Abstract
The goal of systems biology is to access and integrate information about the parts (e.g., genes, proteins, cells) of a biological system with a view to computing and predicting the behavior of the system. The past decade haswitnessed technological revolutions in the capacity to make high throughput measurements about the behavior of genes, proteins, and cells. Such technologies are widely used in biological research and in medicine, such as toward prognosis and therapy response prediction in cancer patients. More recently, systems biology is being applied to vaccinology, with the goal of: (1) understanding the mechanisms by which vaccines stimulate protective immunity, and (2) predicting the immunogenicity or efficacy of vaccines. Here, we review the recent advances in this area, and highlight the biological and computational challenges posed.
INTRODUCTION
The primary goal of vaccination is to confer long-term protection in populations susceptible to infection and disease. Despite their tremendous success, most vaccines were designed empirically, with limited understanding of the immunological mechanisms by which they mediate protection.1 Unfortunately, such approaches have been less successful with respect to vaccine development against global pandemics such as HIV, malaria, and tuberculosis. A lack of understanding of the relevant correlates of protection against such pathogens, as well as the unique mechanisms by which they have evolved to evade host immunity, pose formidable challenges to vaccine development.2
Recent advances in nanotechnology, robotics, optics, and electronics have revolutionized the way scientists study the molecules of living organisms. High-throughput techniques commonly used by many laboratories can now assess entire genomes, sets of transcripts (transcriptome), proteins (proteome), and metabolites (metabolome) of cells and tissues. Systems biology utilizes and integrates the large amount of data generated by these techniques in order to describe the complex interactions between all parts of a biological system, with the ultimate goal of predicting the behavior of the system.3,4 The application of systems biology to vaccinology could potentially fill many fundamental gaps in our knowledge concerning the mechanisms of action of the current successful vaccines, as well as enable the rational design and testing of novel vaccines.5 Here, we review the emerging field of systems vaccinology and describe key bioinformatic analyses as well as computational and biological challenges that permeate this new field.
SYSTEMS VACCINOLOGY
Much has been learned about vaccines using conventional immunology and reductionistic methods such as cellular and molecular biology. Though these methods are invaluable, they have limited power when it comes to analyzing many features of a system in parallel. Instead, they focus on parts (i.e., specific cell subsets, proteins or genes) of the immune system. Systems biology approaches, however, can characterize complex vaccine responses by analyzing the system as a whole. These approaches include a wide range of technologies, such as DNA microarrays,6–9 modern mass spectrometry,10–12 antibody microarrays,13,14 and pathogen proteome microarrays15 (Figure 1).
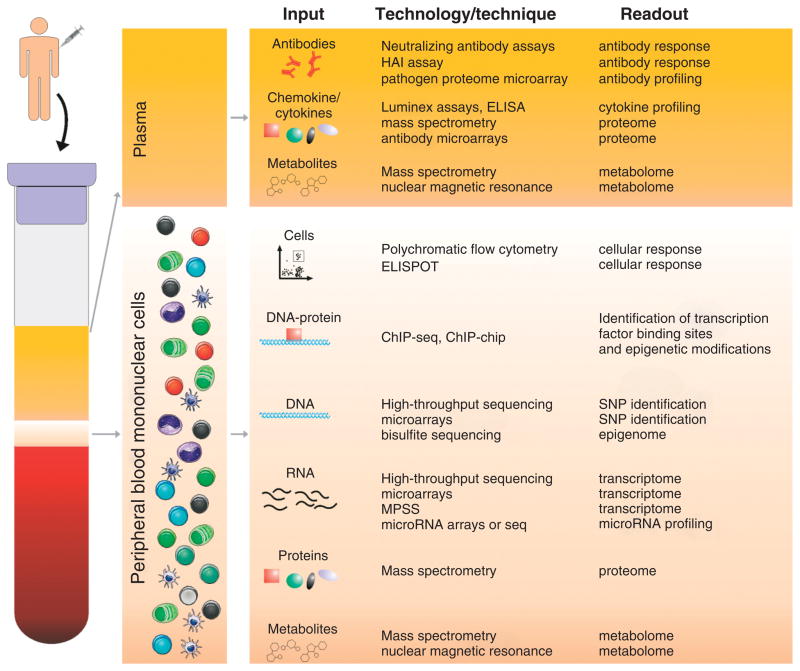
FIGURE 1.
