Abstract
Serotonergic neurons differentiate in the neurogenic animal plate ectoderm of the sea urchin embryo. The regulatory mechanisms that control the specification or differentiation of these neurons in the sea urchin embryo are not yet understood, although, after the genome was sequenced, many genes encoding transcription factors expressed in this region were identified. Here, we report that zinc finger homeobox (zfhx1/z81) is expressed in serotonergic neural precursor cells, using double in situ hybridization screening with a serotonergic neural marker, tryptophan 5-hydroxylase (tph) encoding a serotonin synthase that is required for the differentiation of serotonergic neurons. zfhx1/z81 begins to be expressed at gastrula stage in individual cells in the anterior neuroectoderm, some of which also express delta. zfhx1/z81 expression gradually disappears as neural differentiation begins with tph expression. When the translation of Zfhx1/Z81 is blocked by morpholino injection, embryos express neither tph nor the neural marker synaptotagminB in cells of the animal plate, and serotonergic neurons do not differentiate. In contrast, Zfhx1/Z81 morphants do express fez, another neural precursor marker, which appears to function in the initial phase of specification/differentiation of serotonergic neurons. In addition, zfhx1/z81 is one of the targets suppressed in the animal plate by anti-neural signals such as Nodal as well as Delta-Notch. We conclude that Zfhx1/Z81 functions during the specification of individual anterior neural precursors and promotes the expression of tph and synaptotagminB, required for the differentiation of serotonergic neurons.
Introduction
The presence of serotonergic neurons in anterior neuroectoderm, as in a brain or an apical organ, is conserved in all metazoans except for sponges and ctenophores (Hay-Schmidt, 2000). Although a number of previous studies have revealed some of the regulatory mechanisms involved in serotonergic neuron development (reviewed in Cordes, 2005), the whole pathway from specification to terminal differentiation still needs to be elucidated, especially in invertebrates. Because the regulatory state of ectoderm in absence of signals supports neural differentiation in vertebrates and sea urchin embryos (Levine and Brivanlou, 2007: Tropepe et al., 2001; Vallier et al., 2004; Watanabe et al., 2005), researchers have focused more on the mechanisms of how this state is protected from anti-neural signals like BMP (De Robertis and Kuroda, 2004; Bradham et al., 2009; Lapraz et al., 2009; Yaguchi et al., 2010a). However, in order to understand how specific neurons differentiate within the neuroectoderm, it is important to decipher the underlying regulatory mechanisms that promote it.
In sea urchin embryos, the two early neurogenic ectoderm territories are the anterior neuroectoderm, which includes animal plate and adjacent cells, and the ciliary band ectoderm (reviewed in Angerer et al., 2011). Each of these is specified separately and patterned by combined functions of maternal factors and different zygotic signaling molecules. Under the control of those factors, a number of neurons differentiate at specific locations in each region. The first neurogenic territory to be specified is the anterior neuroectoderm. Within this region, serotonin-positive neurons appear at the aboral edge of animal plate of late gastrula (Bisgrove and Burke, 1986; 1987). They progressively increase in number and at pluteus stage their axons extend to form a plexus (Yaguchi et al., 2000). In embryos, in which all signals are shut down by injecting Δcadherin or discarding the vegetal half (Logan et al., 1999; Wikramanayake and Klein, 1997; Duboc et al., 2004), most of the prospective ectoderm becomes the animal plate and consequently many serotonergic neurons differentiate throughout it but, unlike in the normal embryo, they are scattered without any orderly pattern (Yaguchi et al., 2006). These findings suggest that the state of sea urchin embryo blastomeres in the absence of Wnt/β-catenin or Nodal/BMP2/4 signaling supports differentiation of anterior neuroectoderm, which contains the animal plate. Subsequently Wnt/β-catenin signals convert blastomere fates to endoderm, mesoderm and, within the ectoderm, eliminates anterior neuroectoderm fates except at the animal pole.
After the animal plate is restricted to the animal pole at early blastula stage, the differentiation of serotonergic neurons is prevented on the oral side by Nodal signals. In contrast to the process of ciliary band formation (Yaguchi et al., 2010a), Nodal is not involved in the specification of the animal plate (Yaguchi et al., 2006) but in patterning the region along oral-aboral axis (Yaguchi et al., 2007). In the absence of Nodal signaling, serotonergic neurons develop radially around the animal plate, while in its presence they are restricted to the aboral edge (Yaguchi et al., 2006, 2007). However, it is yet unclear how this patterning leads to serotonergic neurons differentiating only at the aboral edge of the animal plate. Here we show that Zinc finger homeobox (Zfhx1/Z81) is the earliest known transcription factor to be expressed specifically in individual serotonergic neural precursor cells in the animal plate, to be required for their differentiation and to be repressed on the oral side by Nodal signaling. Furthermore, it is co-expressed with Delta and repressed by Delta/Notch-mediated lateral inhibition. We show that Zfhx1/Z81 is required for synthesis of serotonin and that it depends on FoxQ2, which is essential for animal plate formation. This work establishes an important layer of regulatory control for the development and precise patterning of serotonergic neurons in the anterior neurogenic ectoderm of sea urchin embryos.
Materials and Methods
Animals and embryo culture
Embryos of Hemicentrotus pulcherrimus collected around Shimoda Marine Research Center, University of Tsukuba, and around Marine and Coastal Research Center, Ochanomizu University were used. The gametes were collected by intrablastocoelar injection of 0.5 M KCl and the embryos were cultured by standard methods with filtered natural seawater (FSW) at 15 °C.
Whole-mount in situ hybridization and immunohistochemistry
Whole-mount in situ hybridization was performed as described previously (Minokawa et al., 2004; Yaguchi et al., 2010b). Immunohistochemistry for detecting serotonin, synaptotagminB (synB), and c-myc was performed as described previously (Yaguchi et al., 2006). The primary antibodies were detected with secondary antibodies conjugated with Alexa-568 and Alexa-488 (Life Technologies, Carlsbad, CA, USA). The specimens were observed with a Zeiss Axio Imager.Z1 equipped with Apotome system, and optical sections were stacked and analyzed with ImageJ and Adobe Photoshop. Panels and drawings for figures were made with Microsoft PowerPoint.
Microinjection of morpholino antisense oligonucleotides (MO)
Microinjection into fertilized eggs and one blastomere of two-cell stage were performed as described previously (Yaguchi et al., 2006; Yaguchi et al., 2010b). We used the following morpholinos (Gene Tools, Philomath, OR, USA) at the indicated concentrations in 24% glycerol in injection needles: Two different morpholinos blocking expression of Zfhx1/Z81 [Zfhx1/Z81-MO1 (2.0 mM), Zfhx1/Z81-MO2 (1.9–3.8 mM)] were used to confirm the specificity of Zfhx1/Z81 function. The phenotypes obtained with FoxQ2-MO (200 µM; Yaguchi et al., 2010b), Delta-MO (2.0 mM), Nodal-MO (200 µM; Yaguchi et al., 2010b), Lefty-MO (400 µM; Yaguchi et al., 2010b), BMP2/4-MO (400 µM; Yaguchi et al., 2010b) were the same as published previously in H. pulcherrimus or other species (Duboc et al., 2004; Duboc et al., 2008; Yaguchi et al., 2008; Lapraz et al., 2009). The morpholino sequences were the following: Zfhx1/Z81-MO1: 5’- ACGTAGGTATGTTCCAAAACACAAG -3’, and Zfhx1/Z81-MO2: 5’- CAGAAGGCAGAGTCCCACAGTCCCA -3’. mRNAs were synthesized from linearized plasmids using the mMessage mMachine kit (Life Technologies, Carlsbad, CA, USA), and injected at the indicated concentrations in 24% glycerol: Δ-cadherin (0.3–0.6 µg/µl; Logan et al., 1999), myc-mRNA (0.1 µg/µl).
