Abstract
Stage-specific rearrangement of immunoglobulin heavy and light chain genes poses an enigma because both processes utilize the same recombinational machinery, but the heavy chain locus is accessible at the pro-B cell stage, while the light chain loci become accessible at the pre-B cell stage. Transcription factor STAT5 is a positive-acting factor for rearrangement of distal Vh genes but attenuation of IL7 signaling and loss of activated STAT5 at the pre-B cell stage corresponds with Igκ locus accessibility and rearrangement, suggesting that STAT5 plays an inhibitory role at this locus. Indeed, loss of IL7 signaling correlates with increased activity at the Igκ intron enhancer. However, the κE3’ enhancer must also be regulated as this enhancer plays a role in Igκ rearrangement. We show here that STAT5 can repress κE3’ enhancer activity. We find that STAT5 binds to a site that overlaps the κE3’ PU.1 binding site. We observed reciprocal binding by STAT5 and PU.1 to the κE3’ enhancer in primary bone marrow cells, STAT5 and PU.1 retrovirally transduced pro-B cell lines, or embryonic stem cells induced to differentiate into B lineage cells. Binding by STAT5 corresponded with low occupancy of other enhancer binding proteins, whereas PU.1 binding corresponded with recruitment of IRF4 and E2A to the κE3’ enhancer. We also find that IRF4 expression can over-ride the repressive activity of STAT5. We propose a novel PU.1/STAT5 displacement model during B cell development, and this, coupled with increased IRF4 and E2A activity regulates κE3’ enhancer function.
Introduction
Immunoglobulins (Ig) are composed of two polypeptide chains, the heavy chain and the light chain (either kappa or lambda). Proper expression of Ig heavy and light chain genes requires a somatic rearrangement process that links together either V, D, and J segments (heavy chain), or V and J segments (light chain) to produce functional Ig genes (1). The rearrangement of Ig genes is a highly ordered process in which heavy chains rearrange prior to light chains at the pro-B cell stage, whereas light chains rearrange at the pre-B cell stage. The stage-specific rearrangement of heavy and light chain genes poses an enigma because both processes utilize the same recombinational machinery. To explain the developmentally controlled specificity of light chain and heavy chain rearrangement, it has been proposed that the heavy chain locus is accessible to the recombinational machinery at the pro-B cell stage, while the light chain loci become accessible or active later at the pre-B cell stage (1, 2).
Developmentally controlled differences in histone H3 and H4 acetylation and methylation have been observed at the Ig loci and these modifications are believed to be part of the locus accessibility mechanism (3, 4). At the pro-B cell stage when IgH rearrangement commences, VH genes are associated with hyperacetylated histones H3 and H4. Histone acetylation appears to occur in a stepwise fashion with distal VH genes requiring IL-7 signaling for rearrangement (3, 5). Similarly, there are developmentally associated changes in the histone acetylation status at the Igκ and Igλ light chain loci (6). Changes in locus accessibility also correlate with the expression of transcripts from the Ig germline V, D, J, and constant regions (7, 8).
The various Ig enhancers are directly involved in the Ig rearrangement processes (9–11). For instance, deletion of either the Igκ intron (Eκi) or 3’ (κE3’) enhancers reduces recombination, and deletion of both enhancers ablates recombination (10, 12). Locus accessibility is apparently controlled through the function of DNA binding transcription factors. Positive acting factors such as E2A and NF-κB are believed to play important roles in positively regulating activity of the Igκ intron enhancer, while PU.1, IRF4, and E2A (as well as other factors) positively regulate the κE3’ enhancer and subsequent Igκ rearrangement (13–17). However, mechanisms must exist that maintain the Igκ locus in an inactive state in pro-B cells when the IgH locus is fully accessible and is actively undergoing rearrangement. A silencer element (Sis) that lies between the Jκ segments and the first Vκ gene (Vκ21G) appears to control the frequency of Vκ rearrangement, but this element does not control the developmental timing of Vκ rearrangement (18).
Transcription factor STAT5 has been proposed to be important for rearrangement of distal VH genes (19). STAT5 can be recruited to the VH gene promoters in pro-B cells by physical interaction with transcription factor Oct1 (19). STAT5a and STAT5b are closely related genes involved in mammary gland and hematopoietic development (20–23). Two STAT5 knock-out alleles have been generated. A hypomorphic STAT5 knock-out allele leads to hematopoietic defects, with a preferential deficiency in early pro-B and pre-B cell development (21, 24). A completely null STAT5 knock-out results in severely impaired lymphoid development with cells accumulating at the prepro-B cell stage (25). Therefore, STAT5 function is clearly crucial for early B cell development, although others have shown that Bcl-2 can compensate for loss of STAT5 in knock-out animals to restore IgH rearrangement to wild-type levels (15).
In pro-B cells, STAT5 is phosphorylated in response to IL7 signaling resulting in dimer formation and functional activation. At the pre-B cell stage, IL7 signaling diminishes leading to loss of activated STAT5 coincident with activation of the Igκ locus for rearrangement. STAT5 has been noted to have both activation and repression activities (26–28), and can recruit co-repressor proteins such as SMRT and HDAC1 to DNA (26–28). An attractive hypothesis is that STAT5 activates distal VH gene accessibility at the IgH locus in pro-B cells, but simultaneously inhibits activity of the Igκ locus. Loss of STAT5 in pre-B cells would then remove this repressive mechanism.
Consistent with the above idea, Johnson et al (14) identified two distinct pathways for regulating Igκ locus rearrangement. First, attenuated IL-7 signaling (i.e., reduced STAT5 activity) in pre-B cells led to increased E2A DNA binding and histone acetylation at the Igκ intron enhancer and subsequent Igκ locus recombination (14). Second, transcription factor IRF4, which is elevated in pre-B cells compared to pro-B cells was able to induce Igκ rearrangement and to stimulate histone acetylation at the Igκ 3’ enhancer (13, 14). Similar results were obtained by Mandal et al (29) who showed that STAT5 inhibited Igκ locus transcription and prevented E2A binding to the Igκ intron enhancer. In addition, Malin et al (15) showed that STAT5 reduced Igκ germline transcripts and Igκ rearrangement in pro-B cells indicating that STAT5 suppresses premature Igκ rearrangement in pro-B cells (15, 29, 30). Thus, STAT5 is able to repress function of the Igκ intron enhancer.
However, while IL7 signaling represses the Igκ intron enhancer at the pro-B cell stage, factors that repress the κE3’ enhancer are unknown. Regulation of both Igκ enhancers is important because inhibition of the Igκ intron enhancer alone is insufficient to inhibit Igκ rearrangement since deletion of this enhancer fails to ablate rearrangement apparently due to compensatory activity of the κE3’ enhancer (10, 12).
We show here that the STAT5 repressive mechanism extends to the Ig κE3’ enhancer. We found that STAT5 can inhibit κE3’ enhancer activity and that STAT5 repression occurs through the same DNA motif that we, and others, previously found yields synergistic PU.1-IRF4 DNA binding and transcriptional activation (16, 31–35). We found that STAT5 repression at the κE3’ enhancer occurs by displacing PU.1, thereby reducing its occupancy on the enhancer DNA. This represents a powerful mechanism for regulating 3’ enhancer activity because loss of PU.1 DNA binding will also result in concomitant loss of IRF4 DNA binding to its adjacent site (16, 31, 33–35). We present a model in which at the pro-B cell stage, the Igκ locus is maintained in a repressed state by STAT5 causing reduced E2A binding to the intron enhancer, while simultaneously inhibiting PU.1, IRF4, and E2A DNA binding to the κE3’ enhancer. As cells mature to the pre-B cell stage, reduced STAT5 activity and increased IRF4 expression and E2A activity results in increased cooperative PU.1 and IRF4 DNA binding at the Igκ 3’ enhancer as well as increased cooperative E2A-IRF4 DNA binding (36, 37). Consistent with this model, we previously observed elevated PU.1 and IRF4 binding to the 3’ enhancer in pre-B cells compared to pro-B cells (38). Therefore, activation of PU.1/IRF4 and E2A/IRF4 motifs in pre-B cells, along with concomitant loss of STAT5, promotes κE3’ enhancer activation.
Materials and Methods
DNA constructs
Reporter plasmids containing the Eκi enhancer, κE3’ enhancer core sequence, Eμ intron enhancer, multimerized PU.1 binding motif (4 copies), and multimerized E47 binding motif (4 copies) were described previously (16, 17, 31, 39). MigRI-PU.1 and MigR1-STAT5 retroviral constructs were prepared by placing the PU.1 and STAT5a cDNA sequences at the vector EcoR1-Not1 and HpaI sites, respectively.