The tool kit of Systems Vaccinologists. Low and high-throughput technologies that can be used by systems vaccinologists to investigate the mechanisms of vaccines. Antibody response, cytokine profiling, metabolome, and proteome of vaccinees can be assessed in the blood plasma. T and B cell responses can be determined by flow cytometry and ELISPOT in the fraction containing peripheral blood mononuclear cells. Different separation methods can be used to collect from these cells the DNA, RNA, proteins, or metabolites which can be screened by high-throughput techinologies, such as microarrays and mass spectrometry. ELISA, enzyme-linked immunosorbent assay; ELISPOT, enzyme-linked immunosorbent spot; HAI, haemagglutination inhibition; ChIP-seq, chromatin-immunoprecipitation followed by high-throughput sequencing; ChIP-chip, chromatin-immunoprecipitation followed by hybridization to microarrays; MPSS, Massively Parallel Signature Sequencing; SNP, single-nucleotide polymorphism.
Systems vaccinology has recently emerged as an interdisciplinary field that combines systems-wide measurements, networks and predictive modeling in the context of vaccinology.5 It aggregates the key properties of systems biology,3,4 which are: perturbation of the system (e.g., vaccine administration), monitoring responses at the systems level (e.g., blood transcriptomics, serum proteomics or metabolomics), data integration, network modeling and development of predictive rules that describe the system’s response to individual perturbations (e.g., prediction of vaccine responses). Therefore, its application does not rely simply on data collection from high-throughput techniques, but also on the integration of different types of data (Figure 1) in order to generate hypotheses and new insights that may explain the mechanism of vaccines.
Two major goals of systems vaccinology can be reached using microarray data from vaccine studies. Identifying genes and pathways whose expression is significantly altered in most of the vaccinees provides the immunological ‘flavor’ of the host response to the vaccine, and may help to unravel the molecular mechanisms of action of such vaccines. The second goal tries to identify signatures that correlate or predict various measurements of vaccine immunogenicity and or protection. This is especially relevant in situations where vaccination induces suboptimal immunity in certain populations, such as the elderly or immunocompromised individuals. In such cases, it is important to prospectively identify individuals in whom the vaccination did not confer protective immunity, and who are thus likely to be at risk of infection. In addition, the ability to prospectively predict vaccine efficacy would be useful in rapidly evaluating immunogenicity of vaccines in clinical trials.
YELLOW FEVER VACCINE AS A PROOF OF CONCEPT
The first examples of the application of systems biology to understanding vaccine induced immune responses came from studies with the yellow fever vaccine YF-17D, one of the most successful vaccines ever developed.16 YF-17D is a live-attenuated virus vaccine which is known to confer over 90% efficacy and with which a single dose confers protection for up to 40 years.16 Neutralizing antibodies are the primary correlate of protection against infection with yellow fever virus,17 but recent evidence shows that CD8+ cytotoxic T cells may also play an important role in this protection.18 Using mouse model and in vitro experiments, we showed that YF-17D activates multiple Toll-like receptors (TLR2, 7, 8, and 9) in dendritic cells to elicit innate and adaptive immune responses.19 To further explore on a systems level the mechanisms by which YF-17D induces protective immunity, we6 and others7 have independently applied systems biology approaches to profile the blood of subjects vaccinated with YF-17D. Although different microarray platforms were used by each laboratory, the gene expression signatures induced by YF-17D were largely consistent between both studies. A prominent antiviral response, activated type I interferon pathway, was manifested during the two weeks following vaccination.6,7 Genes involved in the complement pathway and inflammasome were also induced, as were a broad spectrum of regulatory genes involved in innate sensing, including TLR7, RIG-I, MDA5, and LGP2, and signaling, such as JUN, STAT1, IRF7, and RNF36.
