβRI, and FRP-tag (all from Abcam); phosphorylated 14-3-3 (Affinity BioReagents); Lys63-specific ubiquitin (Millipore); BMPRII and 14-3-3ε (both from Santa Cruz Biotechnology Inc.); ACVR2A and ACVR2B (both from Abnova); rabbit monoclonal antibodies against phosphorylated p38 and phosphorylated SMAD2 (both from Cell Signaling Technology); and mouse monoclonal antibodies against SMAD2/3 (BD Biosciences), mouse monoclonal antibodies against SMAD4 and TβRIII (both from Santa Cruz Biotechnology Inc.); mouse monoclonal antibodies against β-spectrin and TGF-β2 (both from Abcam), mouse monoclonal antibody against His-tag (Invitrogen), and mouse monoclonal antibody against GAPDH (Chemicon).
Immunoprecipitation.
Extracts in cell lysis buffer containing 10 mM Tris (pH 7.5), 100 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail (Complete, Invitrogen) were lysed by sonication. Lysates containing an equal amount of protein (total volume 1 ml) were centrifuged at 1,000 g for 5 minutes at 4°C to remove debris. The supernatant was precleared with 10 μl pure goat anti-rabbit or anti-mouse IgG gel and specific immobilized antibodies (EY Laboratories Inc.). 10 μl of beads and 2 μg of anti-TβRI, TRAF6 (both from Abcam), TβRIII, β-spectrin (both from Santa Cruz Biotechnology Inc.), TAK1, or His-tag (both from Cell Signaling Technology) antibody were then added to the lysate, and the mixture was rotated for 12 hours at 4°C. The immunoprecipitate/gel complex was washed 5 times with ice-cold cell lysis buffer. The complex was boiled for 5 minutes in SDS sample buffer in the presence of β-mercaptoethanol to elute proteins and centrifuged at 1,000 g for 5 minutes at 4°C. For the ubiquitin assay, cell lysates were heated in the presence of 1% SDS and β-mercaptoethanol to disrupt noncovalent protein-protein interaction and then diluted with lysis buffer (1:10). The supernatant was subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and analyzed by immunoblotting with indicated antibodies (see the legend for Figure 3).
Affinity cross-linking.
Cells were incubated with I125-labeled TGF-β2 (1 ng/ml) in a binding buffer (128 mM NaCl, 5 mM KCl, 1.2 mM CaCl2, 5 mM MgSO4, 50 mM HEPES buffer [pH 7.4]). After incubation for 30 minutes on ice, cross-linker solution (200 μM disuccinimidyl suberate in binding buffer) was added for 30 minutes on ice. The reaction was stopped with a detachment buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Tris buffer [pH 7.4]). The precipitate was dissolved in SDS-PAGE sample buffer, boiled for 5 minutes, and subject to SDS-PAGE analysis.
Quantitative RT-PCR.
Total RNA was isolated from mouse embryonic palates dissected at each developmental stage with the QIAshredder and RNeasy Mini extraction kit (QIAGEN), as described previously (54). PCR primers were purchased from Santa Cruz Biotechnology Inc. Two-tailed Student’s t test was applied for statistical analysis of qPCR data. A P value of less than or equal to 0.05 was considered statistically significant. For all graphs, data are represented as mean ± SD.
siRNA transfection.
MEPM cells (2 × 106 cells) were plated in a 6-well cell culture plate until the cells reached 60%–80% confluence. siRNA duplex and reagents were purchased from Invitrogen and Santa Cruz Biotechnology Inc., respectively. siRNA mixture in transfection medium was incubated with cells for 7 hours at 37°C in a CO2 incubator, as described previously (54). A total of 5 × 103 cells was cultured for 30 minutes with or without TGF-β2 (10 ng/ml).
Palatal shelf organ culture.
Timed-pregnant mice were sacrificed at E13.5. Genotyping was carried out as described above. The palatal shelves were microdissected and cultured in serum-free chemically defined medium, as previously described (13). After 24 hours in culture treated with TAK1 inhibitor (5Z)-7-Oxozeaenol (80 nM), TβRI kinase inhibitor A83-01 (120 nM) or SB431542 (1 μM), p38 MAPK inhibitor SB203580 (10 μM), or NAb for TGF-β2 (20 μg/ml) or TβRIII (20 μg/ml), palates were harvested, fixed in 4% paraformaldehyde/0.1 M phosphate buffer (pH 7.4), and processed. At least 3 samples were analyzed for each experiment. Error bars represent SD.
Whole-mount skeletal staining.
The 3-dimensional architecture of the skeleton was examined using a modified whole-mount Alcian blue–Alizarin red S staining protocol, as previously described (13, 54).
