Abstract
Domain swapping is a mechanism for forming protein dimers and oligomers with high specificity. It is distinct from other forms of oligomerization in that the binding interface is formed by reciprocal exchange of polypeptide segments. Swapping plays a physiological role in protein-protein recognition and it can also potentially be exploited as a mechanism for controlled self assembly. Here, we demonstrate that domain-swapped interfaces can be engineered by inserting one protein into a surface loop of another protein. The key to facilitating a domain swap is to destabilize the protein when it is monomeric but not when it is oligomeric. We achieve this condition by employing the ‘mutually exclusive folding’ design to apply conformational stress to the monomeric state. Ubiquitin is inserted into one of six surface loops of barnase. The 38 Å amino-to-carboxy terminal distance of ubiquitin stresses the barnase monomer, causing it to split at the point of insertion. The 2.2 Å X-ray structure of one insertion variant reveals that strain is relieved by intermolecular folding with an identically-unfolded barnase domain, resulting in a domain-swapped polymer. All six constructs oligomerize suggesting that inserting ubiquitin into each surface loop of barnase results in a similar domain-swapping event. Binding affinity can be tuned by varying the length of the peptide linkers used to join the two proteins, which modulates the extent of stress. Engineered, swapped proteins have the potential to be used to fabricate ‘smart’ biomaterials, or as binding modules from which to assemble heterologous, multi-subunit protein complexes.
Nature uses 3D domain swapping to mediate protein-protein recognition when high specificity is required (1–4). For example, cell surface-expressed cadherins are responsible for cell-cell adhesion and the development of tissue architecture (5). By binding via β-strand exchange between monomers, classical cadherins ensure that cells expressing a particular cadherin will adhere to cells expressing the same cadherin subtype and not another (6–9). The strictest definition of a domain swap requires that both monomeric and oligomeric states of the protein be observed, a polypeptide segment of the monomer exchange with the same segment of another monomer, and the structures of the monomer and oligomer be identical except at the points of strand exchange (10). This interaction can result in dimers, closed ring-shaped oligomers, or polymers of indefinite length. Indeed, runaway swapping occurs in protein deposition diseases such as prion amyloidosis and serpinopathies (11). Aside from its pathogenic manifestations, however, swapping can potentially be exploited as a mechanism for controlled self assembly. It has been suggested that homopolymeric hydrogels with targeted bulk properties can be fabricated from domain-swapped proteins or peptides (12, 13). Taking this idea one step further, it may be possible to use domain swapping as a means to create self-assembling macromolecular structures of defined subunit composition, stoichiometry, and quaternary structure; e.g. multi-subunit enzyme complexes that efficiently catalyze otherwise difficult reactions. For this application domain-swapping proteins could serve as genetically-encoded tags, fused to the target subunits, to facilitate assembly of the heterologous complex.
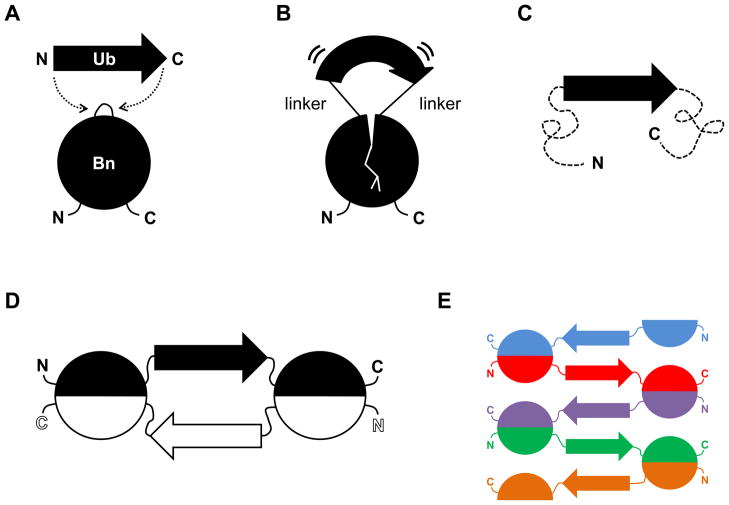
Here we demonstrate that domain swapping can be induced by inserting one small protein (ubiquitin, 76 amino acids) into surface loops of another small protein (barnase, 110 amino acids). Any protein seems capable of undergoing a domain swap, but few actually do (there are ~60 examples in the PDB (1)). Logically, the key to facilitating a domain swap is to destabilize the protein when it is monomeric but not if it is oligomeric (4, 6, 14). We achieve this unusual condition by employing the ‘mutually exclusive folding’ design (15) to apply selective stress to the monomeric state, as illustrated schematically in Fig. 1. We previously inserted ubiquitin (Ub) into a surface loop of barnase (Bn) at position 66 to generate barnase-ubiquitin 66 (BU66; Fig. 1A). Ub (38 Å N-to-C terminal distance) stretchesBn at the point of insertion while Bn simultaneously compresses the ends of Ub (Fig. 1B). This antagonistic interaction is parameterized by a coupling free energy which depends primarily on the length of the peptides used to link the two proteins (16, 17). Very long linkers (10 Gly each) decouple the molecular tug-of-war and the coupling free energy is zero. If the linkers are sufficiently short (<2 amino acids), then the coupling free energy exceeds the folding free energy (ΔG) of the Ub or Bn domain and the more stable protein is predicted to unfold the less stable protein (Fig. 1C). This relationship can be represented by a thermodynamic box consisting nominally of four states [Bn(unfolded)-Ub(unfolded), Bn(folded)-Ub(unfolded), Bn(unfolded)-Ub(folded), and Bn(folded)-Ub(folded)] linked by coupling and folding free energy terms (16).