Cell culture, transfection, and infection
Mouse plasmacytoma cell line S194 was cultured in DMEM with 10% horse serum. Virus packing cell line BOSC and NIH3T3 cells were was grown in DMEM supplemented with 10% fetal bovine serum. 38B9 cells were grown in RPMI 1640 plus 10% fetal bovine serum and 50µM β-mercaptoethanol. Bone marrow cells were cultured in IMDM medium supplemented with 10% heat inactivated fetal bovine serum, 1% Penn/Strep, 1% glutamine, 1% non-essential amino acids, 50µM β-mercaptoethanol, and 10ng/ml IL-7. In vitro culture resulted in 90% of cells obtaining a pro-B cell phenotype (B220+IgM−CD43+). For withdrawal of IL-7, the cells were cultured with 0.1ng/ml IL-7 for 18 hours. ES cells were grown and maintained in an undifferentiated state in DMEM supplemented with 15% Fetal bovine serum, 50 µm 2-mercaptoetanol, 0.1 mM nonessential amino acids, 50 µg/ml Penicillin/Streptomycin and 5 ng/ml Leukemia inhibitory factor (LIF, CHEMICON). Undifferentiated ES cells were grown on mouse primary embryonic fibroblast cells (CHEMICON). The Bone marrow stromal cell line (OP9) was cultured as a monolayer in αMEM medium supplemented with 20% FBS (ES grade and lot tested; Hyclone). For retroviral packaging, subconfluent BOSC cells were cotransfected with retroviral DNA, plus Gag-Pol and envelope plasmids using Lipofectamine 2000 (Invitrogen). After 48–72 hours conditioned medium was harvested, supplemented with polybrene(4 µg/mL, Sigma-Aldrich), and used for infection of 38B9 cells. S194 cells were transfected by the DEAE-dextran procedure, and CAT assays were performed as previously described (17). NIH3T3 cells were transfected by the Lipofectamine 2000 method (Invitrogen). To induce differentiation of ES cell/OP9 co-cultures, single cell suspensions of undifferentiated ES cells were plated onto an OP9 monolayer in αMEM medium. Medium was changed at day 3 and by day 5 nearly 100% of ES colonies differentiated into mesoderm like colonies. At day 5 the co-cultures were trypsinized and the single cell suspension was pre-plated for 30 minutes. Nonadherent cells were reseeded onto a new confluent OP9 monolayer along with 5 ng/ml Flt3L and 5 ng/ml IL-7 (both from R&D Systems). On day 8 loosely adherent cells were gently plated on a new OP9 monolayer with the addition of fresh Flt3L and IL-7. This process of adding fresh cytokines and replating cells onto new OP9 layers was repeated at days 8, 12, 16, 21 and 28.
Flow Cytometry
Anti-mouse CD45R and CD19 antibodies were purchased from BD Pharmingen. Cells were stained in 100 µl staining buffer (PBS + 1% BSA) for 30 minutes on ice and washed twice before analysis.
Stained cells were analyzed with a CANTO flow cytometer. Data analysis was performed using FLOWJO software.
Reverse Transcription
RNA was isolated from ES cell/OP9 co-cultures using the Trizol method.
cDNA was prepared from 5 µg RNA by Oligo dT -priming using the Superscript cDNA Synthesis kit from Strategene. Real-Time PCR was performed with primers specific for the transcripts shown in Table I.
Table I.
Primers for RT-PCR, EMSA probes, and ChIP
| RT-PCR Primers | |
| IRF4-Forward | GACTGCCGGCTGCATATCTGC |
| IRF4 –Reverse | GCCGATCGCTGCACAGTGCCAG |
| E47 –Forward | CGAGAAGCCGCAGACCAAACT |
| E47- Reverse | TCCCCGACCACGCCAGAC |
| GAPDH-Forward | CTGAGGACCAGGTTGTCTCC |
| GAPDH-Reverse | GCCTCTCTTGCTCAGTGTCC |
| STAT5-Forward | CGCTGGACTCCATGCTTCTC |
| STAT5-Reverse | GACGTGGGCTCCTTACACTGA |
| PU.1-Forward | CCCCTGGAGGTGTCTGATGGAG |
| PU.I-Reverse | CTTCTTGCGGTTGCCCTTCTGGAT |
| EMSA Probes | |
| κE3’PU.1-STAT5 | CTTTGAGGAACTGAAAACAGAACCT |
| SV40 PU.1 | TGAAATAACCTCTGAAAGAGGAACTTGGTTAGGTA |
| κE3’ Enhancer Conventional PCR ChIP Primers | |
| Forward | ATATTATCAGCCAACATCTTCTAC |
| Reverse | CATTGGGACTGGGGACTAACC |
| κE3’ Enhancer QPCR ChIP Primers | |
| Forward | TGGTGACTGGCCTGAGAAGAT |
| Reverse | GAGGAGGATGGGAGCGAAA |
Electrophoretic mobility shift assays
EMSA was performed with 0.1 ng of labeled DNA probe (10,000 cpm) in a 20 µl reaction mixture containing 2 µg poly(dI-dC), 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 5% glycerol, and protein made by in vitro translation. PU.1 and STAT5 proteins were made by in vitro transcription and translation using a coupled transcription and translation kit (Promega) with T3 or T7 RNA polymerase and equimolar amounts of PU.1 and STAT5 were used for EMSA. The PU.1:STAT5 binding site from the κE3’ enhancer and the SV40 enhancer-derived PU.1 probe are shown in Table I. Samples were electrophoresed on 4% polyacrylamide gels in 6.7 mM Tris-HCl (pH 7.5), 3.3 mM NaAc, and 1 mM EDTA.
Western Blots
ES cells or primary bone marrow cells grown in either 10ng/ml or 0.1 ng/ml IL-7 were harvested in RIPA buffer (50mM Tris-HCl, pH 7.4; 1% NP-40; 0.25% sodium deoxycholate; 150mM NaCl; 1mM EDTA; 1mM PMSF) and subjected to the western blot procedure with antibodies against PU.1 (Santa Cruz Biotechnologies SC-352), STAT5 (Santa Cruz Biotechnologies SC-835), or IRF4 (32), using anti-alpha-Tubulin (Sigma) as a loading control. For immunoprecipitation of primary bone marrow IL-7 cultures, 1 mg of RIPA lysate containing phosphatase inhibitor cocktail (Sigma-Aldrich P-5726) was incubated with anti-STAT5 antibody overnight at 4°C and immune complexes were captured with Protein A-agarose beads. After three washes with lysis buffer, bound protein was eluted with 2× Laemmli buffer, and subjected to the western blot procedure using anti-phosphotyrosine antibody 4G10 (Millipore 05–321). Blots were incubated with donkey anti-rabbit antibody (Amersham) and signals were detected with Super Signal West Pico Chemiluminescent Substrate (Pierce).
Chromatin Immunoprecipitation (ChIP) Assays
ChIP was carried out as previously described (40) using antibodies against STAT-5 (sc-835X; Santa Cruz Biotechnologies), E2A (sc-349; Santa Cruz Biotechnologies), IRF4 (32), and PU.1 (sc-352; Santa Cruz Biotechnologies). Quantitative PCR analyses were performed by real time PCR using a Roche Lightcycler 1.5 and repeated at least three times. Primers for the κE3’ enhancer are shown in Table I. Relative fold increases were calculated based on preimmune ChIP sample quantitative PCR values. Relative enrichments for each region are presented as the percentage of input. In some experiments imatinib (Gleevec; Novartis) prepared as a 10mM stock solution (1000× in water and 10mM HCl) was added to 38B9 cells to a final concentration of 10 µM and cells were harvested 12 hours later for ChIP evaluation.
Vκ-Jκ rearrangement
Bone marrow samples from either wild-type C57BL/6 or constitutively active STAT5b-ca mice (21) were subjected to purification on 8A MACS columns (Miltenyi Biotech) followed by FACS to isolate pre-B cells. DNA isolated from primary pre-B cells was subjected to PCR using of 5ng, 10ng, or 20ng of input DNA. Degenerate primers for various Vκ genes and a reverse primer from the Jκ-Cκ intron were used for PCR (41). Amplified products were evaluated by southern blot using a 3.9 kb Xba1-BamH1 DNA probe containing the kappa intron and constant region sequences. Primers that amplify the Cκ constant region were used as a loading control for normalization. Quantitation of rearrangement was performed using a Storm Scan phosphoimager and Image Quant GE software. Animal studies were reviewed and approved by the IACUC at the University of Pennsylvania.