Results
Expression of zfhx1/z81 during development
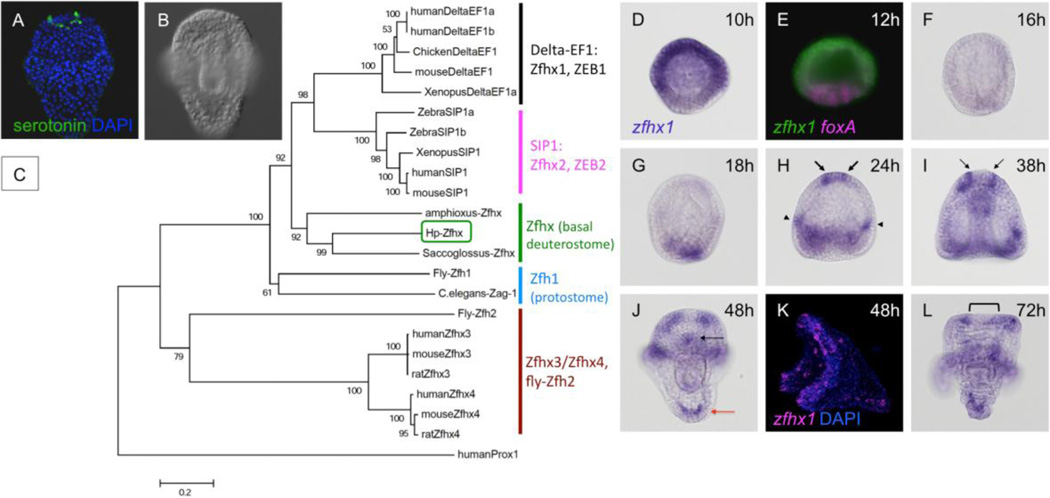
During the annotation of the sea urchin genome sequence (Sodergren et al., 2006), the spatial patterns of expression of a number of predicted genes encoding putative transcription factors were determined. Among those that were expressed in the anterior neuroectoderm (ANE) was one encoding a zinc finger-containing protein, called Z81 (Materna et al., 2006). Further studies showed that its expression in the ANE depended on Six3, a factor required for neural development (Wei et al., 2009). This gene (Z81; SPU_022242) was initially annotated as zfh-1 (Sodergren et al., 2006) and has subsequently been called Smad Interacting protein, Sip1 or SmadIP (Saudemont et al., 2010) or SpSip1 (Su et al., 2009). As shown below, we confirmed previously reported expression patterns in other species (Howard-Ashby et al., 2006; Materna et al., 2006; Saudemont et al., 2010) in Hemicentrotus pulcherrimus and observed that this gene is expressed in individual cells of the ANE arranged in a pattern suggesting they could be serotonergic precursors (Fig. 1A, B). Because revealing the transcription factor activities required for specification or differentiation of serotonergic neurons in sea urchin embryos is the primary goal, we selected this gene for further study. We cloned and sequenced it using a Japanese sea urchin, H. pulcherrimus, employed 5’RACE to determine the 5’ end of the ORF (accession number: AB630322), and found that it lacks the first two exons included in the predicted sequence, SPU_022242. We analyzed its phylogenetic position in detail and found that the gene belongs to the E-box binding zinc finger protein family including delta-EF and smad-interacting protein1 (SIP1). Based on the phylogenetic tree, it belongs to neither of these but is very closely related to non-vertebrate zinc finger homeobox proteins (Saccoglossus-Zfhx and Amphioxus-Zfhx: Fig. 1C, supplemental Fig. 1). Among the 4 classes of vertebrate Zfhx proteins, this non-vertebrate, deuterostome group type is more closely related to Zfhx1 (Delta-EF; ZEB1) and Zfhx2 (SIP1; ZEB2) than to Zfhx3 and Zfhx4. Among other invertebrate proteins, Fly-Zfh-1 and C. elegans Zag-1 are the closest. Therefore, we named it Hp-Zfhx1/Z81 (Zfhx1/Z81 hereafter in this paper).
Figure 1.
zfhx1/z81 is expressed in serotonergic neurons in the animal plate. The animal pole of embryos in each microscopic image is at the top unless otherwise indicated. (A) Serotonergic neurons in a prism larva of the sea urchin, Hemicentrotus pulcherrimus (green). (B) DIC image of (A). (C) Phylogenetic tree drawn using MEGA 5 (Tamura et al., 2011) shows that Hp-Zfhx1/Z81 belongs to basal deuterostome-type Zfhx/Zfh branch. ZEB1 and ZEB2, zinc finger E-box binding protein 1 and 2, respectively. SIP1, smad-interacting protein 1. humanProx, prospero-related homeobox of human. Numbers on the branches show the bootstrap value (%; 1,000 replicates). The scale bar indicates 0.2 amino acid substitutions per position in sequence. (D–L) Expression of zfhx1/z81 at the following stages. (D) unhatched blastula, 10-hpf (10h). (E) double fluorescent in situ hybridization with zfhx1/z81 (green) and foxA (magenta) in unhatched blastula, 12-hpf (12h). (F) hatched blastula, 16-hpf (16h). (G) mesenchyme blastula, 18-hpf (18h). (H) early gastrula, 24-hpf (24h). Arrows and arrowheads show zfhx1/z81 expression in the animal plate and future ciliary band region, respectively. (I) prism larva, 38-hpf (38h). The arrows indicate the outer edge of the central part of animal plate, where zfhx1/z81 is missing. (J) pluteus larva, 48-hpf (48h). Black and red arrow shows zfhx1/z81 gene expression in lower lip region and posterior mesenchyme cells, respectively. (K) lateral view of pluteus larva, fluorescent in situ hybridization. (L) 72-hpf pluteus stage (72h).
zfhx1/z81 is not expressed maternally (Wei et al., 2006), but just before embryo hatching, the mRNA appears in a broad region except at the vegetal plate, which expresses foxA (Fig. 1D, E). The function of Zfhx1/Z81 at this early time is discussed in elsewhere (Su et al., 2009). Expression in this domain disappears when the embryo hatches (Fig. 1F), and appears in a new set of cells in the endomesoderm region at mesenchyme blastula stage (Fig. 1G). Adding to the vegetal expression, when the gut begins to invaginate, zfhx1/z81 is expressed in a few cells in the animal plate region as well as a few cells in the lateral ectoderm, where the lateral ganglion will form (Fig. 1H, arrows and arrowheads, respectively; Howard-Ashby et al., 2006). At later stages, zfhx1/z81 is expressed in a pattern like that of the future ciliary band neurons (Fig. 1J–L; most clearly revealed in the fluorescent in situ hybridization in panel K)(Bisgrove and Burke, 1986; Nakajima et al., 2004). Here we focus only on zfhx1/z81 expression in the animal plate because the pattern of its expression is similar to that of serotonergic neurons (Fig. 1A). At the prism stage, zfhx1/z81 continues to be expressed in similar regions as those in gastrulae, but disappears from the central part of the animal plate (Fig. 1I, between arrows). In pluteus larvae, the gene expression patterns of the ciliary band are the same as those in prism stage, and lower lip cells and mesenchymal cells at the vertex begin to express zfhx1/z81 (Fig. 1J–L; black and red arrow, respectively). In contrast, the expression in animal plate region begins to disappear at this stage (Fig. 1L, bracket).
zfhx1/z81 expression is transient in neural precursor cells, disappearing after tryptophan 5-hydroxylase expression begins
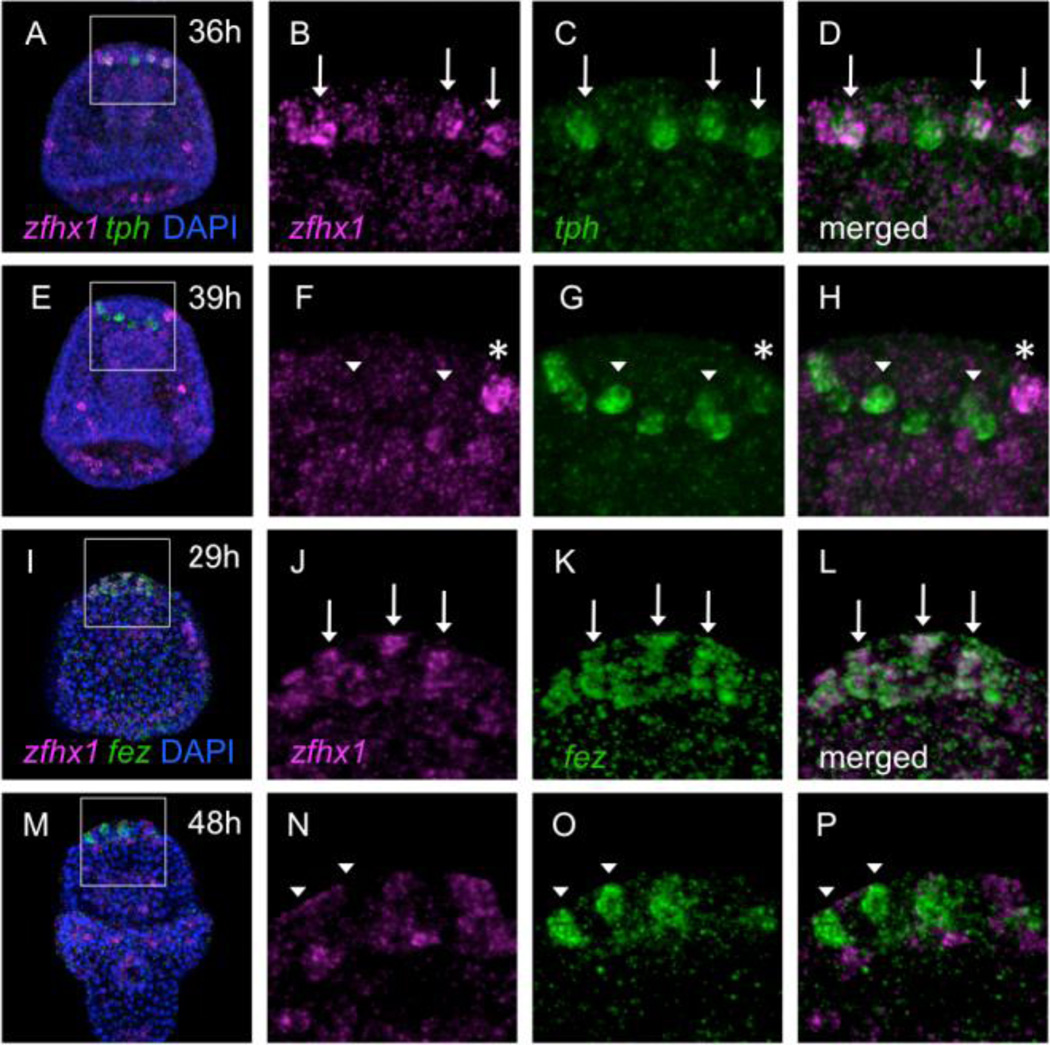
To investigate when and where zfhx1/z81 is expressed in the animal plate region in detail, we performed double fluorescent in situ hybridization detecting zfhx1/z81 and tryptophan 5-hydroxylase (tph), which encodes the rate-limiting enzyme in serotonin synthesis and therefore is a differentiation marker specific for serotonergic neurons in the sea urchin embryo (Yaguchi and Katow, 2003). zfhx1/z81-expressing cells in the animal plate (as described in Figure 1) begin to express tph at late gastrula stage (36 hours post fertilization (hpf); Fig. 2A–D, arrows). This indicates that zfhx1/z81 is expressed in serotonergic neural precursor cells. However, although these neural precursors express both genes at 36-hpf (Fig. 2A–D), at 39-hpf most of them lack zfhx1/z81 transcripts (Fig. 2E–H, arrowheads), suggesting that zfhx1/z81 expression precedes tph. At this stage, a cell appears which expresses zfhx1/z81 strongly but tph weakly and is likely to be a new serotonergic precursor cell (Fig. 2E–H, asterisk). Next, we compared distributions of zfhx1/z81 and fez, forebrain embryonic zinc finger, which we recently reported as being expressed in the entire animal plate during blastula stages and subsequently in serotonergic neurons and their precursors (Yaguchi et al., 2011). When the blastula-stage expression of fez begins to fade and is progressively replaced by stronger signals in a few individual cells in the animal plate region at mid-gastrula stage (Fig. 2K), zfhx1/z81 mRNA is present in the same cells (Fig. 2I–L, arrows). Afterward, zfhx1/z81 transcripts disappear by the prism stage, whereas fez mRNA remains in the serotonergic neurons (Fig. 2M–P, arrowheads). Taken together, zfhx1/z81 is expressed in neural precursors at beginning of gastrulation and disappears soon after these cells begin to differentiate, as indicated by tph expression at late gastrula stage.