Figure 1.
Schematic of mutually exclusive folding-induced domain swapping. (A) Ub (38 Å N-to-C distance) is inserted into a surface loop of Bn (10 Å distance between the termini of the loop at position 66). (B) If the linkers used to join Ub and Bn are sufficiently short, the Ub domain stretches the Bn domain and the Bn domain compresses the Ub domain. (C) The more stable Ub domain stretches the Bn domain to the point where it unfolds (dashed lines). This state is expected to be stable only at low protein concentrations. (D) The Bn domain refolds by domain swapping with an identical monomer to generate a closed dimer. This species is anticipated to be populated at intermediate protein concentrations. (E) Runaway swapping is predicted to occur at high protein concentrations, producing a long, linear polymer.
Enzymatic, thermodynamic, and CD structural results (17, 18) as well as molecular dynamics simulations (19), however, called the four-state model into question. Those data suggested that inserting Ub (or the GCN4 DNA binding domain) into position 66 of Bn unfolded the latter, but Bn was able to bind to and refold with another copy (or copies) of itself to regenerate the active enzyme (Fig. 1D and Fig. 1E). In this study we directly test that hypothesis and determine whether inserting Ub into the other surface loops of Bn evokes a similar response.
Results
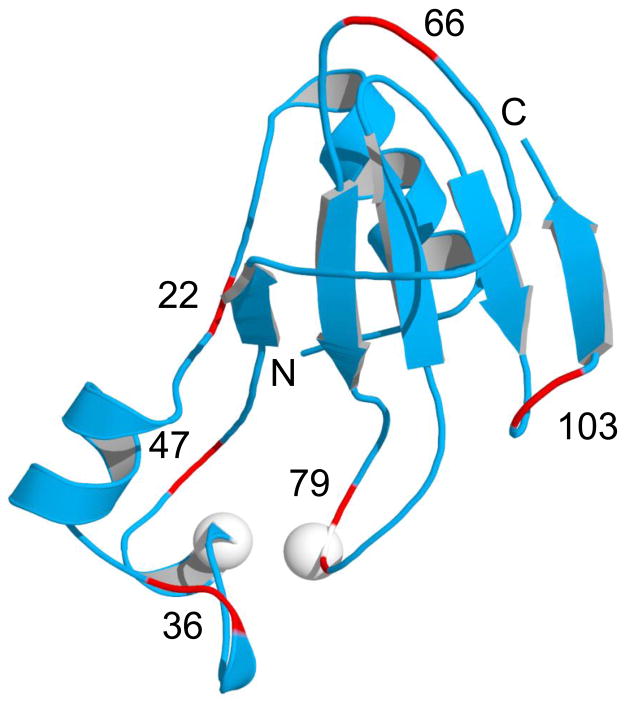
The conformational stress model predicts that inserting an unstructured or unstable protein into Bn will not dramatically destabilize Bn and thus not induce domain swapping. Accordingly, we first placed a destabilized Ub mutant (V26G) into each of the six surface loops and turns of Bn (centered at positions 22, 36, 47, 66, 79, and 103; see Fig. 2 for locations and nomenclature), to test whether Bn can tolerate insertion while remaining folded and principally monomeric. With a ΔG value of 1.8 kcal/mol (20), V26G Ub is expected to exert relatively little stress on the more stable Bn domain (ΔG = 11.5 kcal/mol for WT Bn; stability of the Bn domain is lower in the BU variants due to loop-closure entropy loss (16)). Table 1 summarizes stability parameters measured by Trp fluorescence. The Ub domain does not contain any Trp residues so fluorescence reports only on the conformation of the Bn domain. The Bn domain is destabilized, as anticipated, but it remains folded in all six constructs (Table 1). BU66 (V26G) is the most stable. One possible reason is that the 66-loop, being the largest loop in Bn, can accommodate insertion with the least perturbation to flanking Bn residues. Size exclusion chromatography finds that all V26G variants are predominantly monomeric, although minor peaks corresponding to oligomeric states up to tetramers are observed (Fig. 3A). Bn therefore seems robust in its ability to accept unstable or unfolded proteins into its surface loops while remaining folded.