Results
STAT5 represses activity of the κE3’ enhancer
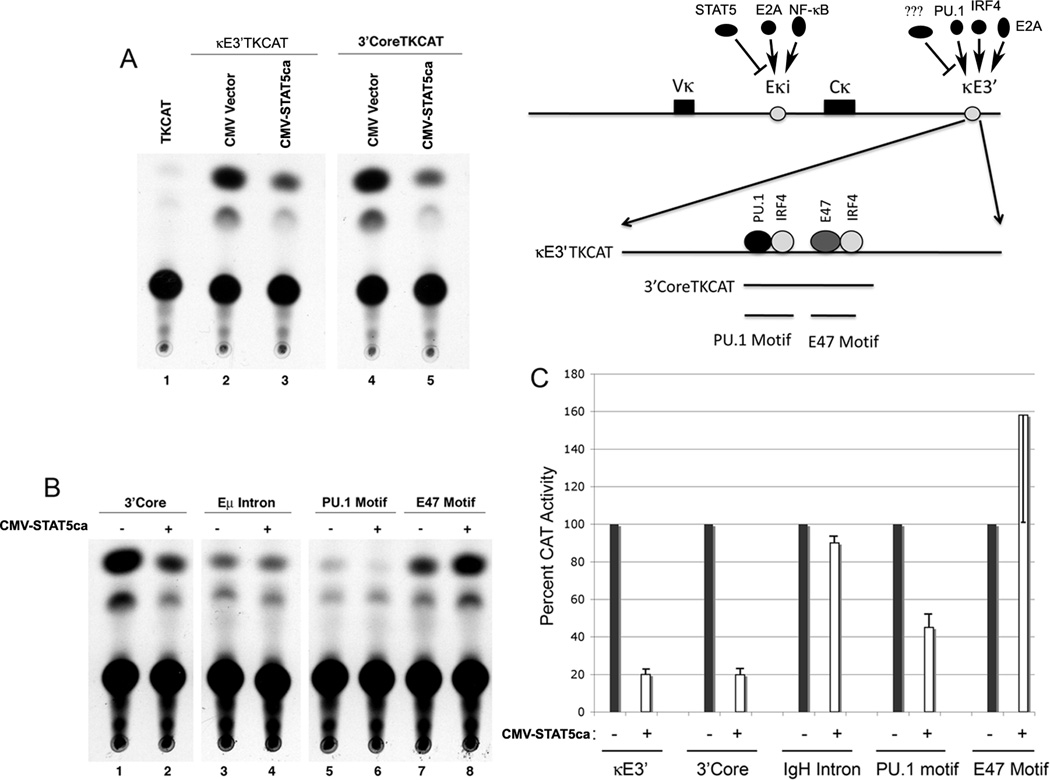
To explore whether STAT5 can inhibit κE3’ enhancer activity, we first performed transient expression assays in S194 plasmacytoma cells. A construct containing the entire κE3’ enhancer (Fig. 1A) linked to the thymidine kinase promoter driving the chloramphenicol acetyltransferase gene was transfected with empty CMV vector, or a vector expressing a constitutively active form of STAT5a (CMV-STAT5ca). Interestingly, expression of STAT5ca reduced enhancer activity five fold to 20% activity observed with empty CMV vector (Fig. 1A, lanes 1–3; 1C). A reporter plasmid containing 129bp that constitutes the κE3’ enhancer core sequence was similarly repressed (Fig. 1A, lanes 4 and 5, 1C). This repression was specific because the IgH Eκ intron enhancer was not repressed by STAT5 expression (Fig. 1B, lanes 1–4). To determine the target site of repression in the κE3’ enhancer we explored the impact of STAT5 expression on two functional κE3’ enhancer motifs, the PU.1 motif and the E2A (E47) motif (Fig. 1A). Whereas STAT5 repressed activity of the PU.1 binding site motif, there was no repression of the E2A motif (Fig. 1B, lanes 5–8). In fact, activity of this element increased, perhaps due to squelching of histone deacetylases by STAT5. These data are quantitatively shown in Fig. 1C. Thus, we conclude that STAT5 expression can lead to repression of the κE3’ enhancer, and this repression may target the PU.1 binding region of the enhancer.
FIGURE 1. STAT5 represses Ig κE3’ enhancer activity through the PU.1 binding site.
(A) The κE3’ enhancer is repressed by STAT5 expression. A map of the Igκ locus showing factors that regulate intron (Eκi) and 3’ (κE3’) enhancers is shown on the right. The intron enhancer is positively regulated by E2A and NF-κB, but is negatively regulated by STAT5. The κE3’enhancer is positively regulated by PU.1, IRF4, and E2A. Existence of a negative regulatory factor (???) is unknown. S194 plasmacytoma cells were transfected with either TKCAT, Eκ3’TKCAT or 3’CoreTKCAT reporter plasmids and either empty CMV or constitutively active STAT5a encoding expression plasmids. Cell extracts were evaluated for chloramphenicol acetyltransferase activity using 14C-chloramphenicol and products were evaluated by thin layer chromatography. A representative of three experiments is shown. (B and C) STAT5 repression occurs through the PU.1 binding region. Reporter plasmids shown above the lanes were co-transfected with either empty expression vector (−) or constitutively active STAT5a expressing vector (+). Cell extracts were evaluated for chloramphenicol acetyltransferase activity using 14C-chloramphenicol and products were evaluated by thin layer chromatography. Data is quantitated in panel C from three experiments and error bars show standard deviation of the mean.
STAT5 binds to the κE3’ enhancer and can displace PU.1 binding in vitro
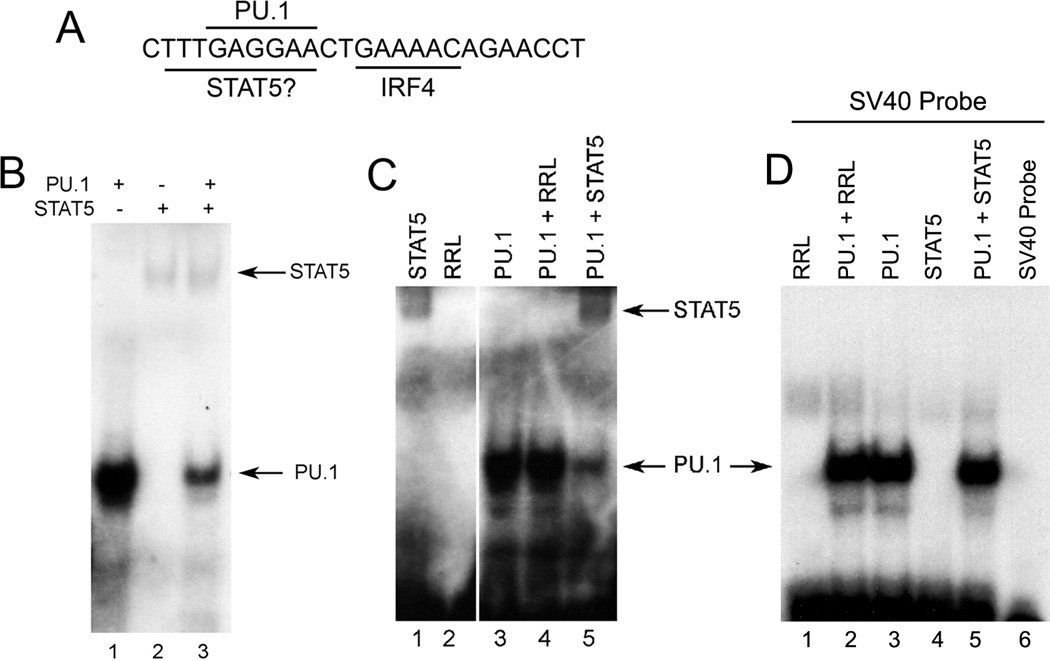
Repression of κE3’ enhancer activity by STAT5 could be due to direct DNA binding or by an indirect mechanism. We scanned the Igκ 3’ enhancer sequence for potential STAT5 binding sites and found one site that contained a 5 out of 6 match to the specific nucleotide sequences of the STAT5 consensus binding motif (TTCNNNGAA). Interestingly, this site overlaps the PU.1 binding site (GAGGAA) (Fig. 2A) that is crucial for maximal enhancer activity (16). We used this DNA sequence (shown in Fig. 2A) as a probe in electrophoretic mobility shift assays (EMSA) as we have used this probe extensively in the past to measure PU.1 DNA binding (16, 31, 32). This probe bound to recombinant PU.1 as well as to recombinant STAT5 prepared by in vitro translation (Fig. 2B, lanes 1 and 2). Mixture of the two proteins led to a dramatic reduction in PU.1 binding (Fig. 2B, lane 3). This loss was specifically due to STAT5 because incubation with unprogrammed rabbit reticulocyte lysate had no impact on PU.1 DNA binding (Fig. 2C). In addition, we used a probe derived from the SV40 enhancer that binds PU.1 but not STAT5. As expected, this probe bound PU.1 but addition of STAT5 had no impact on PU.1 binding (Fig. 2D). Thus, we conclude that STAT5 can bind to the κE3’ enhancer via a site that overlaps the PU.1 binding site.