One additional goal of our study was to identify early gene expression signatures that predict adaptive immune outcomes (neutralizing antibody and CD8+ T cell responses). Principal component analysis of 839 genes whose expression correlated with CD8+ T cell responses divided the subjects of the first YF17D vaccine trial into two groups: a low responder group comprised of subjects with less than 3% activated CD38+ HLA-DR+ CD8+ T cells after immunization, and a high responder group, comprised of those with 3% or more activated cells.6 The challenge was then to identify which of these genes, if any, could also correctly classify a second and independent YF17D vaccine trial. Two different classification methods were tested using the first trial as a training set and the second trial as a validation set. Classification to the nearest centroid, ClaNC20 and discriminant analysis via mixed integer programming, DAMIP,21 correctly classified 80 and 90% of the vaccinees, respectively. However, DAMIP program achieved this precision using gene sets comprised of a few genes (the so called ‘predictive rules’).6 DAMIP analysis was also performed to predict B cell neutralizing antibody responses.
New mechanistic insights are emerging from these studies.6,7 A gene that ismost frequently found in the predictive signatures of the CD8+ T cell response against YF-17D is EIF2AK4 (also known as GCN2). In response to certain types of cellular stresses (e.g. amino acid starvation), GCN2 is phosphorylated and in turn phosphorylates eIF2a, which results in the shutdown of housekeeping mRNA genes and the formation of stress granules.22–24 In fact, it was observed that phosphorylation of eIF2a and formation of stress granules are induced by YF-17D in human PBMC and baby hamster kidney cells.6 Furthermore, EIF2AK4 knockout mice, when vaccinated with YF-17D, have impaired CD8+ T cell responses (Khan et al., manuscript in preparation). In the case of antibody responses, TNFRSF17, a key gene in the signatures that predict the magnitude of the neutralizing antibody response, encodes for a protein, (also known as BCMA), which is a member of the BAFF/BLyS family of receptors, known to promote B cell survival and enhance responses to BCR and TLR signaling.25 Although further experiments are needed, our study suggests that this gene may play a critical role in mounting antibody responses after YF17D and influenza immunizations.26 Overall, these results provide the first successful demonstration of how systems vaccinology can predict vaccine immunogenicity and help bring about new biological insights regarding the mechanism of vaccines.
Recently, our group has extended this approach to other types of vaccines, including the inactivated seasonal influenza vaccine.26 Thus, we compared the global expression changes that occur after vaccination with a live attenuated virus vaccine (LAIV) and the trivalent inactivated virus vaccine (TIV).26 Like YF17D vaccine, LAIV induced a robust interferon response in the blood of vaccinees, while TIV induced a plasma B cell response signature. Since the immune responses induced by vaccination against seasonal influenza in adults are recall responses, due to prior vaccination or infection, it was not clear that systems biological approaches could result in the identification of molecular signatures that predict recall responses. However, our work demonstrates that the approach was successful not only in predicting recall responses, but also in providing new mechanistic insights about vaccine induced immunity.26
GATHERING AND MINING THE DATA
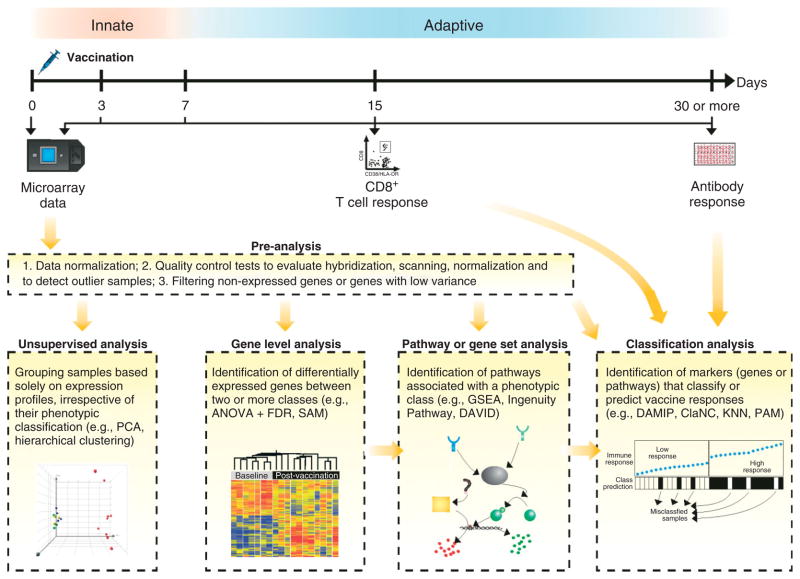
Current high-throughput technologies allow scientists to measure the expression of all genes transcribed by cells of the immune system during and after perturbation (e.g., vaccination or infection). Highthroughput sequencing (RNA-seq)27 is a powerful approach capable of measuring the transcriptome of cells with great genome coverage. A typical RNA-Seq experiment converts RNAs into a library of cDNAs, which, after sequencing adaptors are added to each end of a cDNA fragment, are subjected to one of the deep sequencing technologies. The copy number of initial RNA species can then be counted digitally.28 In addition, sequence variations can be directly detected, opening up new opportunities in immunology, where individual sequences are informative, for example, sequences of HLA, TCR, and MHC classes. As the cost of deep sequencing is going down quickly, this approach will potentially replace the hybridization based technologies such as DNA microarrays. However, the costs of running samples from many vaccinees collected at several time points are still prohibitive to most labs. For this reason, and due to the flexibility and the availability of many easy-to-use analytical software and programs, microarrays are still the preferable high-throughput screening tool for most systems vaccinologists29 (Table 1). In Figure 2 we illustrate how microarray analyses can be applied to study and predict vaccine responses.