Figure 2.
Structure of WT Bn (PDB 1A2P) showing surface loops (red) where Ub is inserted. The Bn-Ub fusion proteins constructed in this study (BU22, BU36, BU47, BU66, BU79, and BU103) are named according to the Bn residue numbers indicated in the figure. White spheres denote positions of Cys residues in the BU66 (A43C+S80C) double mutant.
BU genes were created by inserting the Ub gene into the Bn gene at the positions indicated, following the procedure of Geiser et al. (24). The genes were fused using nucleotides encoding Gly-Gly and Gly as the first and second linkers, respectively, and the Ub gene lacked a codon for the N-terminal Met. BU proteins were purified as described (17).
Table 1.
Stability parameters of the Bn domain of BU variants. Cm is the midpoint of urea-induced denaturation. Protein concentration is 5 μM (monomer). Errors are standard deviations of three measurements. Urea denaturation studies were carried out in 10 mM sodium phosphate (pH 7.0), 0.1 M NaCl, 10 °C. Unfolding of the Bn domain was monitored by Trp fluorescence (Ub does not contain any Trp residues), and unfolding data were fit to the two-state linear extrapolation equation ΔG(urea) = ΔG – m[urea] as described [reference 16].
| Variant | Cm (M) | ΔG (kcal/mol) | m (kcal/mol/M) |
|---|---|---|---|
| BU22 (V26G) | 1.27 ± 0.08 | 3.19 ± 0.80 | 2.49 ± 0.56 |
| BU36 (V26G) | 2.79 ± 0.02 | 7.83 ± 0.23 | 2.80 ± 0.06 |
| BU47 (V26G) | 1.70 ± 0.01 | 4.42 ± 0.13 | 2.60 ± 0.10 |
| BU66 (V26G) | 3.42 ± 0.02 | 9.54 ± 1.25 | 2.81 ± 0.37 |
| BU79 (V26G) | 2.75 ± 0.06 | 7.31 ± 0.07 | 2.65 ± 0.07 |
| BU103 (V26G) | 3.1 ± 0.04 | 8.64 ± 0.64 | 2.79 ± 0.22 |
| BU22 | 0.22 ± 0.04 | 0.54 ± 0.07 | 2.43 ± 0.24 |
| BU36 | 1.66 ± 0.03 | 3.42 ± 0.28 | 2.05 ± 0.15 |
| BU47 | 0.63 ± 0.06 | 1.06 ± 0.12 | 1.69 ± 0.16 |
| BU66 | 2.70 ± 0.07 | 4.30 ± 0.30 | 1.59 ± 0.16 |
| BU79 | 1.69 ± 0.10 | 3.68 ± 0.42 | 2.13 ± 0.36 |
| BU103 | 2.62 ± 0.03 | 5.45 ± 0.98 | 2.09 ± 0.38 |
Figure 3.
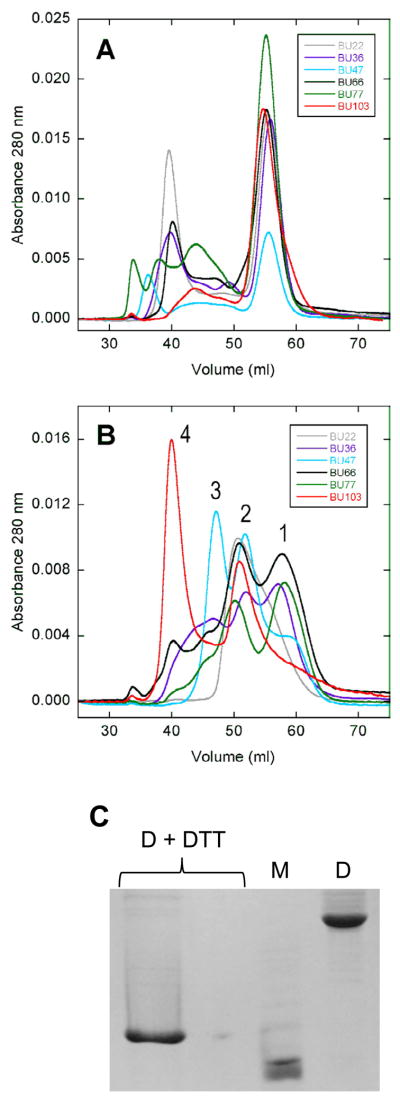
Chacterization of BU insertion variants by size exclusion chromatography and disulfide crosslinking. (A) and (B) show gel filtration chromatograms of V26G Ub and WT Ub constructs, respectively. Peaks numbered 1, 2, 3, and 4 indicate monomer, dimer, trimer, and tetramer. (C) SDS-PAGE analysis of BU66 (A43C+S80C) dimer (D) and monomer (M) peaks purified by size exclusion chromatography (DTT, dithiothreitol). M migrates faster than the fully-reduced, monomeric control (D+DTT) because the former contains an intramolecular disulfide bond. A ten-fold dilution of the (D+DTT) sample is shown in lane 2. The gel is stained with Coomassie Brilliant Blue.