FIGURE 2. PU.1 and STAT5 compete for in vitro binding to the κE3’ enhancer.
(A) The κE3’ enhancer PU.1 and IRF4 binding sites and the potential STAT5 binding site are shown. (B and C) EMSA was performed with the κE3’ enhancer DNA probe shown in panel A with equimolar amounts of PU.1 and STAT5 proteins prepared by in vitro transcription and translation. Addition of STAT5 resulted in reduced PU.1 DNA binding. RRL: unprogrammed rabbit reticulocyte lysate. A representative of three experiments is shown. (D) STAT5 has no impact on PU.1 binding to the SV40 enhancer PU.1 binding site. EMSA was performed with the PU.1 binding site from the SV40 enhancer which lacks an overlapping STAT5 binding site. A representative of two experiments is shown.
STAT5 and PU.1 compete for DNA binding in vivo
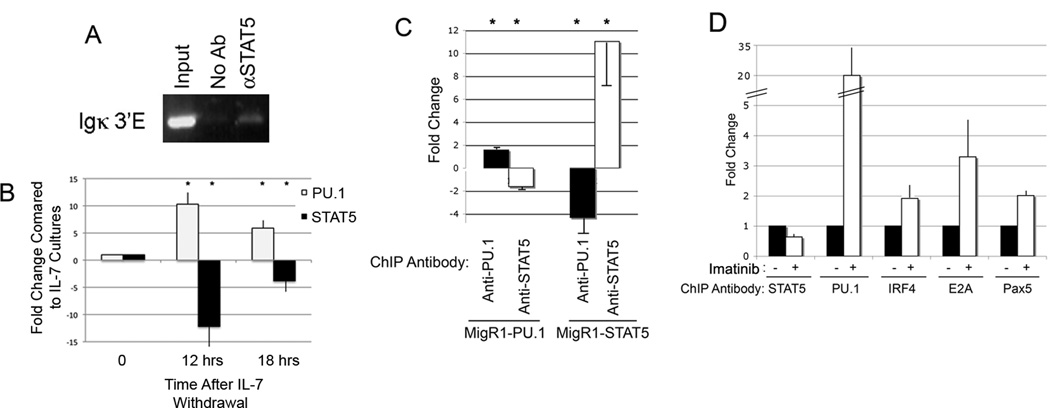
To determine whether STAT5 can bind to the κE3’ enhancer in vivo, we performed chromatin immunoprecipitation (ChIP) studies using chromatin isolated from primary bone marrow IL7 cultures that are mainly composed of primary pro-B cells (90% of bone marrow cells became B220+IgM−CD43+ after one week of culture). Indeed, we found that STAT5 bound to the κE3’ enhancer in vivo (Fig. 3A). We then wanted to determine whether reduction of STAT5 binding would increase PU.1 occupancy at the 3’ enhancer. Removal of IL7 from the medium of bone marrow cultures results in reduced STAT5 activation and DNA binding (42). We found that removal of IL-7 from the media increased PU.1 expression whereas STAT5 overall levels remained unchanged (Supplemental Fig. 1), consistent with transcript levels in pro-B and pre-B cells (http://www.immgen.org). However, the level of phosphorylated STAT5 dropped upon IL-7 removal (Supplemental Fig. 1). This is consistent with the well characterized phosphorylation of STAT5 in response to IL7 signaling (21, 43–45). To determine relative STAT5 and PU.1 occupancy at the κE3’ enhancer in IL7-high and IL7-low cultures, we performed ChIP assays with chromatin isolated from bone marrow cultures grown in 10 ng/ml IL7 as compared to 0.5ng/ml IL7. As expected, relative STAT5 binding decreased dramatically upon reduction of IL7 in the culture media (Fig. 3B). Strikingly, this was accompanied by a concomitant dramatic increase in PU.1 DNA occupancy suggesting competitive DNA binding between STAT5 and PU.1 (Fig. 3B). The increased PU.1 binding is consistent with our previous work showing elevated PU.1 binding in primary pre-B cells compared to primary pro-B cells (38). To determine whether increased PU.1 or STAT5 levels would alter DNA binding by the reciprocal protein, we transduced 38B9 pro-B cells with retroviruses expressing either PU.1 or STAT5 (MigR1-PU.1 or MigR1-STAT5, respectively). As expected, transduction with MigR1-PU.1 resulted in elevated PU.1 occupancy at the κE3’ enhancer (Fig. 3C). In addition, increased PU.1 binding was accompanied by a concomitant reduction in STAT5 occupancy (Fig. 3C). Similarly, transduction with MigR1-STAT5 resulted in increased STAT5 occupancy and reduced PU.1 DNA binding (Fig. 3C).
FIGURE 3. PU.1 and STAT5 compete with one another for binding to the κE3’ enhancer in vivo.
(A) STAT5 can bind to the κE3’ enhancer in pro-B cells. ChIP assays were performed on bone marrow cells grown in IL7 (90% of bone marrow cells were B220+IgM−CD43+) using antibody to STAT5 and DNA primers to the κE3’ enhancer. The κE3’ enhancer primers used for conventional PCR lie approximately 150 bp upstream and downstream of the 132 bp enhancer core defined previously (17) and yield a product of 448bp. A representative of four experiments is shown. (B) Withdrawal of IL7 from bone marrow cultures and loss of STAT5 activation results in reduced STAT5 DNA binding and elevated PU.1 binding as measured by ChIP assay. The fold change in DNA binding is shown relative to the level of binding before IL7 withdrawal. Asterisks show statistically significant differences where p<0.05 of three independent experiments. (C) Elevated PU.1 or STAT5 levels lead to concomitant reduction in STAT5 and PU.1 binding, respectively. 38B9 pro-B cells were transduced with control MigR1 retroviral vector, or vectors expressing PU.1 or STAT5. ChIP assays were performed with PU.1 or STAT5 antibodies and fold change of PU.1 and STAT5 binding was calculated relative to vector control cells. Asterisks show statistically significant differences where p<0.05 from 3 experiments. (D) Imatinib reduces STAT5 occupancy leading to increased PU.1 DNA binding and increased binding by IRF4, E2A, and Pax5. 38B9 cells were treated with imatinib then processed for ChIP with the indicated antibodies and primers to the κE3’ enhancer. The κE3’ enhancer primers used for real-time PCR flank the 132 bp enhancer core and yield a product of 172 bp. Asterisks show statistically significant differences where p<0.05 from three experiments.
To further test our model of competitive STAT5-PU.1 DNA binding, we used the small molecular compound, imatinib, which inhibits the ABL kinase in pro-B cells resulting in reduced STAT5 activation (46). Treatment of cells with imatinib should therefore lead to reduced STAT5 occupancy and elevated PU.1 binding at the κE3’ enhancer. Imatinib treatment also leads to increased IRF4 expression (13) and this could also increase PU.1 binding due to cooperative PU.1-IRF4 interactions. Indeed, treatment of 38B9 cells with imatinib resulted in lowered occupancy by STAT5 (Fig. 3D). As expected, reduced STAT5 binding and elevated IRF4 expression was accompanied by elevated PU.1 DNA binding (Fig. 3D). We previously showed that PU.1 DNA binding at the κE3’ enhancer functions to nucleate binding of other enhancer factors (16, 31, 39). Therefore, we assayed whether loss of STAT5 and increased PU.1 occupancy would lead to increased binding by κE3’ enhancer binding factors IRF4, E2A, and Pax5. As anticipated, elevated PU.1 binding lead to increased occupancy by IRF4, E2A, and Pax5 (Fig. 3D).