TABLE 1.
RNA Array Hybridization versus RNA-seq
| DNA Array Hybridization | RNA-seq | |
|---|---|---|
| Platforms | Affymetrix, Aglient, Illumina, Nimblegen (Roche), custom arrays | Roche 454, Illumina Solexa, Life SOLiD, Helicos BioSciences, Pacific Biosciences |
| Dynamic range1 | Up to a few hundred fold | >8000 fold |
| Required amount of RNA | Low (RNA amplification) | Low |
| Gene sequences required | Yes | No |
| Readout | Analog hybridization signal | Digital count, single base resolution |
| Cost | Microarrays are dropping to the $100 range. | High setup cost; cost can be split by tagged multiplexing. |
| Infrastructure | Microarray facilities are widely available | New technologies in rapid development |
On the basis of Wang et al.28
FIGURE 2.
Identifying molecular signatures of vaccination. Multiple microarray analyses can be performed to identify genes and pathways whose expression is modulated in response to vaccination or to identify signatures that classify subjects based on their immune responses.
Although microarray experiments can provide a valuable ‘snapshot’ of the transcriptional levels of all genes inside the cells, the interactions among those genes cannot be captured if microarrays were solely used to generate lists of genes or pathways associated to a phenotypic class. It is precisely the functional relationships among genes and also among proteins and metabolites that may help us to better understand biological processes and cellular responses. Molecular network modeling aims to study the structure, function and dynamics of these interactions,30 and may have a profound impact on data interpretation, maximizing knowledge discovery if incorporated into study design.31–34
Transcriptomic Analyses at Genes and Pathways Levels
Reports of blood transcriptomics have offered important insights about the contribution of genes and pathways in the immune response to vaccines.6–8 A central question in such studies is: which genes change in expression after immunization? Although multiple ways of answering this can be considered, the identification of differentially expressed genes ultimately depends on several factors. It begins with an appropriate study design that takes into account the sample size and the immune status of the vaccinees (i.e., age, overall health, or treatments) as well as the immunogenicity of the vaccine. Next, the transcriptomic data should be correctly normalized and processed (removing non-informative genes using variance or expression filters to reduce noise in the analysis). Finally, the statistical method and the degree of stringency applied will determine the list of genes that are affected by vaccination. Changes in the expression of each gene can be tested individually using univariate statistics such as t-test and ANOVA; however, as microarrays contain thousands of genes, correction for multiple tests, for example, false discovery rate35,36 becomes necessary. Using this approach we identified genes differentially expressed post-YF17D vaccination.6 Ramilo et al. used nonparametric univariate Mann–Whitney test to rank genes on the basis of their ability to discriminate patients with distinct acute infections.37 SAM,38 a popular software program in this field, borrows the variation between genes to assess the overall sample variation, which is valuable for small sample sizes, but less important for large ones.39
Next, gene set and pathway analyses can be performed to put results in a biological context and even to gain statistical power. The goal is to evaluate the association of a phenotypic class (before or after vaccination for example) with predefined gene sets, for example, genes belonging to the same biochemical pathways or being co-expressed in previous experiments.40 GSEA,41 DAVID (http://david.abcc.ncifcrf.gov/), and GS-SAM42 are able to perform these analyses using different statistical tests or gene sets. Some of them utilize gene sets curated and deposited in databases such as KEGG,43 Reactome,44 MsigDB,41 Gene Ontology,45 and Nature/NCI pathway database (http://pid.nci.nih.gov). Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com) was performed by us6 and others7 to reveal the interacting network of many interferon and antiviral genes affected by vaccination with YF-17D. Another example of gene set analysis is found in the study of Vahey et al.8 Transcriptome analysis of blood from subjects before and after the last dose of adjuvanted RTS, S malaria vaccine revealed 32 genes in the proteasome degradation pathway associated with protection against challenge (which was determined by experimental challenge with mosquito-borne Plasmodium falciparum malaria).8 These genes were involved with the efficient processing of the MHC peptides, suggesting a potential role of the vaccine in conferring major histocompatibility complex class 1–mediated protection.8