Oligomerization experiments were performed by denaturing BU variants in 2 M guanidine hydrochloride, then refolding the samples by rapid 20-fold dilution to a final protein concentration of 20 μM. Samples were then injected onto a Superdex-75 gel filtration column (GE Healthcare). Disulfide crosslinking experiments were performed as above except 10 mM dithiothreitol was present during denaturation. After dilution, the samples were dialyzed to remove reductant and allowed to oligomerize under oxidizing conditions prior to injection onto the Superdex-75 column.
We next intensified the conformational stress exerted by Ub and determined whether Bn unfolds or oligomerizes. Disruptive force was generated by replacing V26G Ub with WT Ub (ΔG = 6.9 kcal/mol (16)) at the same insertion points. In agreement with the thermodynamic model, stabilizing the Ub domain destabilizes the Bn domain by an additional 2.7 – 5.2 kcal/mol (average ΔΔG = −3.8 ± 0.9 kcal/mol; Table 1). Still, the Bn domain remains folded and stable in all variants except for BU22. For BU22, the midpoint of denaturation (Cm) is 0.22 M urea and ~20 % of the protein is unfolded in the absence of denaturant. Size exclusion chromatography, however, reveals that the constructs now exist predominantly as dimers, trimers, and tetramers (Fig. 3B). The distribution of oligomeric states varies among mutants. BU103 is mainly tetrameric with a smaller population of dimer. BU22 is almost exclusively dimeric, and BU36, BU47, BU66, and BU79 exist as a mixture of trimers, dimers, and monomers. Because of the multiplicity of oligomeric states, the observed folding transitions are likely not two state (although all curves fit satisfactorily to the two-state linear extrapolation equation (not shown)). Free energies reported in Table 1 are therefore apparent values that depend on protein concentration (5 μM in the present experiments).
We employed disulfide cross linking to ask whether the BU66 dimers observed in Fig. 3B associate via domain swapping. We took advantage of the Bn double-cysteine mutant (A43C+S80C), which is known to readily form an intramolecular disulfide bond (Fig. 2) (21). This bond cross links the two fragments of Bn that the model predicts to be split in BU66. If BU66 dimers bind in a conventional manner, i.e. via complementary patches on the monomer surface, then the C43-C80 disulfide bond is expected to form intramolecularly and the dimers will dissociate to monomers on reductant-free SDS-PAGE. If BU66 dimers are domain swapped, then C43 of one monomer is predicted to form an intermolecular S-S bridge with C80 of the second monomer (and vice versa), thereby crosslinking the black and white semicircles in Fig. 1D. This species will migrate on the gel as a covalent dimer. BU66 (A43C+S80C) was denatured and reduced in guanidine hydrochloride/dithiothreitol then allowed to oligomerize and oxidize in the absence of denaturant and reductant. SDS-PAGE shows that the dimer peak recovered from size exclusion chromatography is the intermolecularly cross-linked dimer, whereas the monomer peak is the intramolecularly cross-linked monomer (Fig. 3C). This result suggests that BU66 (A43C+S80C) monomers bind via a domain swap, yielding the closed, symmetrical dimer depicted in Fig. 1D.