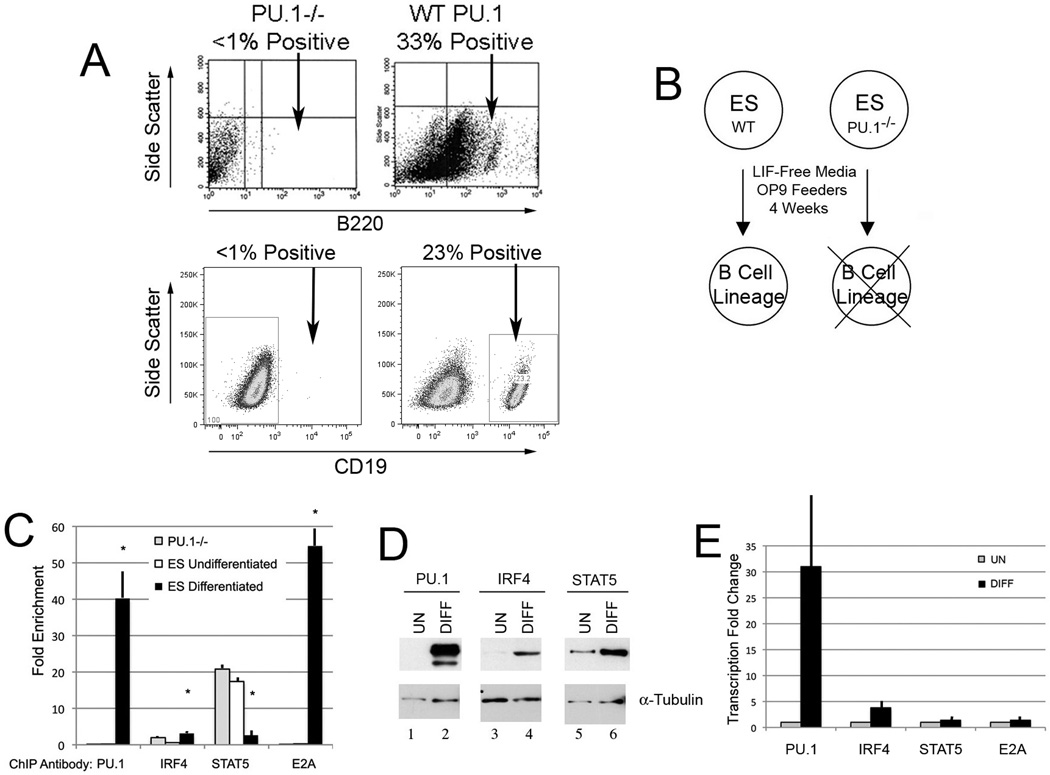
Finally, we tested competitive PU.1/STAT5 binding in an additional B cell developmental system. Embryonic stem (ES) cells can be induced to differentiate into B lineage cells if grown in leukemia inhibitory factor (LIF) negative media on OP9 feeder cells (47–49). We tested whether PU.1-null ES cells (50, 51) would differentiate into B lineage cells under the same conditions and found that they were unable to yield B lineage cells (Fig. 4A and B). We placed wild-type ES cells and PU.1-null ES cells in B cell differentiation media (LIF−) on OP9 feeders for four weeks and harvested cells to prepare chromatin for ChIP assays. When comparing undifferentiated ES cells to cells induced to undergo B cell differentiation, we observed a dramatic difference in protein occupancy at the κE3’ enhancer (Fig. 4C). In undifferentiated ES cells, STAT5 binding was elevated at the κE3’ enhancer whereas there was minimal occupancy by PU.1, IRF4, or E2A. On the contrary, after induction of B cell differentiation, PU.1 binding increased dramatically while STAT5 binding was reduced (Fig. 4C). Increase PU.1 binding was accompanied by elevated IRF4 and E2A protein occupancy. The lost STAT5 binding was not the result of reduced STAT5 expression, but correlated with increased PU.1 and IRF4 expression (Fig. 4D and E). As predicted, PU.1-null ES cells maintained high STAT5 binding at the κE3’ enhancer with little PU.1, IRF4, or E2A binding (Fig. 4C). Therefore, we conclude that PU.1 and STAT5 compete for DNA binding to the κE3’ enhancer and their occupancy correlates with the level of binding by other enhancer binding factors.
FIGURE 4. Competitive PU.1/STAT5 binding in an ES cell-B cell development system.
(A) Wild-type or PU.1-null (PU.1−/−) ES cells were grown in LIF− media on OP9 feeders. After 4 weeks inculture, one fourth to third of cells from the wild-type sample had differentiated into B220 positive or CD19 positive B lineage cells, whereas PU.1-null cells remained B220 negative. (B) Diagram of B cell differentiation capacity of wild-type and PU.1-null ES cells. (C) ChIP assays using chromatin from undifferentiated ES, or wild-type ES cells in differentiation medium, or PU.1-null cells in differentiation medium. Fold enrichment of DNA is plotted using the antibodies shown in the panel. Asterisks show p<0.05 comparing differentiated levels to undifferentiated and PU.1-null samples from three experiments. (D) Western blots were performed on either undifferentiated or differentiated wild-type ES cells using the antibodies shown above the lanes. Blots probed with α-tubulin antibody served as loading controls. (E) Quantitative real-time PCR was performed for PU.1, IRF4, STAT5, and E2A transcripts using RNA isolated from undifferentiated or differentiated wild-type ES cells. Data is shown from three experiments.
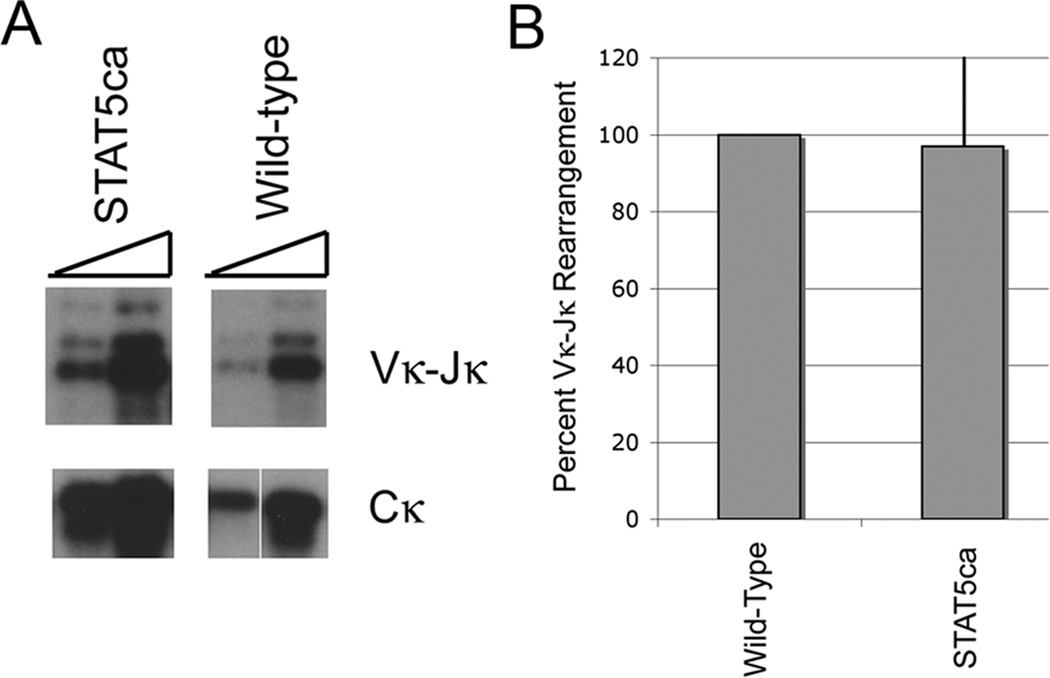
Igκ rearrangement is not inhibited in STAT5ca transgenic animals
The repressive effect of STAT5 on Igκ locus enhancer activity and germline transcription might lead to reduced Igκ rearrangement in mice expressing a constitutively active STAT5 transgene. Alternatively, increased IRF4 expression in pre-B cells might override repressive effects of STAT5, as was previously observed in bone marrow cultures even in the presence of high levels of IL7 (14). To explore this issue, we assessed Igκ rearrangement in pre-B cells isolated from either wild-type mice, or mice containing a constitutively active STAT5 transgene (44). Isolated DNA was subjected to PCR to detect Vκ-Jκ rearrangement using degenerate primers for various Vκ sequences in association with a primer downstream of the Jκ region. DNA was normalized to the level of product observed with Cκ primers. We observed no statistically significant difference between the level of Vκ-Jκ rearrangement in wild-type compared to STAT5ca transgenic pre-B samples (Fig. 5A and B). In this case, consistent with previous work ex vivo (14), elevated IRF4 expression in pre-B cells appears to be sufficient to overcome the repressive effects of STAT5, perhaps because IRF4 synergizes with both PU.1 and E2A bound to the κE3’ enhancer.