Figure 2.
zfhx1/z81 is transiently expressed in serotonergic neural precursor cells. (A–H) Double fluorescent in situ hybridization detecting zfhx1/z81 and tph in 36-hpf (A–D) and 39-hpf (E–H) embryos. (A) zfhx1/z81 is expressed in the animal plate region. A square region is magnified in (B–D). (B) zfhx1/z81 is expressed in a few cells (arrows). (C) tph at the same region (arrows). (D) Merged image of (B) and (C). Arrows show the cells expressing both zfhx1/z81 and tph. (E) Most of zfhx1/z81 disappears from the animal plate in 39-hpf embryo. A square shows the region that is magnified in (F–H). (F) zfhx1/z81 is not expressed in tph-positive cells (arrowheads). Asterisk shows zfhx1/z81-positive cell. (G) tph expression in the same region. Arrowheads indicate the cells expressing tph strongly. Asterisk shows a cell expressing tph weakly. (H) Merged image of (F) and (G). (I–P) Double fluorescent in situ hybridization detecting zfhx1/z81 and fez in 29-hpf (I–L) and 48-hpf (M–P) embryos. (I) zfhx1/z81 is expressed in the animal plate region in 29-hpf. The square shows the region that is magnified in (J–L). (J) zfhx1/z81-expressing cells in the animal plate (arrows). (K) fez-expressing cells in the same region. (L) Merged image of (J) and (K). (M) zfhx1/z81 is down regulated in a 48-hpf embryo. (N) zfhx1/z81 is not detected in the cells in which fez is expressed (arrowheads). (O) fez expression in the same region. (P) Merged image of (N) and (O). zfhx1/z81-positive cells (magenta) in (M–P) are non-serotonergic neurons in the animal plate.
Zfhx1/Z81 is required for the differentiation of serotonergic neurons
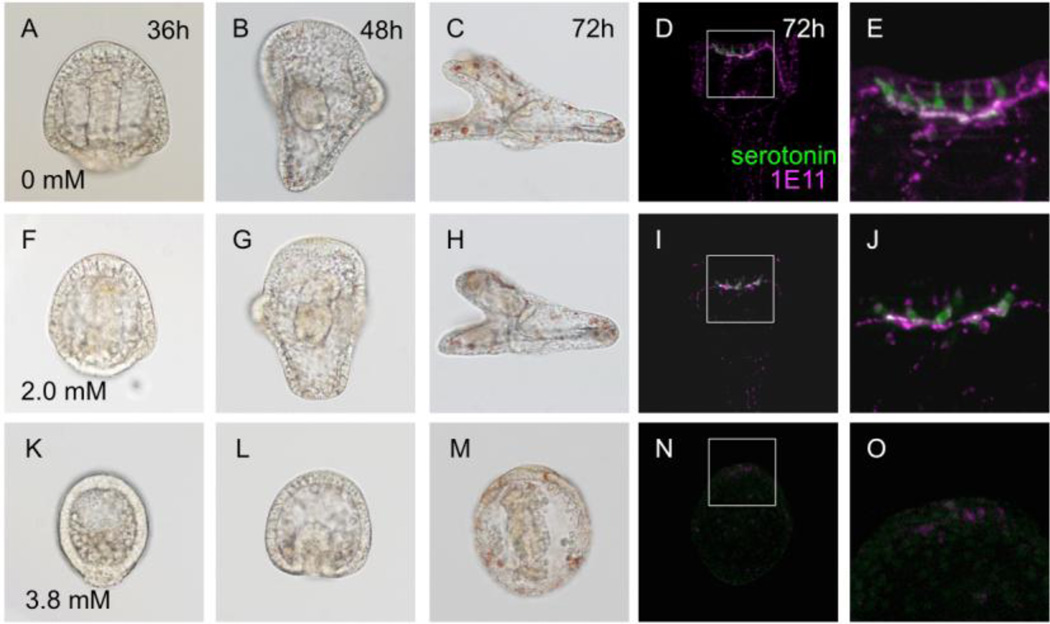
The spatial and temporal expression pattern of zfhx1/z81 suggests that it might be involved in the specification and/or differentiation of serotonergic neurons in the sea urchin embryo. To examine this, we blocked the translation of zfhx1/z81 by injecting morpholino anti-sense oligonucleotide (MO; Zfhx1/Z81-MO represents Zfhx1/Z81-MO2 throughout this study otherwise indicated). In embryos injected with Zfhx1/Z81-MO at 2 mM, gastrulation is delayed (Fig. 3F) and their body size becomes smaller than normal (Fig. 3A–C, F–H). The number of serotonergic neurons decreases in morphants, but those that do form still extend axons to form a complex in the animal plate region as they do normal embryos (Fig. 3D, E, I, J). Although serotonergic neurons do not appear in 3.8 mM Zfhx1/Z81-MO-injected embryo as well as in the 2.0 mM Zfhx1/Z81-MO1-injected embryo (data not shown), it is unclear whether this effect results directly from blocking Zfhx1/Z81 function in neural precursor cells or because of indirect effects that drastically delay gastrulation and lead to ectoderm patterning defects, including loss of oral-aboral polarity (Fig. 3K–O). Indirect effects are possible because zfhx1/z81 is expressed broadly in ectoderm early (Saudemont et al., 2010) and then in animal and vegetal cells (Howard-Ashby et al., 2006) and is thought to play a role in oral-aboral polarity (Su et al., 2009) (also see Fig. 1),
Figure 3.
Knockdown of Zfhx1/Z81 not only decreases the number of serotonergic neurons but also inhibits normal vegetal tissue development and oral/aboral polarity. (A–E) Control embryos (glycerol-injected). (A) 36-hpf prism stage. (B) 48-hpf pluteus stage. (C) 72-hpf early 4-arm pluteus stage, lateral view. (D) Immuno-fluorescent image of a 72-hpf embryo stained for serotonin and synaptotagminB (1E11); the rectangle shows the region magnified in (E). (E) Seven serotonergic neurons are present in this embryo. (F–J) 2.0 mM Zfhx1/Z81-MO-injected embryos. (F) 36-hpf. (G) 48-hpf. (H) 72-hpf. The length of the body along the anterior-posterior axis is shorter than that of normal embryos (C). (I) The development of the nervous system is incomplete in the morphant. The square shows the region magnified in (J). (J) The number of serotonergic neurons is less than that of control. (K–O) 3.8 mM Zfhx1/Z81-MO-injected embryos. (K) 36-hpf. (L) 48-hpf. (M) 72-hpf. (N) This morphant has no detectable neurons in the animal plate. Square shows the region magnified in (O). (O) Neural development is strongly suppressed in the morphants.