Statistics.
Two-tailed Student’s t test was applied for statistical analysis. For all graphs, data represent mean ± SD. A P value of less than 0.05 was considered statistically significant.
Study approval.
All mouse experiments were conducted in accordance with a protocol approved by the Department of Animal Resources and the Institutional Animal Care and Use Committee of the University of Southern California.
Supplementary Material
Acknowledgments
We thank J. Mayo, G. Crump, X. Feng, H. Slavkin, and H. Sucov for critical reading of the manuscript and discussion and thank P. Bringas Jr., S. Smith, W. Grimes, J. Zhou, the Flow Cytometry Core, Molecular Imaging Center, Center for Electron Microscopy, and the Microarray Core for assistance. P.A. Sanchez-Lara is supported by the Harold Amos Faculty Development Program through the Robert Wood Johnson Foundation and the CHLA-USC Child Health Research Career Development Program (NIH K12-HD05954). This study was supported by grants from the National Institute of Dental and Craniofacial Research, NIH (DE020065 and DE012711) to Y. Chai.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(3):873–885. doi:10.1172/JCI61498.
References
- 1.Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- 2.Mizuguchi T, et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet. 2004;36(8):855–860. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeys BL, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37(3):275–281. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- 4.N Engl J Med. 2008;358(26):2787–2795. doi: 10.1056/NEJMoa0706585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habashi JP, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312(5770):117–121. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalluri R, Han Y. Targeting TGF-beta and the extracellular matrix in Marfan’s syndrome. Dev Cell. 2008;15(1):1–2. doi: 10.1016/j.devcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Lindsay EA. Chromosomal microdeletions: dissecting del22q11 syndrome. Nat Rev Genet. 2001;2(11):858–868. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- 8.Vitelli F, Morishima M, Taddei I, Lindsay EA, Baldini A. Tbx1 mutation causes multiple cardiovascular defects and disrupts neural crest and cranial nerve migratory pathways. Hum Mol Genet. 2002;11(8):915–922. doi: 10.1093/hmg/11.8.915. [DOI] [PubMed] [Google Scholar]
- 9.Wurdak H, et al. Inactivation of TGFbeta signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19(5):530–535. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1(3):169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 12.Stenvers KL, et al. Heart and liver defects and reduced transforming growth factor beta2 sensitivity in transforming growth factor beta type III receptor-deficient embryos. Mol Cell Biol. 2003;23(12):4371–4385. doi: 10.1128/MCB.23.12.4371-4385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito Y, et al. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130(21):5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 14.Dudas M, et al. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev Biol. 2006;296(2):298–314. doi: 10.1016/j.ydbio.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Han J, Ito Y, Bringas P, Jr, Urata MM, Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 2006;297(1):238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Cui XM, Shuler CF. The TGF-beta type III receptor is localized to the medial edge epithelium during palatal fusion. Int J Dev Biol. 2000;44(4):397–402. [PubMed] [Google Scholar]
- 17.Xu X, Han J, Ito Y, Bringas P, Jr, Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15(2):322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Casillas F, Payne HM, Andres JL, Massague J. Betaglycan can act as a dual modulator of TGF-beta access to signaling receptors: mapping of ligand binding and GAG attachment sites. J Cell Biol. 1994;124(4):557–568. doi: 10.1083/jcb.124.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Casillas F, Wrana JL, Massague J. Betaglycan presents ligand to the TGF beta signaling receptor. Cell. 1993;73(7):1435–1444. doi: 10.1016/0092-8674(93)90368-Z. [DOI] [PubMed] [Google Scholar]
- 20.Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem. 2010;285(19):14806–14814. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taya Y, O’Kane S, Ferguson MW. Pathogenesis of cleft palate in TGF-beta3 knockout mice. Development. 1999;126(17):3869–3879. doi: 10.1242/dev.126.17.3869. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299(5606):574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 23.van Heusden GP. 14-3-3 proteins: regulators of numerous eukaryotic proteins. IUBMB Life. 2005;57(9):623–629. doi: 10.1080/15216540500252666. [DOI] [PubMed] [Google Scholar]