FIGURE 5. Impact of STAT5 on Igκ rearrangement.
(A and B) Expression of a constitutively active STAT5b transgene (STAT5b-ca) does not reduce the level of endogenous Igκ V-J rearrangement. Pre-B cells were isolated from wild-type or STAT5b-ca transgenic mice and isolated DNA was assayed for Igκ rearrangement using degenerate primers followed by southern blot. No reduction in Igκ V-J rearrangement was observed in the transgenic animals compared to wild-type. Data are shown from four experiments.
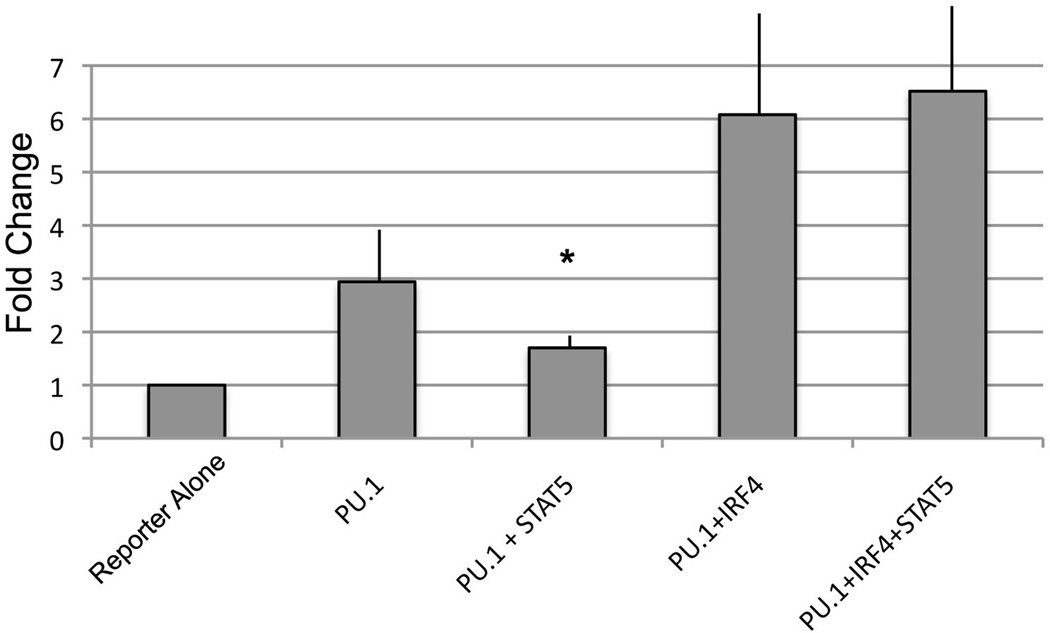
IRF4 expression can overcome STAT5 repression
To test the idea that IRF4 can override STAT5 repression, we performed transfection assays with the PU.1 and STAT5 responsive reporter used in Fig. 1. This reporter binds to PU.1, STAT5, and IRF4 and can therefore be used to determine the ability of IRF4 to counteract STAT5 repression. As expected, in transient expression assays PU1 expression resulted in elevated reporter gene transcription, and addition of STAT5 repressed this PU.1-mediated transactivation (Fig. 6). Cotransfection of PU.1 and IRF4 expressing plasmids resulted in augmented transactivation above that seen with PU.1 alone. However, this augmented PU.1-IRF4 transactivation was no longer sensitive to repression by STAT5 (Fig. 6). Thus, STAT5 repression of PU.1 mediated transactivation can be overcome by IRF4 expression.
FIGURE 6. IRF4 can overcome STAT5 repression of the κE3’ enhancer.
NI3T3 cells were transfected with the κE3’ enhancer PU.1 motif reporter plasmid and the CMV expression plasmids indicated in the figure. The asterisk indicates significant (p< 0. 05) repression of PU.1 by STAT5 when transfected without IRF4. In the presence of IRF4, STAT5 failed to repress transactivation. Error bars show standard deviation of the mean from three experiments.
Discussion
Temporal differences in Ig heavy chain and light chain rearrangement indicate that a mechanism must exist to maintain the Igκ locus in an inactive state in pro-B cells when the heavy chain locus undergoes rearrangement. Attenuation of IL7 signaling and increased E2A and IRF4 activities in pre-B cells correspond with activation of the Igκ locus (13–15, 29). Prior work indicated that the Igκ intron enhancer is a target of increased E2A DNA binding and histone acetylation in response to IL7 attenuation (14). Regulation of Igκ intron enhancer activity alone, however, is insufficient to regulate Igκ locus rearrangement as the κE3’ enhancer can compensate for loss of Igκ intron enhancer activity to drive Vκ-Jκ rearrangement (10, 12). During the pro-B to pre-B cell transition, elevated IRF4 expression causes an increase in H4 acetylation at the κE3’ enhancer (13, 14), but we now show that a mechanism also exists that represses κE3’ enhancer activity in a developmentally controlled fashion. We found that the κE3’ enhancer is also part of the IL7-STAT5 regulatory mechanism. Thus, developmental control of the κE3’ enhancer involves both active stimulation by PU.1, IRF4 and E2A, and repression by STAT5.
We previously showed that the κE3’ enhancer can bind to a variety of transcription factors including c-fos, c-jun, PU.1, IRF4, Pax5, and E2A that regulate enhancer activity (16, 31, 39, 52, 53). Binding by PU.1 is particularly important because PU.1 binding is needed for recruitment of IRF4 to an adjacent binding site (16, 31, 33–35). Deletion of the PU.1 binding site leads not only to lost PU.1 DNA binding, but lost DNA binding by IRF4 as well, and concomitant reduced enhancer activity (39). Thus, regulation of this binding site has profound impact on κE3’ enhancer activity.
We show here that PU.1 binding to the κE3’ enhancer can be modulated by competitive binding by STAT5 to an overlapping site, and this correlates with occupancy by other enhancer binding factors, suggesting that STAT5 binding represents a molecular switch that can control occupancy at the κE3’ enhancer. This, coupled with changes in IRF4 expression and E2A activity can result in precise regulation of κE3’ enhancer function. This mechanism at the κE3’ enhancer is not absolutely essential for regulating Igκ rearrangement. B cell development and Igκ rearrangement can occur in the absence of PU.1 (54–56), although other studies suggest that reduced PU.1 activity can affect B cell development (57). Presumably, in the absence of PU.1 either κE3’ enhancer activity is still sufficiently robust to support Igκ rearrangement, or the process is completely dependent upon the intron enhancer.
Based on the repressive role of STAT5 in κE3’ enhancer activity, we hypothesized that a STAT5ca transgene might reduce Igκ gene rearrangement in vivo. However, we found little difference in Igκ rearrangement when comparing wild-type and STAT5ca transgenic animals. Several explanations are possible. The STAT5b-ca transgene lacks the site required for interaction with SMRT and can only interact with this co-repressor if part of a heterodimer with wild-type endogenous STAT5, potentially leading to a weaker interaction. STAT5b-ca mice do contain fewer pre-B cells relative to pro-B cells, and fewer immature B cells relative to pre-B cells. Thus, these mice may select for cells that rearrange Igκ and differentiate to the immature stage, and cells that fail to rearrange may never differentiate to the pre-B or immature B cell stages thus masking the rearrangement defect. However, our transfection results with IRF4 suggest an additional explanation. Whereas STAT5 was able to repress PU.1 transactivation function, it failed to repress transactivation if IRF4 was present with PU.1. Thus, elevated IRF4 levels in pre-B cells leading to cooperative DNA binding with PU.1 and E2A to the composite PU.1/IRF4 and IRF4/E2A motifs in the κE3’ enhancer is sufficient to override the repressive effect of STAT5.