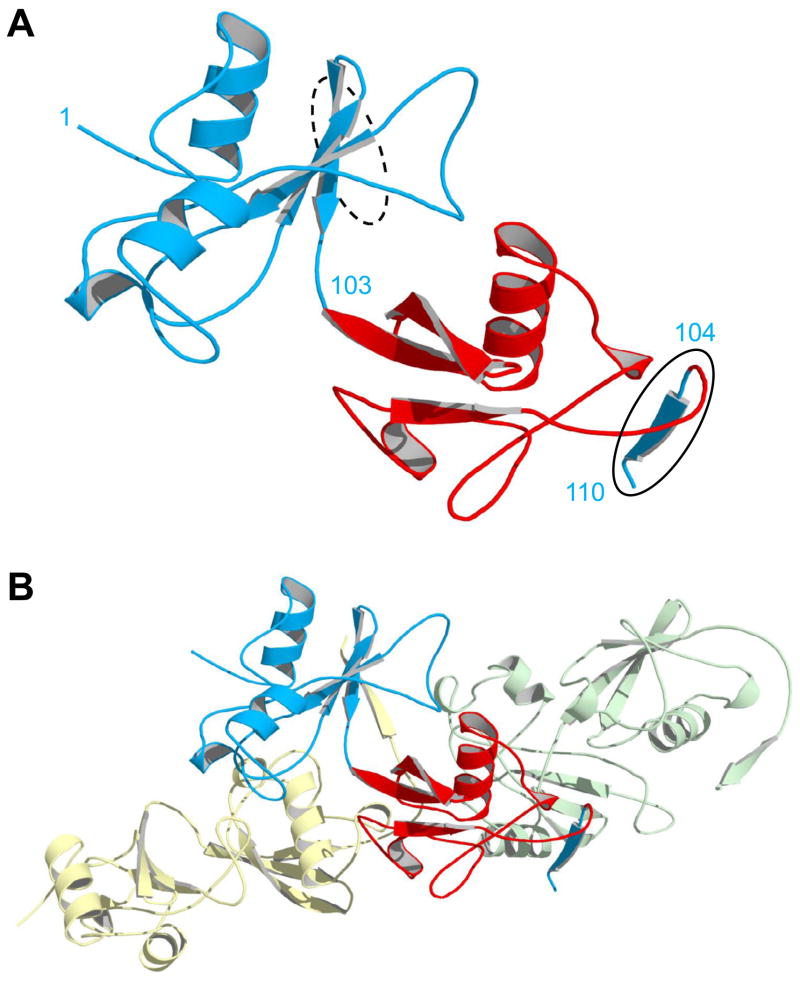
To determine the structural basis for oligomerization, we solved the X-ray structure of BU103 to 2.2 Å resolution. The dimeric species in Fig. 3B was isolated and concentrated to 1.4 mM for crystallization. As a result of the increased concentration, the dimers spontaneously reorganized to form long, linear polymers (Fig. 1E). The asymmetric unit, however, consists of a single BU103 monomer (Fig. 4A). In WT Bn, residues 104–110 form the last strand of the five-stranded β-sheet (Fig. 2). The structure of BU103 shows that Ub has pulled apart Bn, with residues 1–103 and 104–110 of Bn extending from the N- and C-termini of Ub, respectively (Fig. 4A). The binding interface is clearly revealed as a domain swap. The 1–103 and 104–110 fragments of Bn do not contact each other within the same molecule; they are separated by more than 30 Å. Rather, residues 104–110 insert into the BU103 molecule in the next asymmetric unit, replacing the missing fifth β-strand (Fig. 4B). Similarly, the absent fifth strand of the central BU103 molecule in Fig. 4B is supplied by the 104–110 fragment from the monomer in the preceding asymmetric unit. These asymmetric units are related by successive application of the crystallographic three-fold screw operator, generating a helical polymer extending the length of the crystal. While the extent of oligomerization and regular helix observed in the crystal are enforced by high concentration and crystal packing, it seems reasonable to assume that the oligomers seen in solution at lower concentration are connected in the same way. Oligomerization thereby restores native interactions in the Bn domain without conformational stress.
Figure 4.
X-ray structure of domain-swapped BU103. (A) The asymmetric unit consists of a single monomer in which the Ub domain (red) has split apart the Bn domain (blue). Residues are numbered according to the Bn sequence. Bn residues 1–103 and 104–110 (solid oval) extend from the N- and C-termini of Ub at left and right, respectively. The dashed oval indicates where residues 104–110 would normally be located in WT Bn, as the fifth strand of the central β-sheet. (B) The same monomer in panel A is now shown with the molecules in the adjacent asymmetric units. Bn residues 104–110 of the central monomer (blue) insert into the βsheet of the next protomer (green), and the residues 104–110 from the preceding molecule (yellow) complex with residues 1–103 of the central monomer. In this arrangement, all native Bn interactions are restored and conformational strain is relieved.
BU103 was crystallized at 20 °C using the hanging-drop vapor-diffusion method with the mother liquor consisting of 10 mM Tris (pH 8.0), 1 M (NH4)2SO4, 1.5 % isopropanol (v/v). X-ray diffraction data were collected at station A1 at the Macromolecular Diffraction Facility at the Cornell High Energy Synchrotron Source (MacCHESS), reduced using HKL-2000 (25), phased by molecular replacement (1UBQ and residues 3–103 of chain A of 1A2P), and refined using Phenix (26). X-ray statistics are listed in Supplementary Table S1.