- 24.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390(6659):465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 25.Holm TM, et al. Noncanonical TGFbeta signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science. 2011;332(6027):358–361. doi: 10.1126/science.1192149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaguchi K, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science. 1995;270(5244):2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 27.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7(8):758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibuya H, et al. Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 1998;17(4):1019–1028. doi: 10.1093/emboj/17.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorrentino A, et al. The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat Cell Biol. 2008;10(10):1199–1207. doi: 10.1038/ncb1780. [DOI] [PubMed] [Google Scholar]
- 30.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31(6):918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matyas G, et al. Identification and in silico analyses of novel TGFBR1 and TGFBR2 mutations in Marfan syndrome-related disorders. Hum Mutat. 2006;27(8):760–769. doi: 10.1002/humu.20353. [DOI] [PubMed] [Google Scholar]
- 32.Singh KK, et al. TGFBR1 and TGFBR2 mutations in patients with features of Marfan syndrome and Loeys-Dietz syndrome. Hum Mutat. 2006;27(8):770–777. doi: 10.1002/humu.20354. [DOI] [PubMed] [Google Scholar]
- 33.Yetman AT, Beroukhim RS, Ivy DD, Manchester D. Importance of the clinical recognition of Loeys-Dietz syndrome in the neonatal period. Pediatrics. 2007;119(5):e1199–e1202. doi: 10.1542/peds.2006-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stheneur C, et al. Identification of 23 TGFBR2 and 6 TGFBR1 gene mutations and genotype-phenotype investigations in 457 patients with Marfan syndrome type I and II, Loeys-Dietz syndrome and related disorders. Hum Mutat. 2008;29(11):E284–E295. doi: 10.1002/humu.20871. [DOI] [PubMed] [Google Scholar]
- 35.Horbelt D, Guo G, Robinson PN, Knaus P. Quantitative analysis of TGFBR2 mutations in Marfan-syndrome-related disorders suggests a correlation between phenotypic severity and Smad signaling activity. J Cell Sci. 2010;123(pt 24):4340–4350. doi: 10.1242/jcs.074773. [DOI] [PubMed] [Google Scholar]
- 36.Cheifetz S, Hernandez H, Laiho M, ten Dijke P, Iwata KK, Massagué J. Distinct transforming growth factor-beta (TGF-beta) receptor subsets as determinants of cellular responsiveness to three TGF-beta isoforms. J Biol Chem. 1990;265(33):20533–20538. [PubMed] [Google Scholar]
- 37.Clotman F, et al. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 2005;19(16):1849–1854. doi: 10.1101/gad.340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425(6958):577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 39.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118(pt 16):3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 40.Habashi JP, et al. Angiotensin II type 2 receptor signaling attenuates aortic aneurysm in mice through ERK antagonism. Science. 2011;332(6027):361–365. doi: 10.1126/science.1192152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370(6488):341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 42.Chen F, Weinberg RA. Biochemical evidence for the autophosphorylation and transphosphorylation of transforming growth factor beta receptor kinases. Proc Natl Acad Sci U S A. 1995;92(5):1565–1569. doi: 10.1073/pnas.92.5.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge B, et al. MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38alpha. Science. 2002;295(5558):1291–1294. doi: 10.1126/science.1067289. [DOI] [PubMed] [Google Scholar]
- 44.Aitken A, et al. 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem Sci. 1992;17(12):498–501. doi: 10.1016/0968-0004(92)90339-B. [DOI] [PubMed] [Google Scholar]
- 45.Ferl RJ, Manak MS, Reyes MF. The 14-3-3s. Genome Biol. 2002;3(7):REVIEWS3010. doi: 10.1186/gb-2002-3-7-reviews3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baldin V. 14-3-3 proteins and growth control. Prog Cell Cycle Res. 2000;4:49–60. doi: 10.1007/978-1-4615-4253-7_5. [DOI] [PubMed] [Google Scholar]
- 47.McGonigle S, Beall MJ, Feeney EL, Pearce EJ. Conserved role for 14–3–3epsilon downstream of type I TGFbeta receptors. FEBS Lett. 2001;490(1–2):65–69. doi: 10.1016/S0014-5793(01)02133-0. [DOI] [PubMed] [Google Scholar]
- 48.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12(3):167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaty TH, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Oka K, Bringas P, Kaartinen V, Chai Y. TGF-beta type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev Biol. 2008;320(1):19–29. doi: 10.1016/j.ydbio.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci U S A. 2006;103(31):11677–11682. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki T, et al. TGFbeta-mediated FGF signaling is crucial for regulating cranial neural crest cell proliferation during frontal bone development. Development. 2006;133(2):371–381. doi: 10.1242/dev.02200. [DOI] [PubMed] [Google Scholar]
- 53.Sou YS, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19(11):4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwata J, Hosokawa R, Sanchez-Lara PA, Urata M, Slavkin H, Chai Y. Transforming growth factor-beta regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J Biol Chem. 2010;285(7):4975–4982. doi: 10.1074/jbc.M109.035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwata J, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281(7):4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.