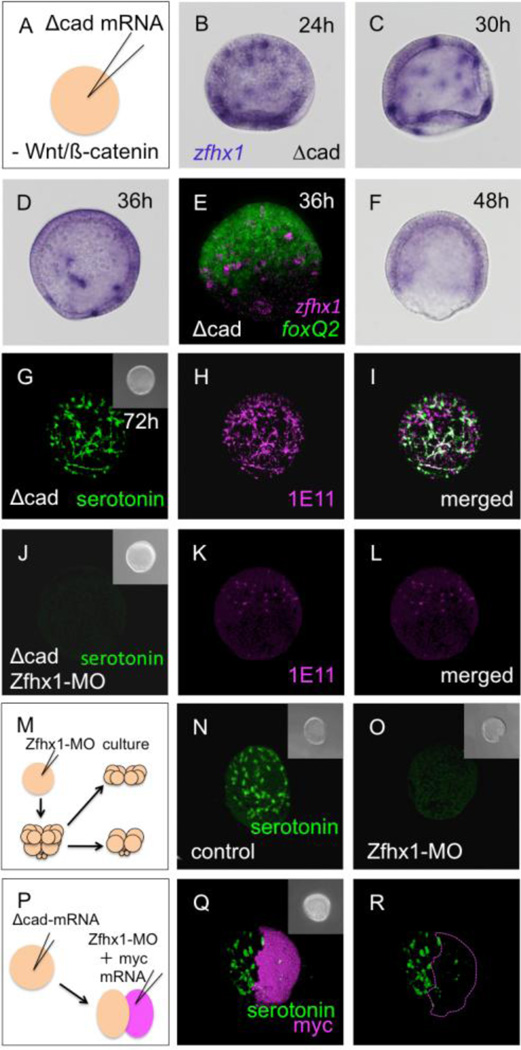
To eliminate possible indirect effects, we examined Zfhx1/Z81 function in two types of embryos that lack vegetal signals that are necessary for endomesoderm development and for Nodal expression that regulates oral-aboral polarity. These are embryos either injected with Δcadherin (Δcad) (Logan et al., 1999; Wikramanayake et al., 1998; Yaguchi et al., 2008) or lacking the vegetal half starting from 8-cell or 16-cell stages (Wikramanayake et al., 1995; Yaguchi et al., 2006; Yaguchi et al., 2008). These two types of embryos are thus far not detectably different as monitored by gene expression and responses to experimental perturbations (Logan et al., 1999; Yaguchi et al., 2006; Yaguchi et al., 2007; Yaguchi et al., 2008; Sasaki and Kominami, 2008). In Δcadherin-injected embryos, the expanded animal plate contains a greatly increased number of serotonergic neurons as reported previously (Yaguchi et al., 2006). As expected, zfhx1/z81-expressing cells are scattered throughout the expanded animal plate of these embryos at 24-hpf (Fig. 4B, C). As development proceeds, the number of zfhx1/z81-positive cells gradually decreases, as observed in normal embryos (Fig. 4A–D), especially, in the central part of the expanded animal plate where foxQ2 is strongly expressed (Fig. 4E). At 2 days after fertilization, the Δcad-injected embryo lacks zfhx1/z81 expression in individual cells completely (Fig. 4F). Therefore, the expression patterns of zfhx1/z81 in the expanded animal plate reflect the behavior of zfhx1/z81 in normal embryos. If Zfhx1/Z81 is knocked down in these embryos, development of serotonergic neurons is strongly inhibited (3.8 mM Zfhx1/Z81-MO2 injection; Fig. 4J–L). This morpholino effect is confirmed by injecting 2.0 mM Zfhx1/Z81-MO1 (data not shown). This is also true in animal-half embryoids (Fig. 4M, O), because loss of Zfhx1/Z81 completely eliminates the large number of serotonergic neurons normally present in them (Yaguchi et al., 2006) (Fig. 4N; cf. with G). To confirm that the requirement for Zfhx1/Z81 for serotonergic neuron differentiation is cell-autonomous, Zfhx1/Z81-MO and mRNA encoding 5 myc epitopes as a lineage tracer were injected into one blastomere of 2-cell embryos already containing Δcad-mRNA (Fig. 4P). In these embryos, the serotonergic neurons differentiate normally in the myc-negative, Zfhx1/Z81-positive side but not in the myc-positive, Zfhx1/Z81-negative region (Fig. 4Q, R). The lack of serotonergic neurons at the border of first cleavage plane next to Zfhx1/Z81-positive cells strongly supports the idea that Zfhx1/Z81 is not required for even short-range signals promoting serotonergic neuron differentiation, but rather acts cell-autonomously. Together, these results indicate that Zfhx1/Z81 is required for the differentiation of serotonergic neurons in the anterior neuroectoderm.
Figure 4.
Zfhx1/Z81 is required for the differentiation of serotonergic neurons. (A) Microinjection to inhibit canonical Wnt signaling. (B–F) The expression patterns of zfhx1/z81 in Δcad-injected embryos. (B) zfhx1/z81-positive neural precursors are scattered in the expanded 24-hpf embryo. (C) 30-hpf embryo. (D) 36-hpf embryo; the number of zfhx1/z81 cells decreased. (E) Double fluorescent in situ hybridization shows that zfhx1/z81 disappears from the central part of the animal plate. (F) zfhx1/z81 is down regulated in 48-hpf Δcad-injected embryos. The apparent staining in this embryo is background diffuse staining that is higher in the thickened ectoderm of these embryos. (G) Many serotonergic neurons differentiate in the expanded animal plate in Δcad-injected embryo. (H) All of serotonergic and non-serotonergic neurons in the animal plate are synaptotagminB (1E11 antigen)-positive. (I) Merged image of (G) and (H). (J) Δcad-injected Zfhx1/Z81 morphants have no serotonergic neurons at 72-hpf. (K) Serotonin-negative 1E11 neurons begin to differentiate in morphants. (L) Merged image of (J) and (K). (M) Method for creating animal caps from Zfhx1/Z81 morphants. (N) Serotonergic neurons differentiate in the glycerol-injected control animal cap. (O) No serotonergic neurons differentiate in the animal cap of Zfhx1/Z81 morphants. (P) Method to inject Zfhx1/Z81-MO and myc mRNA into one of two blastomeres derived from a Δcad-injected egg. (Q) Nearly all of the serotonergic neurons differentiate in the myc (i.e. Zfhx1/Z81-MO)-negative half of the embryo. (R) Only the outline of myc-positive, Zfhx1/Z81-deficient region of (Q) is shown. Insets are DIC images for each panel.
Zfhx1/Z81 is required for the expression of tph but not early neuronal genes
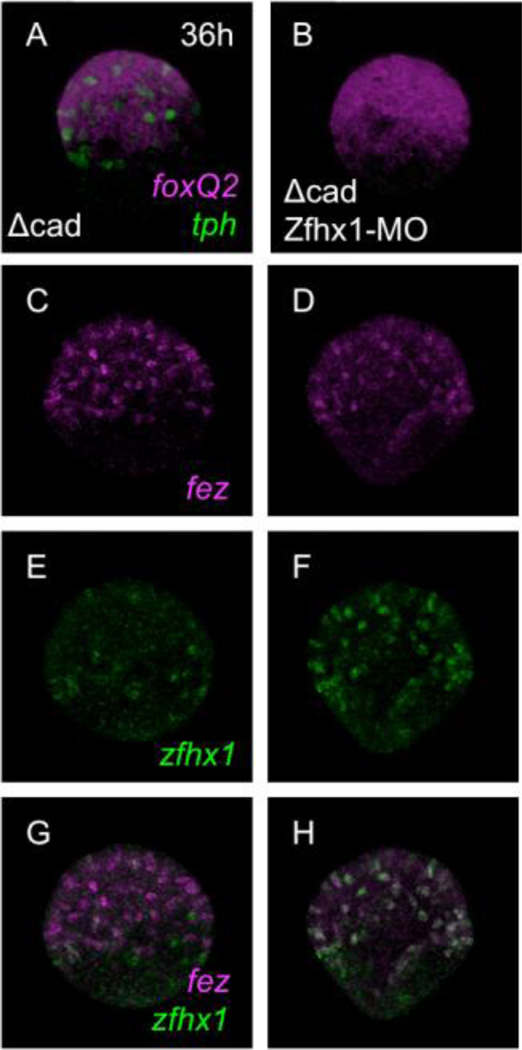
To examine at which step Zfhx1/Z81 is involved during the specification and differentiation of serotonergic neurons, we examined Zfhx1/Z81 morphants for expression of foxQ2, normally in all cells of the animal plate, tph, and fez, an early serotonergic neural marker (Yaguchi et al., 2011). We again used Δcad-injected embryos to eliminate indirect effects caused by Zfhx1/Z81 functions at earlier stages in other regions of the embryo. In Δcad-injected Zfhx1/Z81 morphants foxQ2 is expressed throughout the expanded animal plate as in control Δcad alone-injected embryos (cf. Fig. 5A with B) but tph is not expressed at all (Fig. 5B), indicating that Zfhx1/Z81 is required for tph expression but not for foxQ2. As well, fez, another serotonergic neural marker, is expressed in Δcad-injected Zfhx1/Z81 morphants as in control embryos, indicating that Zfhx1/Z81 is not required for neuron-specific expression of fez (Fig. 5C, D). Conversely, zfhx1/z81 expression does not require Fez (Supplemental Figure 2), indicating that these two genes, while co-expressed in individual cells at the animal plate of early gastrulae, function in parallel pathways. As shown in Figure 2, zfhx1/z81 transcripts gradually start to disappear from the animal plate in control Δcad alone-injected embryos (Fig. 5E). However, intriguingly in Δcad-injected Zfhx1/Z81 morphants, zfhx1/z81 transcripts remain (Fig. 5F), indicating that zfhx1/z81 is regulated by auto-repression mechanism in these embryos (Fig. 5G, H). These results support the temporal expression data (Fig. 1, 2), which suggests that zfhx1/z81 and fez transcripts appear after foxQ2 is expressed, but before the serotonin synthase tryptophan 5-hydroxylase gene, tph. Although both zfhx1/z81 and fez depend on FoxQ2 and are co-expressed in cells in the foxQ2-positive animal plate (see below, Fig. 7), they have independent roles in these serotonergic precursors, since Zfhx1/Z81 is required for differentiation of these neurons while Fez is not (Yaguchi et al., 2011).