Another angle to be explored toward systems vaccinology consists of analyzing the transcriptomic changes of blood leukocytes during chronic or acute infections. Very few infection diseases, like measles, mumps, and rubella (MMR), are able to ‘act’ as a vaccine and induce lifelong immunity. Despite this fact, transcriptomic studies have shown that most pathogens induce changes in transcriptional networks commonly affected by vaccination with live-attenuated strains.46–48 For example, the list of the 22 classifier genes up-regulated in influenza A infected patients compared to those infected with bacteria, reported by Ramilo et al.,37 contains nine genes (RSAD2, SPATS2L, IFI27, IFI44, OAS1, ISG15, DDX60, XAF1, and MX1) whose expression is also altered in the blood of YF17D vaccinees6 and 10 genes (IFI35, OAS1, BST2, MX1, SON, TRIM14, APOBEC3C, DDX60, XAF1, and MLEC) whose expression is also altered by vaccination with the live attenuated influenza vaccine.26 A more detailed analysis can be found by the work of Jenner and Young.46 They compiled published transcriptional-profiling data involving 77 different host–pathogen interactions and found that infection with some viruses results in a more pronounced upregulation of the immune response compared with infection by attenuated virus strains, perhaps explaining their pathogenic nature.46
Although extremely useful, gene set analyses still have intrinsic limitations.28 The definition of gene sets in databases like Gene Ontology45 and KEGG43 is limited by human curation. However, genes belonging to the same ontological category or pathway may not be coregulated at the transcriptional level. Even if they are, the results will only emphasize the wellestablished knowledge in a self perpetuating cycle. On the other hand, programs like GSEA41 utilize gene sets defined as genes coregulated in specific experimental conditions but fail to provide ‘meaningful’ canonical pathways. In any case, it is improbable that predefined gene sets will capture all the molecular players related to a biological process or function of specific cell populations in a given experiment. Careful statistical analysis alone cannot replace human intuition in interpreting genome-wide expression data.
Machine Learning Tools for Predicting Immunogenicity and Efficacy
Microarray data sets can be used to generate predictive models of vaccine protection or immunogenicity. In general, the MicroArray Quality Control (MAQC) consortium recommends following 3 basic steps to generate such models: (1) appropriate study design (e.g., sample size, adequate normalization, feature selection, and classification methods); (2) internal validation (e.g., bootstrap or cross-validation); and (3) external validation by using an independent and completely separated test set.49 Following these lines, gene expression values can be directly compared to measurements of vaccine immunogenicity and or protection using Pearson correlation or Spearman’s rank correlation coefficients. Once signatures that correlate to vaccine immunogenicity are delineated, the next task is to identify the minimum set of genes, or a network, that is capable of predicting the response to the vaccine. Our group used DAMIP program21 to generate ‘predictive rules’ (sets of two or three genes) that discriminate vaccinees with low and high neutralizing antibody or CD8+ T cell responses.6 The accuracy of these predictive rules were at least 80%, as determined in two independent vaccine trials.6 This accuracy level may be particularly relevant for when diseases are live-threatening such as Yellow Fever, or even influenza on immunocompromised subjects.
