Abstract
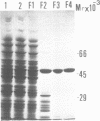
We have cloned the phr gene that encodes DNA photolyase from Salmonella typhimurium by in vivo complementation of Escherichia coli phr gene defect. The S.typhimurium phr gene is 1419 base pairs long and the deduced amino acid sequence has 80% identity with that of E. coli photolyase. We expressed the S.typhimurium phr gene in E.coli by ligating the E.coli trc promoter 5' to the gene, and purified the enzyme to near homogeneity. The apparent molecular weight of S.typhimurium photolyase is 54,000 dalton as determined by SDS-polyacrylamide gel electrophoresis, which is consistent with the calculated molecular weight of 53,932 dalton from the deduced phr gene product. S.typhimurium photolyase is purple-blue in color with near UV-visible absorption peaks at 384, 480, 580, and 625 nm and a fluorescence peak at 470 nm. From the characteristic absorption and fluorescence spectra and reconstitution experiments, S.typhimurium photolyase appears to contain flavin and methenyltetrahydrofolate as chromophore-cofactors as do the E.coli and yeast photolyases. Thus, S.typhimurium protein is the third folate class photolyase to be cloned and characterized to date. The binding constant of S.typhimurium photolyase to thymine dimer in DNA is kD = 1.6 x 10(-9) M, and the quantum yield of photorepair at 384 nm is 0.5.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Eker A. P., Hessels J. K., Dekker R. H. Photoreactivating enzyme from Streptomyces griseus--VI. Action spectrum and kinetics of photoreactivation. Photochem Photobiol. 1986 Aug;44(2):197–205. doi: 10.1111/j.1751-1097.1986.tb03586.x. [DOI] [PubMed] [Google Scholar]
- Eker A. P., Kooiman P., Hessels J. K., Yasui A. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J Biol Chem. 1990 May 15;265(14):8009–8015. [PubMed] [Google Scholar]
- Hamm-Alvarez S., Sancar A., Rajagopalan K. V. Role of enzyme-bound 5,10-methenyltetrahydropteroylpolyglutamate in catalysis by Escherichia coli DNA photolyase. J Biol Chem. 1989 Jun 5;264(16):9649–9656. [PubMed] [Google Scholar]
- Heelis P. F., Payne G., Sancar A. Photochemical properties of Escherichia coli DNA photolyase: selective photodecomposition of the second chromophore. Biochemistry. 1987 Jul 28;26(15):4634–4640. doi: 10.1021/bi00389a007. [DOI] [PubMed] [Google Scholar]
- Husain I., Griffith J., Sancar A. Thymine dimers bend DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2558–2562. doi: 10.1073/pnas.85.8.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Sancar A. Binding of E. coli DNA photolyase to a defined substrate containing a single T mean value of T dimer. Nucleic Acids Res. 1987 Feb 11;15(3):1109–1120. doi: 10.1093/nar/15.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hamm-Alvarez S., Payne G., Sancar G. B., Rajagopalan K. V., Sancar A. Identification of the second chromophore of Escherichia coli and yeast DNA photolyases as 5,10-methenyltetrahydrofolate. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2046–2050. doi: 10.1073/pnas.85.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. P., Jorns M. S. Evidence for a singlet intermediate in catalysis by Escherichia coli DNA photolyase and evaluation of substrate binding determinants. Biochemistry. 1988 Dec 13;27(25):8915–8923. doi: 10.1021/bi00425a007. [DOI] [PubMed] [Google Scholar]
- Jorns M. S., Sancar G. B., Sancar A. Identification of a neutral flavin radical and characterization of a second chromophore in Escherichia coli DNA photolyase. Biochemistry. 1984 Jun 5;23(12):2673–2679. doi: 10.1021/bi00307a021. [DOI] [PubMed] [Google Scholar]
- Jorns M. S., Sancar G. B., Sancar A. Identification of oligothymidylates as new simple substrates for Escherichia coli DNA photolyase and their use in a rapid spectrophotometric enzyme assay. Biochemistry. 1985 Apr 9;24(8):1856–1861. doi: 10.1021/bi00329a008. [DOI] [PubMed] [Google Scholar]