Figure 5.
Zfhx1/Z81 is not required for expression of genes involved in early specification of the animal plate. (A) foxQ2 and tph in a Δcad-injected embryo at 36-hpf. (B) The expression pattern of foxQ2 is not altered in Δcad-injected Zfhx1/Z81 morphants, whereas no tph expression is detected. (C, E, G) Δcad-alone-injected control embryo. (D, F, H) Δcad-injected Zfhx1/Z81 morphant. (C, D) The expression patterns of fez at 36-hpf. (E, F) The expression patterns of zfhx1/z81. (G, H) Merged images of (C) and (E), and (D) and (F), respectively.
Figure 7.
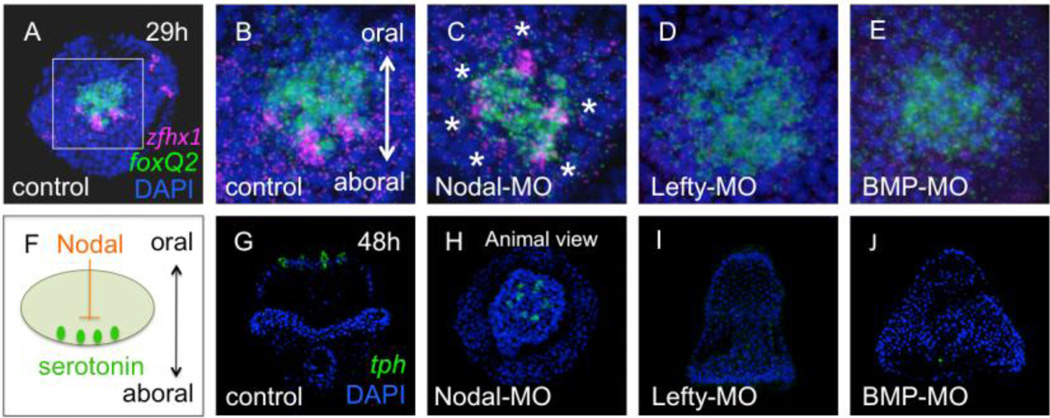
Nodal suppresses the expression of zfhx1/z81 on the oral side of the animal plate. (A) The expression pattern of zfhx1/z81 (magenta) in the animal plate of control (glycerol-injected) embryos is marked by foxQ2 (green) expression. A square shows the region magnified in (B). Animal pole view. (B) zfhx1/z81 is expressed in cells along the aboral edge of the animal plate. (C) zfhx1/z81 is expressed all around the circumference of the animal plate in Nodal morphants (asterisks). (D) zfhx1/z81 is not expressed in Lefty or BMP morphants, in which Nodal expression extends around the animal plate (E). (F) Schematic illustrating that Nodal suppresses the differentiation of serotonergic neurons on the oral side of the animal plate. (G) The expression pattern of tph in the control (glycerol-injected) embryo (green). Oral view. (H) tph is radially expressed in the animal plate in Nodal morphants. Animal pole view. (I, J) tph is not expressed in either Lefty or BMP morphants.
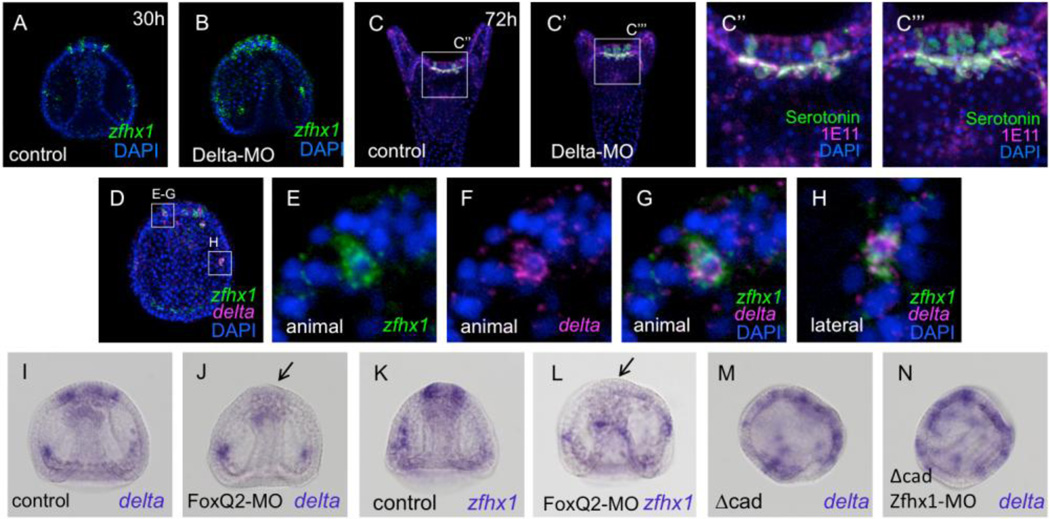
It has been supposed that Delta functions in neurogenesis in the sea urchin embryo based on its expression pattern in ectoderm (Röttinger et al., 2006; Lapraz et al., 2009; Saudemont et al., 2010) and the fact that DAPT, which inhibits Notch signaling and lateral inhibition, results in significant increases in neuron number (Wei et al., 2011; Yaguchi et al., 2011). Further support that it is Delta that mediates lateral inhibition in the anterior neuroectoderm through Notch signaling is that a cluster of contiguous serotonergic neurons develops on the aboral side of the animal plate (Fig. 6C–C’’’), exactly as observed previously in DAPT-treated embryos (Yaguchi et al., 2011). These facts suggest that delta is specifically expressed in neural precursors in sea urchin embryos and could be co-expressed with zfhx1/z81. This is in fact the case since fluorescent double in situ hybridizations showed that it is co-expressed with zfhx1/z81 in serotonergic neuron precursors in the animal plate (Fig. 6D–H; stacks of a few optical sections). In contrast, delta is not expressed in differentiating tph-positive neurons (data not shown). Taken together, these results show that, in animal plate neurons, transient expression of delta and zfhx1/z81 is followed by tph.
Figure 6.
delta is a specific neural marker in the animal plate. (A) zfhx1/z81 expression in the animal plate of 30-hpf (30h) embryo detected with fluorescent in situ hybridization. (B) More cells express zfhx1/z81 and make a cluster in the animal plate of Delta morphants. (C) The normal patterning of serotonergic neurons in 72-hpf embryo. A square shows the region magnified in (C’’). 1E11, a pan-neural marker (magenta); serotonin (green). (C’) A cluster of serotonergic neurons is formed in the animal plate of Delta morphants. A square shows the region magnified in (C’’’). (C’’) Magnified image of the square region in (C). (C’’’) Magnified image of the square region in (C’). (D) Double fluorescent in situ hybridization detects zfhx1/z81 and delta co-expression at gastrula stage. The magnified images are shown in (E–G) for animal plate and (H) for lateral regions. (E) A cell expressing zfhx1/z81 in the animal plate. (F) delta expression. (G) Merged image of (E) and (F). (H) A cell expressing zfhx1/z81 (green) and delta (magenta) in the lateral region. (I) delta expression in the control (glycerol-injected) late gastrula. (J) delta expression in the animal plate is suppressed in FoxQ2 morphants (arrow). (K) zfhx1/z81 in the control late gastrula. (L) zfhx1/z81 expression in the animal plate requires FoxQ2 (arrow). (M) Many delta-expressing cells are present in the expanded animal plate of Δcad-injected embryos. (N) delta expression pattern is unaltered in Δcad-injected Zfhx1/Z81 morphants.