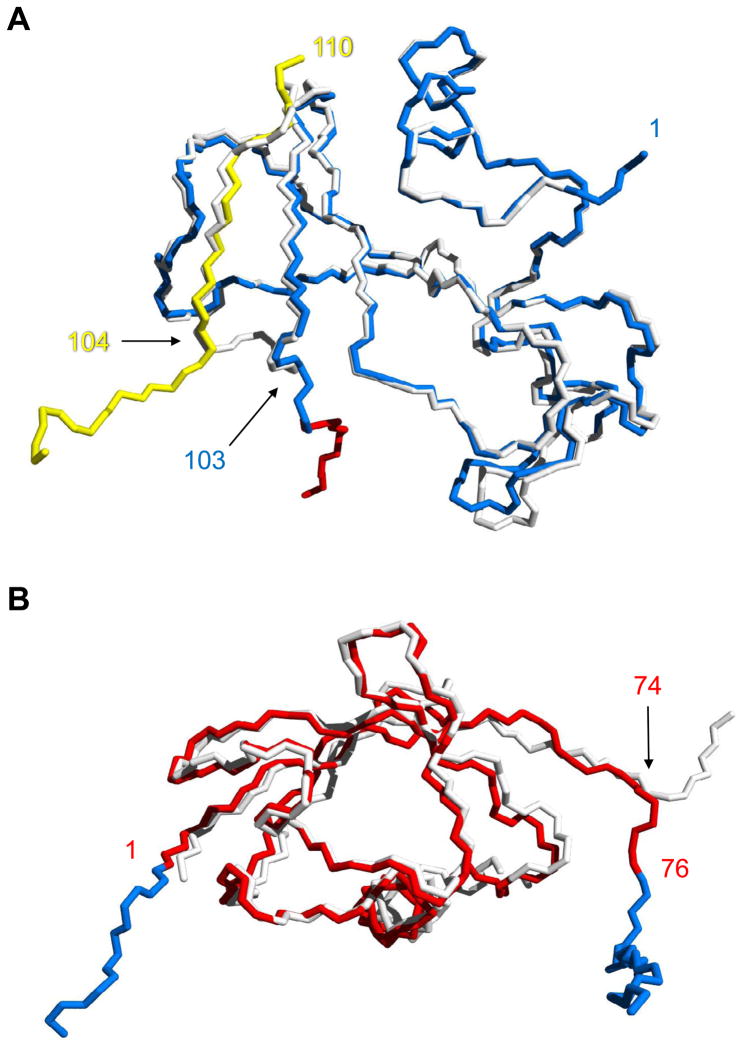
Comparing the Bn domain of BU103 with WT Bn finds the structures of inter- and intramolecularly folded Bn to be very similar. Amino acids 3–102 of WT Bn can be superimposed on the same residues of the Bn domain of BU103 with a Cα root mean-square deviation (RMSD) of 0.57 Å (Fig. 5A). Superposing just that subset of amino acids causes Bn residues 104–109 (from the preceding BU103 molecule) to align on the corresponding residues from WT Bn with a maximum Cα RMSD of 0.94 Å at Q104, and an overall Cα RMSD of 0.66 Å. The swapped β-strand adopts virtually the same structure as the fifth strand in WT Bn, as evidenced by the all-atom RMSD value of 0.75 Å for residues 105–108. BU103 thus satisfies all three criteria for a classically domain-swapped oligomer.
Figure 5.
Alignment of the Bn and Ub domains of BU103 with their respective WT proteins. (A) Comparison of inter- and intramolecularly folded Bn. Backbone atoms of the Bn domain of BU103 (blue and yellow) are superimposed on those of WT Bn (white). Residues are numbered according to the Bn sequence. The point of strand exchange is marked by residue 103 from the blue chain and residue 104 from the protomer in the preceding asymmetric unit (yellow chain). The red residues signify the beginning of the Ub domain. (B) Alignment of the Ub domain of BU103 (red) with WT Ub (white). Residues are numbered according to the Ub sequence. Bn fragments 1–103 and 104–110 (blue) extend from the N-terminus (residue 1) and C-terminus (residue 76) of the Ub domain, respectively.