- Kiener A., Husain I., Sancar A., Walsh C. Purification and properties of Methanobacterium thermoautotrophicum DNA photolyase. J Biol Chem. 1989 Aug 15;264(23):13880–13887. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Heelis P. F., Sancar A. Active site of DNA photolyase: tryptophan-306 is the intrinsic hydrogen atom donor essential for flavin radical photoreduction and DNA repair in vitro. Biochemistry. 1991 Jun 25;30(25):6322–6329. doi: 10.1021/bi00239a034. [DOI] [PubMed] [Google Scholar]
- Li Y. F., Sancar A. Active site of Escherichia coli DNA photolyase: mutations at Trp277 alter the selectivity of the enzyme without affecting the quantum yield of photorepair. Biochemistry. 1990 Jun 19;29(24):5698–5706. doi: 10.1021/bi00476a009. [DOI] [PubMed] [Google Scholar]
- Lorence M. C., Maika S. D., Rupert C. S. Physical analysis of phr gene transcription in Escherichia coli K-12. J Bacteriol. 1990 Nov;172(11):6551–6556. doi: 10.1128/jb.172.11.6551-6556.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe R. S., Kredich N. M. Isolation of Salmonella typhimurium cys genes by transduction with a library of recombinant plasmids packaged in bacteriophage P22HT capsids. J Bacteriol. 1988 Jan;170(1):42–47. doi: 10.1128/jb.170.1.42-47.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G., Heelis P. F., Rohrs B. R., Sancar A. The active form of Escherichia coli DNA photolyase contains a fully reduced flavin and not a flavin radical, both in vivo and in vitro. Biochemistry. 1987 Nov 3;26(22):7121–7127. doi: 10.1021/bi00396a038. [DOI] [PubMed] [Google Scholar]
- Payne G., Sancar A. Absolute action spectrum of E-FADH2 and E-FADH2-MTHF forms of Escherichia coli DNA photolyase. Biochemistry. 1990 Aug 21;29(33):7715–7727. doi: 10.1021/bi00485a021. [DOI] [PubMed] [Google Scholar]
- RUPERT C. S. Photoenzymatic repair of ultraviolet damage in DNA. II. Formation of an enzyme-substrate complex. J Gen Physiol. 1962 Mar;45:725–741. doi: 10.1085/jgp.45.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupert C. S. Cloning of the phr gene and amplification of photolyase in Escherichia coli. Gene. 1978 Dec;4(4):295–308. doi: 10.1016/0378-1119(78)90047-1. [DOI] [PubMed] [Google Scholar]
- Sancar A., Smith F. W., Sancar G. B. Purification of Escherichia coli DNA photolyase. J Biol Chem. 1984 May 10;259(9):6028–6032. [PubMed] [Google Scholar]
- Sancar G. B. DNA photolyases: physical properties, action mechanism, and roles in dark repair. Mutat Res. 1990 Sep-Nov;236(2-3):147–160. doi: 10.1016/0921-8777(90)90002-m. [DOI] [PubMed] [Google Scholar]
- Sancar G. B. Sequence of the Saccharomyces cerevisiae PHR1 gene and homology of the PHR1 photolyase to E. coli photolyase. Nucleic Acids Res. 1985 Nov 25;13(22):8231–8246. doi: 10.1093/nar/13.22.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Heelis P. F. Purification of the yeast PHR1 photolyase from an Escherichia coli overproducing strain and characterization of the intrinsic chromophores of the enzyme. J Biol Chem. 1987 Nov 15;262(32):15457–15465. [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Lorence M. C., Rupert C. S., Sancar A. Sequences of the Escherichia coli photolyase gene and protein. J Biol Chem. 1984 May 10;259(9):6033–6038. [PubMed] [Google Scholar]
- Smith C. M., Koch W. H., Franklin S. B., Foster P. L., Cebula T. A., Eisenstadt E. Sequence analysis and mapping of the Salmonella typhimurium LT2 umuDC operon. J Bacteriol. 1990 Sep;172(9):4964–4978. doi: 10.1128/jb.172.9.4964-4978.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M., Kobayashi T., Oikawa A., Yasui A. Tandem arrangement of photolyase and superoxide dismutase genes in Halobacterium halobium. J Bacteriol. 1989 Nov;171(11):6323–6329. doi: 10.1128/jb.171.11.6323-6329.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. M., Crowne H. M., Pidsley S. C., Sedgwick S. G. Structural characterization of the Salmonella typhimurium LT2 umu operon. J Bacteriol. 1990 Sep;172(9):4979–4987. doi: 10.1128/jb.172.9.4979-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yasui A., Langeveld S. A. Homology between the photoreactivation genes of Saccharomyces cerevisiae and Escherichia coli. Gene. 1985;36(3):349–355. doi: 10.1016/0378-1119(85)90190-8. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]