To establish regulatory relationships between FoxQ2, Delta and Zfhx1/Z81, we carried out a series of morpholino-mediated knock-downs. In FoxQ2 morphants, in which serotonergic neurons fail to differentiate, neither delta nor zfhx1/z81 is expressed in the animal plate region (Fig. 6I–L, arrows). In contrast, both genes are expressed in lateral regions, as expected, since FoxQ2 is not expressed at these sites. Thus, animal plate expression of delta and zfhx1/z81 requires FoxQ2 function. When the translation of delta is blocked by injecting Delta-MO, zfhx1/z81-positive cells increase in number and are immediately adjacent to each other, making a cluster in the animal plate region (cf. Fig. 6A with B; stacks of a few optical sections), as do serotonergic neurons (Fig. 6C–C’’’). These data suggest that Delta functions to inhibit neighboring cells, but not its own expressing cells, from differentiating as Zfhx1/Z81-expressing serotonergic neuronal precursors. Delta expression in animal plate cells does not require Zfhx1/Z81 because it is expressed in the same scattered pattern as serotonergic neurons in Δcad-injected embryos that either contain or lack Zfhx1/Z81 (Fig. 6M, N). Taken together, Zfhx1/Z81 appears in animal plate cells during gastrulation where it is required for tph expression and subsequent serotonin synthesis, but not for the early regulatory genes like foxQ2, fez and delta.
Nodal signaling suppresses zfhx1/z81 expression
Previous studies showed that serotonergic neurons differentiate only at the aboral/lateral edge of the animal plate, and this asymmetry is caused by Nodal signaling from cells on the oral side of the plate (Fig. 7F; Yaguchi et al., 2007). As expected, in normal embryos, zfhx1/z81 is also expressed in cells at the aboral/lateral edge of the foxQ2-positive animal plate region at gastrula stage (Fig. 7A, B), and at prism and pluteus stages the serotonergic neurons expressing tph gene are aligned similarly (Fig. 7G). When the translation of Nodal is blocked by injecting Nodal-MO, zfhx1/z81- and tph-positive cells surround the animal plate (Fig. 7C, asterisks; 7H, respectively). In contrast, when Nodal signaling is enhanced and extends to the aboral side of the animal plate (Duboc et al., 2004; Duboc et al., 2008) by blocking the translation of Lefty, an endogenous antagonist of Nodal signaling, neither zfhx1/z81 nor tph is expressed in the animal plate (Fig. 7D, I). When translation of BMP2/4, another TGF-β member involved in cell fate specification along the aboral side of the embryo, is blocked, the morphants also do not express zfhx1/z81 and tph (Fig. 7E, J). In these morphants Nodal signaling extends further to the aboral side (Yaguchi et al., 2010a), where it suppresses expression of zfhx1/z81 and differentiation of serotonergic neurons. Taken together, Nodal signals in the oral ectoderm suppress the expression of zfhx1/z81 and subsequently tph, leading to development of serotonergic neurons only on the aboral edge of the animal plate.
Discussion
The data presented here show that Zfhx1/Z81 is required cell-autonomously for the differentiation of serotonergic neurons in sea urchin embryos. Most of the transcription factors expressed early throughout the animal plate are required for the specification and differentiation of this territory (Yaguchi et al., 2008; Wei et al., 2009). When the function of those genes is blocked, the animal plate is lost as are the neurons that develop within it as well as the apical tuft (Yaguchi et al., 2010b). Therefore, it was not clear how these early regulatory activities were connected to the specification of individual neurons expressing the terminal differentiation genes, tph and synptotagminB, at late gastrula stage (Yaguchi and Katow, 2003; Burke et al., 2006). Here we show that Zfhx1/Z81 is one of the intermediate factors downstream of genes specifying the early animal plate and upstream of those sponsoring terminal differentiation of serotonergic neurogenesis. Knock-down of either FoxQ2 or Zfhx1/Z81 significantly decreases the number of serotonergic neurons (Yaguchi et al., 2008; this study) and FoxQ2 morphants do not express zfhx1/z81. Furthermore, zfhx1/z81 is co-expressed with delta at early gastrula stage, the first direct demonstration that delta is expressed in neural cells in the animal plate of sea urchin embryos. As in other embryos, we show here that Delta functions in neuronal precursors to limit the number of cells in the animal plate that differentiate as neurons through lateral inhibition. Thus, Delta and Zfhx1/Z81 mark neuronal precursors. As well, the expression pattern and timing of zfhx1/z81 relative to terminal differentiation genes is appropriate for its requirement for the differentiation of serotonergic neurons. Zfhx1/Z81 could be a direct activator of tph since it is co-expressed with tph as serotonergic neurons begin to differentiate. In contrast, delta and tph are rarely co-expressed in normal embryos, consistent with the sequential waves of expression of delta, zfhx1/z81 and tph. Together, the expression patterns and loss-of-function data indicate that FoxQ2 is required for delta and zfhx1/z81 expression in neuronal precursors. Delta/Notch signaling limits the number of these precursors and Zfhx1/Z81 then is required of expression of genes necessary for the terminal differentiation of serotonergic neurons.
The results reported here indicate that Nodal signaling-mediated suppression of serotonergic neural differentiation on the oral side of the animal plate (Yaguchi et al., 2007) must occur downstream of FoxQ2 and at or upstream of zfhx1/z81 expression because here we show that Nodal suppresses zfhx1/z81 expression, but has no detectable effect on foxQ2 expression. Thus, this work fills an important gap in our understanding of the regulatory path that links specification of the neurogenic field to the differentiation of individual neurons in sea urchin embryos.
Zfh/ZEB family members have a characteristic molecular structure; N- and C-terminal zinc finger domains and a central homeodomain (Fortini et al., 1991; Genetta et al., 1994). It has been reported that these transcription factors bind to E-boxes and have been shown to play a role in regulating myogenesis in vertebrates and invertebrates (Postigo et al., 1999). In addition, the vertebrate-type family of ZEB factors includes branches to delta-EF1 and SIP1. They attenuate BMP signaling with Smad-interacting activity (Postigo, 2003), and the Smad-binding domain (SBD) in SIP1 has been already identified (Verschueren et al., 1999). In contrast, the amino acid sequence alignment shows the sea urchin Zfhx1/Z81 as well as fly Zfh-1 have no conserved SDB sequence (supplemental Fig. 1). Although it was annotated as SIP1 after the sea urchin genome was sequenced (Su et al., 2009; Saudemont et al., 2010), there is no evidence that it interacts with the Smad family; instead our phylogenetic analysis suggests that this gene, SPU_022242, does not belong to the SIP1 branches but is most closely related to the invertebrate-type ZEB member, Zfhx (Fig. 1).