Berry et al.50 have performed blood transcriptomic analyses on asymptomatic individuals infected with Mycobacterium tuberculosis and have identified a 393 transcript signature, containing many type I interferon signaling genes, which correlates with radiological extent of tuberculosis disease. In this work, k-nearest neighbors (KNN) algorithm was used to predict subjects with active TB compared to healthy and asymptomatic ones.50 Vahey et al.8 used PAM51 to predict the subjects immunized with malaria vaccine that would be protected after challenge with the pathogen. Additional class predictionmethods applied to genome-wide expression data include LDA,52 ClaNC,20 Support Vector Machines53 and Random Forest.54 Some of these classification methods can be combined with feature selection to identify the best set of genes that predict distinct phenotypic classes.
An extensive evaluation of microarray-based predictive models was recently performed by MAQC consortium.49 Six microarray data sets separated in 13 preclinical and clinical endpoints (e.g., breast cancer, lung or liver toxicity in rodents, neuroblastoma) were analyzed by 36 independent teams to generate predictive models. Their major findings were that the model performance depends largely on the endpoint; there are clear differences in proficiency between teams, and that different approaches can generate models with similar performance.49 These demonstrate that given similar data analysis expertise and approaches, the endpoint, in our case the immune parameter induced by vaccination (e.g., correlate of immunogenicity) will directly affect the performance of the models. Therefore, it is expected that for some vaccines predictive models cannot be reliably generated.
Discovering Gene Networks in Immune Response
Signals are transformed into cellular responses through biomolecules that interact with each other forming complex networks. Transcriptional regulatory networks are the circuits leading to alterations of the expression level of genes. Analyzing the transcription factor binding sites (TFBSs) in the promoters of target genes and correlations between expression of regulator and target genes help us to infer how the transcriptional network is wired.55 Our group analyzed the TFBSs in the promoter region of genes differentially expressed in the blood of subjects vaccinated with Yellow Fever YF17D vaccine using TOUCAN (http://homes.esat.kuleuven.be/~saerts/software/toucan.php).6 Interferon-stimulated response element (ISRE), interferon regulatory factor 7 (IRF7) binding site and sterol regulatory element–binding protein 1 (SREBF1) binding site were TFBSs found to have statistically over-represented frequencies, which support the closely interacting network of 50 interferon and antiviral genes described in our paper.6
An elegant example of network modeling was performed by Amit et al.56 They have reconstructed, in an unbiased way, the transcriptional networks involved in mouse dendritic cell responses to five pathogen-derived components, including the adjuvant CpG, a synthetic single-stranded DNA that binds to toll-like receptor 9. Additionally, a model that associated regulators to their targets was constructed by knocking down the expression of most of candidate regulators in combination with gene expression profiling.56 By systematically perturbing the system and measuring its effect on the expression of gene targets, they found that two key programs related to inflammatory response and antiviral response were controlled by two corresponding regulatory arms, which were integrated into a core network of 24 regulators that balance specific and shared responses through dominant activation and cross-inhibition.56
Inflammation is an essential process of innate immune system. Litvak et al.57 investigated the gene-regulatory networks that govern inflammatory responses using, as model macrophages stimulated by LPS, a Toll-like receptor 4 ligand known to induce inflammatory cytokines such as IL-6 and TNF-α.58 By combining time course microarray analysis, motif-scanning, chromatin immunoprecipitation experiments, and simulation models, they identified a gene circuit, consisted of NF-kB, ATF3, and C/EBPδ that discriminates between transient and persistent LPS-dependent stimulation. These findings can help us to explain the molecular mechanism by which macrophages distinguish real from perceived threats.
Andrea Califano’s lab has spearheaded a series of studies of reconstructing the networks in B cell development and regulation, employing heavily the computational inference of transcriptional regulation.59–61 In general, gene network reconstruction requires data of adequate amount and resolution. This implies that for systems vaccinology, much has yet to be learned through experimental models other than clinical vaccine trials.