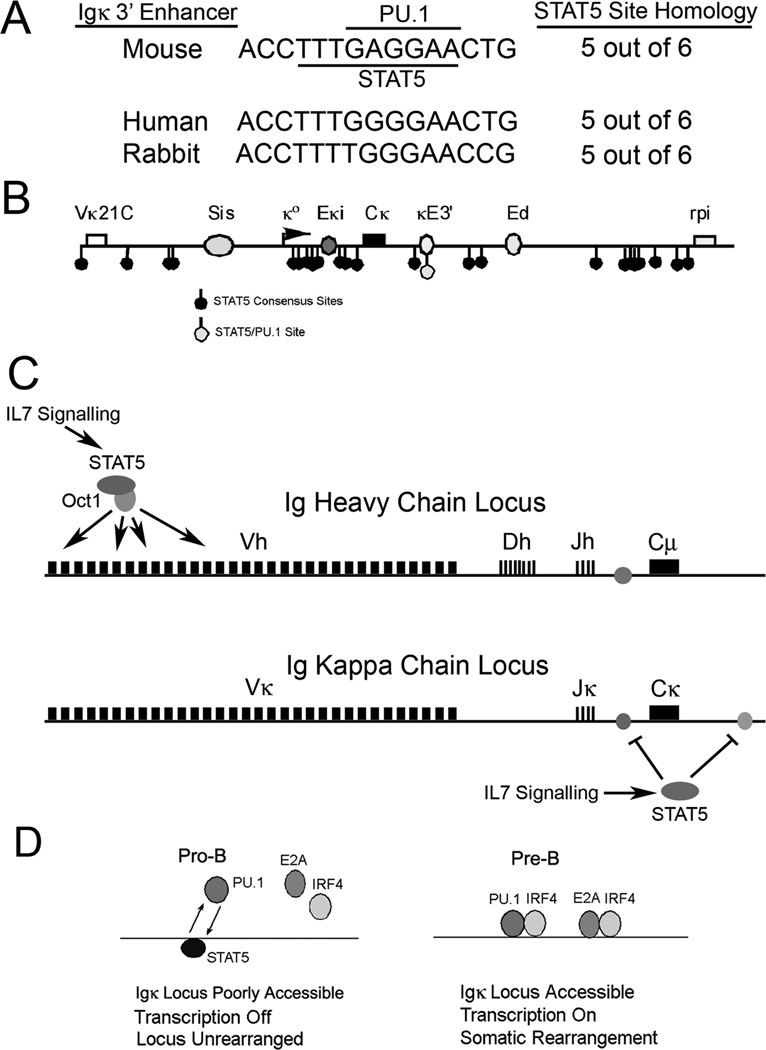
The composite PU.1/STAT5 site is conserved in the Igκ 3’ enhancer sequences from human, mouse, and rabbit (Fig. 7A) indicating likely conservation of mechanisms in these species. While numerous consensus STAT5 sites exist within the mouse Igκ locus, only the site within the κE3’ enhancer is a composite PU.1/STAT5 motif (Fig. 7B). Based on the work of others and our own, we propose that in pro-B cells, IL7 signaling activates STAT5 leading to recruitment of STAT5 to distal Vh promoters via Oct1 (19). This results in enhanced rearrangement of distal Vh genes. Simultaneously, STAT5 binds to the Igκ locus leading to reduced Igκ intron and κE3’ enhancer activities (Fig. 7C), the former by regulating E2A binding (14, 15, 29), and the later by displacing PU.1 from the κE3’ enhancer (Fig. 7D). At the pre-B cell stage when IL7 signaling is attenuated and IRF4 levels increase, the intron enhancer is activated by E2A occupancy. Simultaneously, STAT5 binding to the κE3’ enhancer is reduced and PU.1 binding increases leading to recruitment of IRF4 to the κE3’ enhancer and subsequent enhancer activation (Fig. 7D). Consistent with this model, we previously observed elevated PU.1 and IRF4 binding to the κE3’ enhancer in pre-B cells compared to pro-B cells (38). We also previously showed that E2A and IRF4 cooperatively bind to the κE3’ enhancer leading to enhancer synergy (36, 37). Thus, coupled with intron enhancer activation, both PU.1/IRF4 and E2A/IRF4 motifs in the κE3’ enhancer will yield high activity in pre-B cells thus contributing to enhancer activation and Igκ rearrangement (Fig. 7D).
FIGURE 7. Model for STAT5 inhibition of Igκ locus accessibility.
(A) The overlapping PU.1 and STAT5 binding sites are conserved in κE3’ enhancer sequences from mouse, human and rabbit. (B) Location of putative STAT5 sites within the Igκ locus. Numerous STAT5 consensus sequences are observed within the Igκ locus, but only a single site within the κE3’ enhancer overlaps with a PU.1 binding site. Locations of Vκ21C and rpi genes, and Sis, Eκi, κE3’, and Ed enhancers are indicated. (C) IL7 signaling activates IgH distal V gene rearrangement through STAT5 while simultaneously reducing Igκ locus accessibility through inhibition of Igκ intron and κE3’ enhancers. (D) Model for STAT5 function at the κE3’ enhancer during the pro-B to pre-B cell transition. In pro-B cells IL7 signaling activates STAT5 which competes with PU.1 for binding to the enhancer leading to low IRF4 and E2A occupancy and low κE3’ enhancer activity. In pre-B cells, loss of IL7 signaling and increased IRF4 expression enables PU.1, IRF4 and E2A to bind to the κE3’ enhancer leading to increased activity, and stimulation of Igκ rearrangement.
In total, our results provide a more complete explanation for how the Igκ locus is regulated through both enhancers involved in Igκ rearrangement, and provide a new model for locus activity involving a competitive STAT5-PU.1 DNA binding mechanism.
Supplementary Material
Acknowledgements
We thank T. Kitamura for the STAT5a cDNA clone. We thank E.Scott for PU.1-null ES cells and J.C. Zuniga-Pfucker for OP9 cells.
Footnotes
This work was supported by NIH grants GM071820 and GM082841 to MLA.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Ann. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 2.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv. Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury D, Sen R. Mechanisms for feedback inhibition of the immunoglobulin heavy chain locus. Curr. Opin. Immunol. 2004;16:235–240. doi: 10.1016/j.coi.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Sen R, Oltz E. Genetic and epigenetic regulation of IgH gene assembly. Curr. Opin. Immunol. 2006;18:237–242. doi: 10.1016/j.coi.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Johnson K, Angelin-Duclos C, Park S, Calame KL. Changes in histone acetylation are assoicated with differences in accessibility of Vh gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 2003;23:2438–2450. doi: 10.1128/MCB.23.7.2438-2450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the Igκ locus during B cell development. Nature Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 7.Lennon GG, Perry RP. Cmu containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5'-nontranslatable exon. Nature. 1985;318:475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- 8.Yancopoulous GD, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 9.Inlay MA, Lin T, Gao HH, Xu Y. Critical roles of the immunoglobulin intronic enhancers in maintaining the sequential rearrangement of IgH and Igκ loci. J. Exp. Med. 2006;203:1721–1732. doi: 10.1084/jem.20052310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Stoep N, Gorman JR, Alt FW. Reevaluation of 3'Eκ function in stage- and lineage-specific rearrangement and somatic hypermutation. Immunity. 1998;8:743–750. doi: 10.1016/s1074-7613(00)80579-8. [DOI] [PubMed] [Google Scholar]
- 11.Takeda S, Zou YR, Bluethmann H, Kitamura D, Muller U, Rajewsky K. Deletion of the immunoglobulin κ chain intron enhancer abolishes κ chain gene rearrangement in cis but not l chain gene rearrangement in trans. EMBO J. 1993;12:2329–2336. doi: 10.1002/j.1460-2075.1993.tb05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity. 1996;4:377–385. doi: 10.1016/s1074-7613(00)80251-4. [DOI] [PubMed] [Google Scholar]
- 13.Lazorchak AS, Schlissel MS, Zhuang Y. E2A and IRF-4/Pip promote chromatin modification and transcription of the immunoglobulin κ locus in pre-B cells. Mol. Cell. Biol. 2006;26:810–821. doi: 10.1128/MCB.26.3.810-821.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson K, Hashimshony T, Sawai CM, Pongubala JMR, Skok J, Aifantis I, Singh H. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 2008;28:1–11. doi: 10.1016/j.immuni.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nature Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pongubala JMR, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3' enhancer activity. Mol. Cell. Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pongubala JMR, Atchison ML. Functional characterization of the developmentally controlled immunoglobulin kappa 3' enhancer: Regulation by Id, a repressor of helix-loop-helix transcription factors. Mol. Cell. Biol. 1991;11:1040–1047. doi: 10.1128/mcb.11.2.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Widlak P, Zou Y, Xiao F, Oh M, Li S, Chang MY, Shay JW, Garrard WT. A recombination silencer that specifies heterochromatin positioning and Ikaros association in the immunoglobulin κ locus. Immunity. 2006;24:405–415. doi: 10.1016/j.immuni.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 19.Bertolino E, Reddy K, Medina KL, Pargana E, Ihle J, Singh H. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nature Immunol. 2005;6:836–843. doi: 10.1038/ni1226. [DOI] [PubMed] [Google Scholar]
- 20.Akira S. Functional roles of STAT family proteins: Lessons from knockout mice. Stem Cells. 1999;17:138–146. doi: 10.1002/stem.170138. [DOI] [PubMed] [Google Scholar]
- 21.Goetz CA, Harmon IR, O'Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 recepter-dependent B cell development. J. Immunol. 2004;172:4770–4778. doi: 10.4049/jimmunol.172.8.4770. [DOI] [PubMed] [Google Scholar]