In flies and worms, Zfh-1 and Zfh-2 were reported to possess both zinc fingers and homeodomains, and both are expressed in the nervous system. Zfh-2 contains 17 zinc-finger domains and 3 homeodomains, and in Drosophila it binds to a regulatory region of the DOPA decarboxylase gene, which is essential for the second step of biosynthesis of dopamine and serotonin (Lundell and Hirsh, 1992). The homolog of vertebrate zfh-2 in sea urchins is atbf1 (SPU_017348), suggesting that Zfhx-1, the gene studied here, and Zfh-2 also have different functions in the sea urchin. The function of Zfh-1 in flies is not well understood but it is expressed in the serotonergic lineage in their central nervous system where its expression is regulated by Notch signaling and Eagle transcription factor (Lai et al., 1991; Lee and Lundell, 2007). In C. elegans, a homolog of Zfh-1, Zag-1, is expressed several neuronal lineages including those leading to head and tail ganglia, dorsal and ventral cords, and some of them express tph and synthesize serotonin (Sze et al., 2002; Wacker, et al., 2003). Among those serotonergic neurons, the HSN serotonergic motor neurons require Zag-1 for expression of tph (Clark and Chiu, 2003). However, because tph expression in the head region is not affected in zag-1 mutants, the function of Zfh-1/Zag-1 in the serotonergic neuron-lineage in the anterior neuroectoderm of an ecdysozoan invertebrate differs from the role of Zfhx-1 in this region of sea urchin embryos. Whether Zfhx proteins are involved in development of serotonergic neurons in other deuterostomes is not yet known, although predictions from genome sequences of hemichordate and amphioxus reveal that they have the same invertebrate-type Zfhx (XM_002740578.1; XM_002592121.1, Putnam et al., 2008),
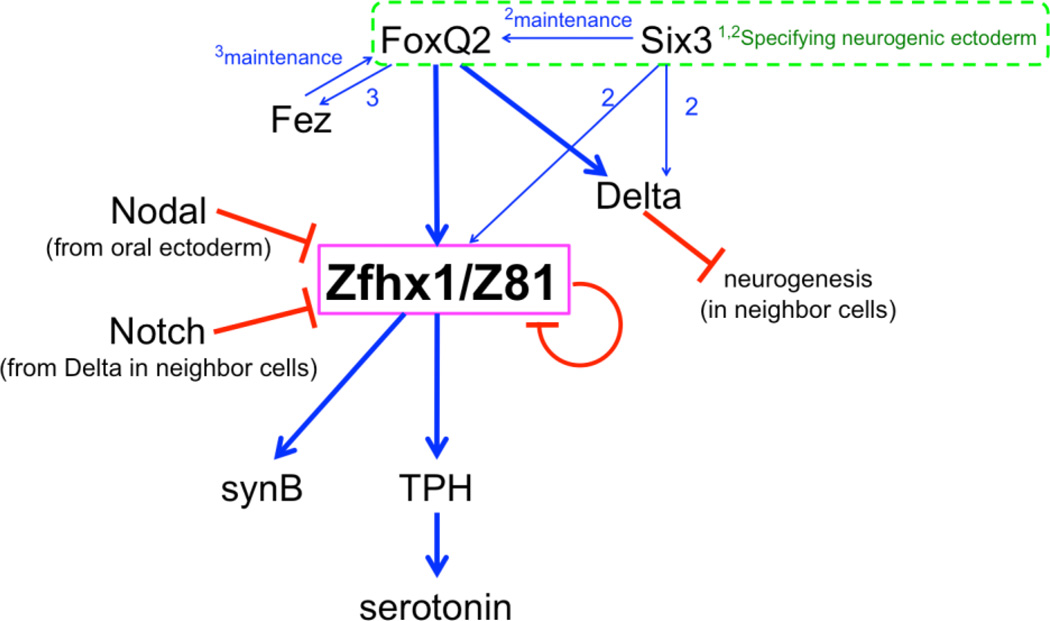
A diagram summarizing the mechanism and timing of Zfhx1/Z81 function is presented in Figure 8. At the beginning of neurogenesis in the animal plate of the sea urchin embryos, FoxQ2 and Six3 are required for formation of the animal plate and expression of downstream genes like fez and nk2.1, which are expressed uniformly in this territory (Yaguchi et al., 2011; Yaguchi et al., 2008; Wei et al., 2009). Whereas Nk2.1 is involved in formation of the long immotile cilia of the apical tuft, (Dunn et al., 2007; Yaguchi et al., 2010b), Fez functions in controlling animal plate size and ultimately the number of serotonergic neurons, but is not required for nerve cell differentiation itself (Yaguchi et al., 2011). delta is expressed in neural precursors in the animal plate starting at late mesenchyme blastula stage and Delta signals through Notch to neighboring cells preventing their differentiation to serotonergic neurons. Shortly thereafter, zfhx1/z81 and fez are expressed in these neural precursors. However, the expression of these three genes, delta, zfhx1/z81 and fez, is regulated by independent mechanisms because knock-downs of each does not affect the expression of other two (Fig. 5, 6; supplemental Fig. 2; Yaguchi et al., 2011).
Figure 8.
Model of the regulatory mechanisms controlling differentiation of serotonergic neurons in the sea urchin embryo. FoxQ2 and Six3 are involved in the specification of the animal plate during early development (1; Yaguchi et al., 2008, 2; Wei et al., 2009). FoxQ2 is required for fez expression and then Fez maintains foxQ2 expression on the aboral side of the animal plate (3; Yaguchi et al., 2011). Both FoxQ2 and Six3 regulate zfhx1/z81 and delta expression and Six3 supports FoxQ2 expression (2). Zfhx1/z81 is required for the expression of tph, which is required for serotonin synthesis, and for synaptotagminB (synB). Delta-Notch signaling limits the number of differentiating neurons by lateral inhibition and Nodal inhibits their development on the oral side of the animal plate. Zfhx1/Z81 suppresses its own expression.
At least, three independent signaling cascades regulate the differentiation of serotonergic neurons: Wnt/β-catenin positions the animal plate at the anterior end of the embryo where serotonergic neurons develop and Delta/Notch and Nodal determine, respectively, the number and position of these neurons. zfhx1/z81 expression exclusively in serotonergic neuron precursors in the animal plate depends on at least one or two positive inputs (FoxQ2 and Six3), and three negative inputs (Nodal, Notch and Zfhx1/Z81 itself). zfhx1/z81 expression depends on Six3 (Wei et al., 2009) and FoxQ2 (this work). The fact that Six3 is important for maintaining foxQ2 (Wei et al., 2009), may explain these observations (Fig. 6). Although FoxQ2 could provide direct inputs into regulating zfhx1/z81 transcription, this would occur well after initial formation of the animal plate. Furthermore, it is clearly not sufficient to control its spatial pattern since zfhx1/z81 is expressed in only a subset of animal plate cells. The mechanism that activates expression of zfhx1/z81 and delta in this subset is not yet understood. Negative regulation of serotonergic neural development by Nodal from the oral side or by Delta/Notch-mediated lateral inhibition in the animal plate acts at or upstream of zfhx1/z81. Finally, Zfhx1/Z81-mediated negative auto-regulation of zfhx1/z81 transcription implies tight regulation of Zfhx1/Z81 levels is required in these neural cells. All of these mechanisms help to ensure zfhx1/z81 expression in a few neural precursors on the aboral side of the animal plate, where it activates expression of genes required for serotonergic differentiation. The regulatory relationships established here provide an important framework for the eventual construction of the serotonergic neural gene regulatory network in the sea urchin embryo.
Highlights.
> Serotonergic neurons differentiate at aboral edge of the anterior neuroectoderm in sea urchin embryos. > zinc-finger homeobox (zfhx1/z81) is co-expressed with serotonin synthase gene, tryptophan 5-hydroxylase (tph). > Zfhx1/Z81 is required for tph expression. > zfhx1/z81 depends on FoxQ2, which is required for formation of the anterior neuroectoderm. > We conclude that Zfhx1/Z81 is an important intermediate regulator in serotonergic neurogenesis.
Supplementary Material
Acknowledgements
We thank Robert Angerer for fruitful comments on this manuscript and Robert Burke, Yoko Nakajima, and David McClay for essential reagents. We thank Masato Kiyomoto and Mamoru Yamaguchi for providing the adult sea urchins and Mrs. Y. Tsuchiya, T. Sato, H, Shinagawa, and Y. Yamada, Shimoda Marine Research Center, for collecting and keeping the adult sea urchins. This work was supported, in part, by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (MEXT), by Grant-in Aid for Young Scientists (B No. 21770227 and No. 23770241), and Takeda Science Foundation to S.Y., in part by MEXT (No. 22370023) to K.I., and in part by the Intramural Program of the National Institutes of Health, National Institute for Dental and Craniofacial Research, to L.M.A. J. Y. was a Predoctoral Fellows of JSPS with research grant (23-3584).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angerer LM, Yaguchi S, Angerer RC, Burke RD. The evolution of nervous system patterning: insights from sea urchin development. Development. 2011;138:3613–3623. doi: 10.1242/dev.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Burke RD. Development of serotonergic neurons in embryos of the sea urchin, Strongylocentrotus purpuratus. Development Growth & Differentiation. 1986;28:569–574. doi: 10.1111/j.1440-169X.1986.00569.x. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Burke RD. Development of the nervous system of the pluteus larva of Strongylocentrotus droebachiensis. Cell and Tissue Research. 1987;248:335–343. [Google Scholar]
- Burke RD, Angerer LM, Elphick MR, Humphrey GW, Yaguchi S, Kiyama T, Liang S, Mu X, Agca C, Klein WH, Brandhorst BP, Rowe M, Wilson K, Churcher AM, Taylor JS, Chen N, Murray G, Wang D, Mellott D, Olinski R, Hallbook F, Thorndyke MC. A genomic view of the sea urchin nervous system. Dev Biol. 2006;300:434–460. doi: 10.1016/j.ydbio.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Chiu C. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development. 2003;130:3781–3794. doi: 10.1242/dev.00571. [DOI] [PubMed] [Google Scholar]