Connecting the Networks by Interactome
Although a description of the component parts (mRNA, proteins, metabolites) represent a first step, it is critical to understand the nature of the interactions between these parts, the so-called interactome. Interactions between proteins can be identified by high-throughput assays, such as yeast two-hybrid screens and co-affinity purification coupled to mass spectrometry.62 Interactions between transcription factors and their respective target genes can be determined by chromatin immunoprecipitation followed by microarray identification (ChIP-chip) or sequencing (ChIP-seq).30 Finally, siRNA screening and data mining of genome-wide experiments are effective strategies for mapping genetic networks.30 This information offers a new opportunity to fill the gap between genotype and expression data, and to discover new mechanisms beyond the canonical pathways.
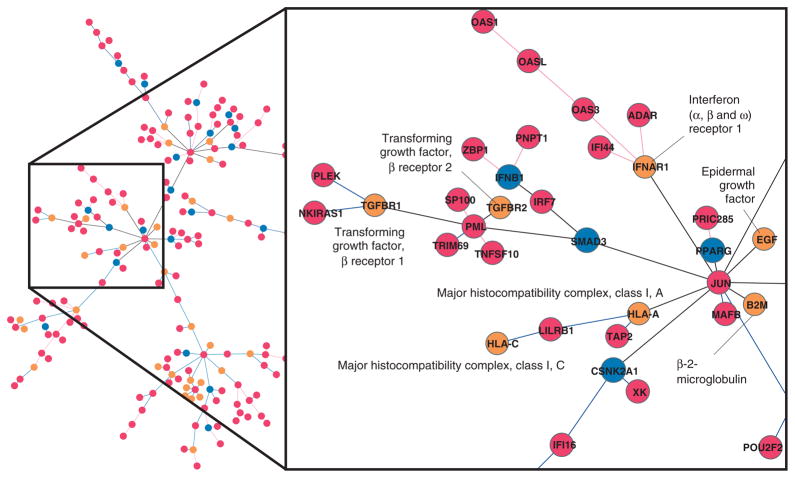
A preliminary transcriptomic analysis of YF17D vaccination demonstrates how protein-protein interaction data can be used to reveal ‘hidden’ components of gene networks involved in immune responses (Figure 3). In general, highly stringent blood transcriptomic analyses yield relatively few differentially expressed genes (red circles in Figure 3). Although related to immune responses, this small number of genes may not be sufficient for a proper gene network analysis. Molecular interactions obtained from the interactome data can be used to fill in the gaps of gene networks (light brown circles in Figure 3). This approach can reveal additional genes of interest, and may generate novel mechanistic hypotheses.
FIGURE 3.
Yellow fever vaccine response network. Microarray analysis of blood of subjects before and after YF17D vaccination generated a list of differentially expressed genes (labeled in red). Genes were subsequently added to the network based on their interactions with those differentially expressed genes and their connections in the interactome. Among these additional genes, the yellow ones still have p-value < 0.05 in the transcriptomic analysis; Genes in blue have no statistical significance in the transcriptomic analysis, but may be required for gene regulation.
Notwithstanding, it is important to mention that interactome data are largely incomplete and full of false positive interactions.62 Additionally, true interactions found in one physiological condition or cell type, may not be true for another biological context. Only with more research we will be able to navigate through these caveats and maximize its benefits.