Like the Bn domain of BU103, the structure of the Ub domain is nearly identical to that of its WT counterpart. Amino acids 1–70 of WT Ub superimpose on the corresponding residues of the Ub domain of BU103 with an all-atom RMSD of 0.47 Å (Fig. 5B). Significant differences are limited to the C-termini. Beginning at R74 and continuing to the end of Ub at G76, the direction of the polypeptide backbone diverges in the two molecules. This result indicates that the C-terminus of Ub is flexible. Some flexibility at one or both of Ub-Bn junction points may be desirable in order to accommodate any rigid-body adjustments that may need to be made at the binding interface.
Discussion
The above results suggest that it may be possible to engineer a self-assembly mechanism based on mutually exclusive folding-induced domain swapping. The general approach is to insert a ‘lever’ protein into a surface loop of an ‘assembler’ protein. As long as the N-to-C and loop-termini distances of the lever and assembler exceed the aforementioned ratio, the global free energy minimum of the system (above a threshold protein concentration) is an oligomer in which the lever and assembler are both folded with the latter protein domain swapped (Fig. 1E). This mechanism for self-assembly is distinct from those developed by others (e.g. coiled-coils, naturally-occurring repeat proteins) in two respects. First, our mechanism is based on conformational stress. In that respect it is reminiscent of the naturally-occurring polymerization reaction of the serine protease inhibitor α1-antitrypsin (22). The native fold of α1-antitrypsin is metastable. Proteolytic cleavage of the inhibitor’s reactive center loop by the target protease permits the cleaved loop to adopt a β-strand conformation and insert into the central β-sheet of the same α1-antitrypsin molecule, thereby allowing it to attain its global free energy minimum. In a spontaneous process that is aided by several disease-causing mutations, the reactive center loop can inappropriately insert into another α1-antitrypsin molecule, triggering a daisy chain reaction that generates highly stable fibers. The basis for the metastable nature of monomeric α1-antitrypsin is not well understood. In contrast, mutually exclusive folding-induced conformational stress—and thus the affinity of the binding reaction—can be controlled by known physical and thermodynamic principles. One straightforward method is to vary the length of the linkers that join lever to assembler. We previously showed that very long linkers fully decouple the mutually exclusive folding interaction between the Ub and Bn domains (17). Linkers of ten Gly each result in the BU66 monomer being fully relaxed; no oligomerization is detected. Shortening the linkers one residue at a time gradually increases stress and causes the dissociation constant (Kd) for dimerization to decrease. When Ub and Bn are fused without any additional linkers, BU66 dimerizes with sub-micromolar Kd(Kd cannot be determined accurately due to the presence of higher-order oligomers) (17).
Another method for tuning binding affinity is to modulate thermodynamic stability of the lever protein. Figure 3A and Fig. 3B demonstrate that a more stable Ub domain is better able to unfold Bn and induce it to oligomerize. Since protein stability is coupled to ligand binding as well as to solution conditions, the domain-swapped binding interaction can in principle be switched on and off by the presence of ligands or changes in temperature, pH, or salt concentration. It is important to recognize that, since neither of the above two modes of affinity tuning modifies the binding interface, they are not expected to compromise the inherent specificity of the domain-swapped interaction. Indeed, it is the combination of high specificity and moderate affinity that likely explains why nature chose domain swapping to mediate cell-cell interactions (6).
The second distinction of the present design is that it may be modular. There is no reason to expect that Bn is unique among potential assemblers in its ability to domain swap upon forced unfolding. This view is supported by the observation that Bn responds in the same way—by forming oligomeric complexes—as a result of being pulled apart at different locations. This is not the case, however, for all mutually exclusive folding constructs. Peng & Li created a mutually exclusive folding chimera in which the 27th Ig domain of titin was inserted into a surface loop of the GB5 protein (23). They were able to directly observe folding of the Ig domain and concomitant unfolding of the GB5 domain in real time. Subsequent refolding/domain swapping of GB5 was not detected. Thus, we still do not fully understand the principles that govern the extent to which assembler proteins swap in response to stress. As to the lever, any stable protein with a moderately long N-to-C distance should be able to perform the same stretching function as Ub. Additional lever-assembler combinations are needed to test the generality of the domain swapping mechanism.
Conclusions
The structures of both domains in the domain-swapped oligomer of BU103 are virtually identical to those of WT Ub and WT Bn. This finding, together with gel filtration and disulfide crosslinking results from the other BU insertion variants, suggests that it may be feasible to create self-assembling oligomers and polymers that retain and integrate the functions of the parent molecules. Our mechanism allows for precise control of both the structural details of protein-protein binding interfaces as well as the strength of their interaction.
Supplementary Material
Acknowledgments
We thank Rebecca Oot for help with structural refinement and David Schuller and Scott Smith at MacCHESS for assistance with X-ray data collection and processing. This work was supported by NIH grant R01GM069755 to S.N.L. The MacCHESS resource is supported by NIH/NCRR award RR-01646.