- Cordes SP. Molecular genetics of the early development of hindbrain serotonergic neurons. Clin Genet. 2005;68:487–494. doi: 10.1111/j.1399-0004.2005.00534.x. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Moy VN, Angerer LM, Angerer RC, Morris RL, Peterson KJ. Molecular paleoecology: using gene regulatory analysis to address the origins of complex life cycles in the late Precambrian. Evol Dev. 2007;9:10–24. doi: 10.1111/j.1525-142X.2006.00134.x. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Lai ZC, Rubin GM. The Drosophila zfh-1 and zfh-2 genes encode novel proteins containing both zinc-finger and homeodomain motifs. Mech Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay-Schmidt A. The evolution of the serotonergic nervous system. Proc Biol Sci. 2000;267:1071–1079. doi: 10.1098/rspb.2000.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev Biol. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Fortini ME, Rubin GM. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lundell MJ. Differentiation of the Drosophila serotonergic lineage depends on the regulation of Zfh-1 by Notch and Eagle. Mol Cell Neurosci. 2007;36:47–58. doi: 10.1016/j.mcn.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Brivanlou AH. Proposal of a model of mammalian neural induction. Dev Biol. 2007;308:247–256. doi: 10.1016/j.ydbio.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J. The zfh-2 gene product is a potential regulator of neuron-specific dopa decarboxylase gene expression in Drosophila. Dev Biol. 1992;154:84–94. doi: 10.1016/0012-1606(92)90050-q. [DOI] [PubMed] [Google Scholar]
- Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Dev Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Kaneko H, Murray G, Burke RD. Divergent patterns of neural development in larval echinoids and asteroids. Evol Dev. 2004;6:95–104. doi: 10.1111/j.1525-142x.2004.04011.x. [DOI] [PubMed] [Google Scholar]
- Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Ward E, Skeath JB, Dean DC. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol Cell Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka AJ, Kuhn A, Groth D, Weise V, Yaguchi S, Burke RD, Herwig R, Lehrach H, Panopoulou G. A global view of gene expression in lithium and zinc treated sea urchin embryos: new components of gene regulatory networks. Genome Biol. 2007;8:R85. doi: 10.1186/gb-2007-8-5-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutierrez EL, Dubchak I, Garcia-Fernandez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin IT, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Rottinger E, Croce J, Lhomond G, Besnardeau L, Gache C, Lepage T. Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development. 2006;133:4341–4353. doi: 10.1242/dev.02603. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Kominami T. Specification process of animal plate in the sea urchin embryo. Development Growth & Differentiation. 2008;50:595–606. doi: 10.1111/j.1440-169x.2008.01057.x. [DOI] [PubMed] [Google Scholar]
- Saudemont A, Haillot E, Mekpoh F, Bessodes N, Quirin M, Lapraz F, Duboc V, Rottinger E, Range R, Oisel A, Besnardeau L, Wincker P, Lepage T. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001259. e1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, Coffman JA, Dean M, Elphick MR, Ettensohn CA, Foltz KR, Hamdoun A, Hynes RO, Klein WH, Marzluff W, McClay DR, Morris RL, Mushegian A, Rast JP, Smith LC, Thorndyke MC, Vacquier VD, Wessel GM, Wray G, Zhang L, Elsik CG, Ermolaeva O, Hlavina W, Hofmann G, Kitts P, Landrum MJ, Mackey AJ, Maglott D, Panopoulou G, Poustka AJ, Pruitt K, Sapojnikov V, Song X, Souvorov A, Solovyev V, Wei Z, Whittaker CA, Worley K, Durbin KJ, Shen Y, Fedrigo O, Garfield D, Haygood R, Primus A, Satija R, Severson T, Gonzalez-Garay ML, Jackson AR, Milosavljevic A, Tong M, Killian CE, Livingston BT, Wilt FH, Adams N, Belle R, Carbonneau S, Cheung R, Cormier P, Cosson B, Croce J, Fernandez-Guerra A, Geneviere AM, Goel M, Kelkar H, Morales J, Mulner-Lorillon O, Robertson AJ, Goldstone JV, Cole B, Epel D, Gold B, Hahn ME, Howard-Ashby M, Scally M, Stegeman JJ, Allgood EL, Cool J, Judkins KM, McCafferty SS, Musante AM, Obar RA, Rawson AP, Rossetti BJ, Gibbons IR, Hoffman MP, Leone A, Istrail S, Materna SC, Samanta MP, Stolc V, Tongprasit W, Tu Q, Bergeron KF, Brandhorst BP, Whittle J, Berney K, Bottjer DJ, Calestani C, Peterson K, Chow E, Yuan QA, Elhaik E, Graur D, Reese JT, Bosdet I, Heesun S, Marra MA, Schein J, Anderson MK, Brockton V, Buckley KM, Cohen AH, Fugmann SD, Hibino T, Loza-Coll M, Majeske AJ, Messier C, Nair SV, Pancer Z, Terwilliger DP, Agca C, Arboleda E, Chen N, Churcher AM, Hallbook F, Humphrey GW, Idris MM, Kiyama T, Liang S, Mellott D, Mu X, Murray G, Olinski RP, Raible F, Rowe M, Taylor JS, Tessmar-Raible K, Wang D, Wilson KH, Yaguchi S, Gaasterland T, Galindo BE, Gunaratne HJ, Juliano C, Kinukawa M, Moy GW, Neill AT, Nomura M, Raisch M, Reade A, Roux MM, Song JL, Su YH, Townley IK, Voronina E, Wong JL, Amore G, Branno M, Brown ER, Cavalieri V, Duboc V, Duloquin L, Flytzanis C, Gache C, Lapraz F, Lepage T, Locascio A, Martinez P, Matassi G, Matranga V, Range R, Rizzo F, Rottinger E, Beane W, Bradham C, Byrum C, Glenn T, Hussain S, Manning G, Miranda E, Thomason R, Walton K, Wikramanayke A, Wu SY, Xu R, Brown CT, Chen L, Gray RF, Lee PY, Nam J, Oliveri P, Smith J, Muzny D, Bell S, Chacko J, Cree A, Curry S, Davis C, Dinh H, Dugan-Rocha S, Fowler J, Gill R, Hamilton C, Hernandez J, Hines S, Hume J, Jackson L, Jolivet A, Kovar C, Lee S, Lewis L, Miner G, Morgan M, Nazareth LV, Okwuonu G, Parker D, Pu LL, Thorn R, Wright R. The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwick SM, Paige S, Morgan CA, 3rd, Bremner JD, Krystal JH, Charney DS. Neurotransmitter alterations in PTSD: catecholamines and serotonin. Semin Clin Neuropsychiatry. 1999;4:242–248. doi: 10.153/SCNP00400242. [DOI] [PubMed] [Google Scholar]
- Su YH, Li E, Geiss GK, Longabaugh WJ, Kramer A, Davidson EH. A perturbation model of the gene regulatory network for oral and aboral ectoderm specification in the sea urchin embryo. Dev Biol. 2009;329:410–421. doi: 10.1016/j.ydbio.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Zhang S, Li J, Ruvkun G. The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development. 2002;129:3901–3911. doi: 10.1242/dev.129.16.3901. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony, methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropepe V, Hitoshi S, Sirard C, Mak TW, Rossant J, van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol. 2004;275:403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, Smith JC, Huylebroeck D. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5'-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Wacker I, Schwarz V, Hedgecock EM, Hutter H. zag-1, a Zn-finger homeodomain transcription factor controlling neuronal differentiation and axon outgrowth in C. elegans. Development. 2003;130:3795–3805. doi: 10.1242/dev.00570. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM. A database of mRNA expression patterns for the sea urchin embryo. Dev Biol. 2006;300:476–484. doi: 10.1016/j.ydbio.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM. Direct development of neurons within foregut endoderm of sea urchin embryos. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018513108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Yaguchi J, Yaguchi S, Angerer RC, Angerer LM. The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Development. 2009;136:1179–1189. doi: 10.1242/dev.032300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Klein WH. Multiple signaling events specify ectoderm and pattern the oral-aboral axis in the sea urchin embryo. Development. 1997;124:13–20. doi: 10.1242/dev.124.1.13. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Kanoh K, Amemiya S, Katow H. Initial analysis of immunochemical cell surface properties, location and formation of the serotonergic apical ganglion in sea urchin embryos. Development Growth & Differentiation. 2000;42:479–488. doi: 10.1046/j.1440-169x.2000.00535.x. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Katow H. Expression of tryptophan 5-hydroxylase gene during sea urchin neurogenesis and role of serotonergic nervous system in larval behavior. J Comp Neurol. 2003;466:219–229. doi: 10.1002/cne.10865. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Angerer RC, Angerer LM. A Wnt-FoxQ2-nodal pathway links primary and secondary axis specification in sea urchin embryos. Dev Cell. 2008;14:97–107. doi: 10.1016/j.devcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Angerer RC, Angerer LM, Burke RD. TGF-beta signaling positions the ciliary band and patterns neurons in the sea urchin embryo. Dev Biol. 2010a;347:71–81. doi: 10.1016/j.ydbio.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Burke RD. Specification of ectoderm restricts the size of the animal plate and patterns neurogenesis in sea urchin embryos. Development. 2006;133:2337–2346. doi: 10.1242/dev.02396. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Burke RD. Sp-Smad2/3 mediates patterning of neurogenic ectoderm by nodal in the sea urchin embryo. Dev Biol. 2007;302:494–503. doi: 10.1016/j.ydbio.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Wei Z, Jin Y, Angerer LM, Inaba K. Fez function is required to maintain the size of the animal plate in the sea urchin embryo. Development. 2011;138:4233–4243. doi: 10.1242/dev.069856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi S, Yaguchi J, Wei Z, Shiba K, Angerer LM, Inaba K. ankAT-1 is a novel gene mediating the apical tuft formation in the sea urchin embryo. Dev Biol. 2010b;348:67–75. doi: 10.1016/j.ydbio.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.