- 22.Snow J, Abraham N, Ma MC, Abbey NW, Herndier B, Goldsmith MA. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood. 2002;99:95–101. doi: 10.1182/blood.v99.1.95. [DOI] [PubMed] [Google Scholar]
- 23.Sexl V, Piekorz R, Moriggl R, Rohrer J, Brown MP, Bunting KD, Rothammer K, Roussel MF, Ihle JN. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl-and bcr/abl-induced transformation are independent of Stat5. Blood. 2000;96:2277–2283. [PubMed] [Google Scholar]
- 24.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 25.Yao Z, Cui Y, Watford WT, Bream J, H, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, Hennighausen L, O'Shea JJ. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol. Cell. Biol. 2003;23:4162–4173. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima H, Brindle PK, Handa M, Ihle JN. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. EMBO J. 2001;20:6836–6844. doi: 10.1093/emboj/20.23.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M, Nie L, Kim S-H, Sun X-H. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPb. EMBO J. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H, Clark MR. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nature Immunol. 2009;10:1110–1117. doi: 10.1038/ni.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malin S, McManus S, Busslinger M. STAT5 in B cell development and leukemia. Curr. Topics Immunol. 2010;22:168–176. doi: 10.1016/j.coi.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Pongubala JMR, Van Beveren C, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 32.Perkel JM, Atchison ML. A two-step mechanism for recruitment of Pip by PU.1. J. Immunol. 1998;160:241–252. [PubMed] [Google Scholar]
- 33.Eisenbeis CF, Singh H, Storb U. PU.1 is a component of a multiprotein complex which binds an essential site in the murine immunoglobulin λ2–4 enhancer. Mol. Cell. Biol. 1993;13:6452–6461. doi: 10.1128/mcb.13.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific PU.1-dependent transcriptional activator. Genes & Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 35.Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes & Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 36.Nagulapalli S, Atchison ML. Transcription factor Pip can enhance DNA binding by E47, leading to transcriptional synergy involving multiple protein domains. Mol. Cell. Biol. 1998;18:4639–4650. doi: 10.1128/mcb.18.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagulapalli S, Goheer A, Pitt L, McIntosh LP, Atchison ML. Mechanism of E47-Pip interaction on DNA resulting in transcriptional synergy and activation of immunoglobulin germ line sterile transcripts. Mol. Cell. Biol. 2002;22:7337–7350. doi: 10.1128/MCB.22.20.7337-7350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDevit DC, Perkins L, Atchison ML, Nikolajczyk BS. The Igκ3' enhancer Is activated by gradients of chromatin accessibility and protein association. J. Immunol. 2005;174:2834–2842. doi: 10.4049/jimmunol.174.5.2834. [DOI] [PubMed] [Google Scholar]
- 39.Pongubala JMR, Atchison ML. PU.1 can participate in an active enhancer complex without its transcriptional activation domain. Proc. Natl. Acad. Sci. USA. 1997;94:127–132. doi: 10.1073/pnas.94.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai Y, Srinivasan L, Perkins L, Atchison ML. Protein acetylation regulates both PU.1 transactivation and Igκ 3' enhancer activity. J. Immunol. 2005;175:5160–5169. doi: 10.4049/jimmunol.175.8.5160. [DOI] [PubMed] [Google Scholar]
- 41.O'Brien DP, Oltz EM, Van Ness BG. Coordinate transcription and V(D)J recombination of the kappa immunoglobulin light-chain locus: NF-κB-dependent and -independent pathways of activation. Mol. Cell. Biol. 1997;17:3477–3487. doi: 10.1128/mcb.17.7.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X-L, Goetz CA, Katerndahl CDS, Sakaguchi N, Farrar MA. A Flt3- and Ras-dependent pathway primes B cell development by inducing a state of IL-7 responsiveness. J. Immunol. 2010;184:1728–1736. doi: 10.4049/jimmunol.0903023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corfe SA, Gray AP, Paige CJ. Generation and characterization of stromal cell independent IL-7 dependent B cell lines. J. Immunol. Meth. 2007;325:9–19. doi: 10.1016/j.jim.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Will WM, Aaker JD, Burchill MA, Harmon IR, O'Neil JJ, Goetz CA, Hippen KL, Farrar MA. Attenuation of IL-7 receptor signaling is not required for allelic exclusion. J. Immunol. 2006;176:3350–3355. doi: 10.4049/jimmunol.176.6.3350. [DOI] [PubMed] [Google Scholar]
- 45.Hirokawa S, Sato H, Kato I, Kudo A. EBF-regulating Pax5 transcription is enhanced by STAT5 in the early stage of B cells. Eur. J. Immunol. 2003;33:1824–1829. doi: 10.1002/eji.200323974. [DOI] [PubMed] [Google Scholar]
- 46.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nature Immunol. 2003;4:31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 47.Cho SK, Webber TD, Carlyle JR, Nakano T, Lewis SM, Zuniga-Pflucker JC. Functional characterization of B lymphocytes generated in vitro from embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1999;96:9797–9802. doi: 10.1073/pnas.96.17.9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho SK, Zuniga-Pflucker JC. Development of lymphoid lineages from embryonic stem cells in vitro. Meth. Enzymol. 2003;365:158–169. doi: 10.1016/s0076-6879(03)65011-1. [DOI] [PubMed] [Google Scholar]
- 49.Nakano T, Kodama H, Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- 50.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 51.Olson MC, Scott EW, Hack AA, Su GH, Tenen DG, Singh H, Simon MC. PU.1 is not essential for early myeloid gene expression but is required for terminal myeloid differentiation. Immunity. 1995;3:703–714. doi: 10.1016/1074-7613(95)90060-8. [DOI] [PubMed] [Google Scholar]
- 52.Pongubala JMR, Atchison ML. Activating transcription factor 1 and cyclic AMP response element modulator can modulate the activity of the immunoglobulin κ 3' enhancer. J. Biol. Chem. 1995;270:10304–10313. doi: 10.1074/jbc.270.17.10304. [DOI] [PubMed] [Google Scholar]
- 53.Maitra S, Atchison M. BSAP can repress enhancer activity by targeting PU.1 function. Mol. Cell. Biol. 2000;20:1911–1922. doi: 10.1128/mcb.20.6.1911-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schweitzer BL, DeKoter RP. Analysis of gene expression and Ig transcription in PU.1/Spi-B-deficient progenitor B cell lines. J. Immunol. 2004;172:144–154. doi: 10.4049/jimmunol.172.1.144. [DOI] [PubMed] [Google Scholar]
- 55.Polli M, Dakic A, Light A, Wu L, Tarlinton DM, Nutt SL. The development of functional B lymphocytes in conditional PU.1 knockout mice. Blood. 2005;106:2083–2090. doi: 10.1182/blood-2005-01-0283. [DOI] [PubMed] [Google Scholar]
- 56.Ye M, Ermakova O, Graf T. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J. Exp. Med. 2005;202:1411–1422. doi: 10.1084/jem.20051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Houston IB, Kamath MB, Schweitzer BL, Chlon TM, DeKoter RP. Reduction in PU.1 activity results in a block to B-cell development, abnormal myeloid proliferation, and neonatal lethality. Exp. Hematol. 2007;35:1056–1068. doi: 10.1016/j.exphem.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.