Footnotes
Accession numbers
Coordinates and structure factors for BU103 have been deposited in the Protein Data Bank with accession number 3Q3F.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gronenborn AM. Protein acrobatics in pairs--dimerization via domain swapping. Curr Opin Struct Biol. 2009;19(1):39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Science. 1995;4(12):2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci U S A. 1994;91(8):3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newcomer ME. Protein folding and three-dimensional domain swapping: a strained relationship? Curr Opin Struct Biol. 2002;12:48–53. doi: 10.1016/s0959-440x(02)00288-9. [DOI] [PubMed] [Google Scholar]
- 5.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7(5):619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 6.Patel SD, Chen CP, Bahna F, Honig B, Shapiro L. Cadherin-mediated cell-cell adhesion: sticking together as a family. Curr Opin Struct Biol. 2003;13:690–698. doi: 10.1016/j.sbi.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Harrison OJ, et al. Two-step adhesive binding by classical cadherins. Nat Struct Mol Biol. 2010;17(3):348–357. doi: 10.1038/nsmb.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciatto C, et al. T-cadherin structures reveal a novel adhesive binding mechanism. Nat Struct Mol Biol. 2010;17(3):339–347. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miloushev VZ, et al. Dynamic properties of a type II cadherin adhesive domain: implications for the mechanism of strand-swapping of classical cadherins. Structure. 2008;16(8):1195–1205. doi: 10.1016/j.str.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennett MJ, Sawaya MR, Eisenberg D. Deposition diseases and 3D domain swapping. Structure. 2006;14:811–824. doi: 10.1016/j.str.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Nagarkar RP, Hule RA, Pochan DJ, Schneider JP. Domain swapping in materials design. Biopolymers. 2010;94(1):141–155. doi: 10.1002/bip.21332. [DOI] [PubMed] [Google Scholar]
- 13.Banta S, Wheeldon IR, Blenner M. Protein engineering in the development of functional hydrogels. Annu Rev Biomed Eng. 2010;12:167–186. doi: 10.1146/annurev-bioeng-070909-105334. [DOI] [PubMed] [Google Scholar]
- 14.Kuhlman B, O’Neill JW, Kim DE, Zhang KY, Baker D. Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc Natl Acad Sci USA. 2001;98:10687–10691. doi: 10.1073/pnas.181354398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radley TL, Markowska AI, Bettinger BT, Ha J-H, Loh SN. Allosteric switching by mutually exclusive folding of protein domains. J Mol Biol. 2003;332:529–536. doi: 10.1016/s0022-2836(03)00925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutler T, Loh SN. Thermodynamic analysis of an antagonistic folding-unfolding equilibrium between two protein domains. J Mol Biol. 2007;371:308–316. doi: 10.1016/j.jmb.2007.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cutler T, Mills BM, Lubin DJ, Chong LT, Loh SN. Effect of interdomain linker length on an antagonistic folding-unfolding equilibrium between two protein domains. J Mol Biol. 2009;386:854–868. doi: 10.1016/j.jmb.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha J-H, Butler JS, Mitrea DM, Loh SN. Modular enzyme design: regulation by mutually exclusive protein folding. J Mol Biol. 2006;357:1058–1062. doi: 10.1016/j.jmb.2006.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills BM, Chong LT. Molecular simulations of mutually exclusive folding in a two-domain protein switch. Biophys J. 2011;100(3):756–764. doi: 10.1016/j.bpj.2010.12.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khorasanizadeh S, Peters ID, Roder H. Evidence for a three-state model of protein folding from kinetic analysis of ubiquitin variants with altered core residues. Nature Struct Biol. 1996;3:193–205. doi: 10.1038/nsb0296-193. [DOI] [PubMed] [Google Scholar]
- 21.Clarke J, Henrick K, Fersht AR. Disulfide mutants of barnase I: changes in stability structure assessed by biophysical methods and X-ray crystallography. J Mol Biol. 1995;254:493–504. doi: 10.1006/jmbi.1995.0568. [DOI] [PubMed] [Google Scholar]
- 22.Bottomley SP. The folding pathway of alpha1-antitrypsin: avoiding the unavoidable. Proc Am Thorac Soc. 2010;7(6):404–407. doi: 10.1513/pats.201004-031AW. [DOI] [PubMed] [Google Scholar]
- 23.Peng Q, Li H. Direct observation of tug-of-war during the folding of a mutually exclusive protein. J Am Chem Soc. 2009;131(37):13347–13354. doi: 10.1021/ja903480j. [DOI] [PubMed] [Google Scholar]
- 24.Geiser M, Cebe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–92. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.