CHALLENGES AND PERSPECTIVES
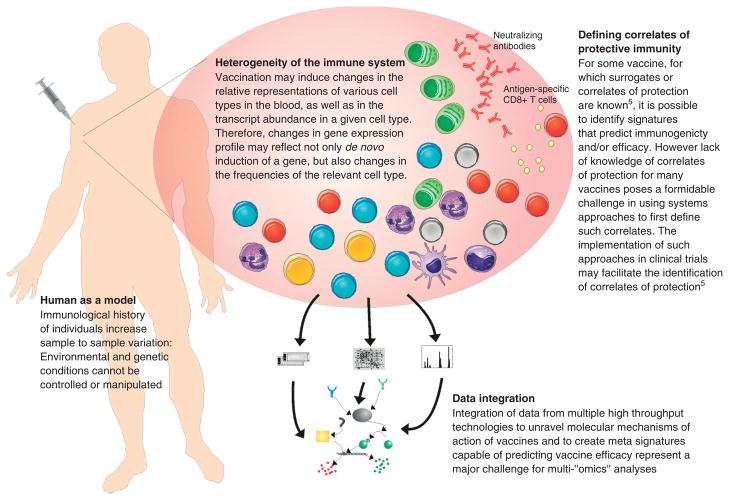
Systems vaccinology faces similar challenges as other systems biology fields in dealing with the huge amount of information derived from high-throughput techniques and the complexity of the system. It is estimated that the 3 billion base pair human genome contains 30,000 genes, of which due to post-transcriptional and post-translational modifications could potentially generate, respectively, 180,000 mRNA molecules and 1,800,000 proteins.63 Added to this diversity, thousands noncoding RNAs (microRNAs, piRNAs, snoRNAs, tRNAs, etc.) present in the human genome and the virtually infinite possible interactions between these different types of features. Studying the dynamics of such complex system, at any level (genes, proteins, metabolites, interactions) seems to be an extraordinarily daunting task. The challenge of systems biology challenge relies in extracting meaningful information from this sea of data. Computers can be used to aid this process, and to integrate and perform analyses and rigorous statistical tests in order to select relevant features (e.g., differentially expressed genes, predictors or networks). However, human knowledge and experimental validation are extremely important factors in the meaningful interpretation of results, and in the creation and validation of imaginative hypotheses. Figure 4 summarizes some of the challenges faced by systems vaccinologists.
FIGURE 4.
Challenges of systems vaccinology.
Human as a Model Organism
Host immunity is not only affected by genetics but also a rich immunological history in its lifespan.64 Therefore, the magnitude of immune response conferred by vaccination depends not only on its composition but also on the state of the host immune system. Previous exposition to pathogens, history of vaccinations, immune senescence (i.e., the age of the vaccinee), chronic or acute diseases and use of medication, in combination with the frequently ‘noisy’ data inherent to high-throughput experiments, contribute to a very high individual variation. Carefully designed systems vaccinology studies try to overcome this intrinsic variation by imposing strict pre-requisites for the volunteers (age range, health condition, prevaccination history); measuring immune parameters at baseline (i.e., before vaccination) and by including the number of vaccinees required to meet a certain statistical significance. Additionally, it is advisable to validate the results on an independently collected set of subjects (e.g., volunteers from a different vaccine season).
Another important aspect of systems vaccinology is the ethical and technological limitations of using human as a model. Systems biology studies aim to develop complex predictive and testable models by performing iterative perturbation and monitoring responses at the systems level. This can be relatively easy to achieve by using model organisms such as yeast or sea urchin embryos. In the context of human studies, however, environmental and genetic conditions cannot be controlled or manipulated. In such cases, once a signature is obtained in a human model, subsequent iterative perturbations must be done in animal models (including mice and monkeys). Alternatively, one could design a series of small human trials, in which there is, within limits, variations on the initial perturbation (e.g., addition of an adjuvant, switching the pathogen strain of a vaccine).
Integrating Multiple Data Types
Integration of multiple layers of information, derived from distinct ‘omics’ analyses (genomics, transcriptomics, proteomics, etc.) provides a better understanding of the functional principles and dynamics of cellular systems.65 Monitoring the levels of not only genes but also proteins and metabolites on blood of vaccinees will therefore provide a bigger picture of the intricate and complex mechanisms of vaccines. However, many technical, computational and biological issues should still be overcome or accounted before multi‘omics’ analyses generate discrete and testable biological hypotheses.65
Heterogeneity of the System
An additional layer of complexity can be applied to systems vaccinology. The immune system itself is comprised by a myriad of cells that are phenotypically and functionally distinct. Vaccination may induce changes in the frequencies of specific cell types and also in the transcript abundance inside the cells. Blood transcriptomics will therefore measure relative changes of both cellular and transcript abundance, making the results hard to interpret.66 It has been attempted to deconvolute microarray signals to individual cell types.67,68 However, cell types themselves are not unambiguously defined, and flow cytometry data and microarray expression were shown to be poorly correlated.69
Chaussabel et al.70 have used transcriptional modules formed by genes coordinately expressed in multiple blood transcriptomic studies, to classify disease activity. As some of these modules were enriched in genes transcribed by plasma B cells, cytotoxic T cells or neutrophils, they could also be used to indicate changes in the frequencies of cell subsets in the blood. We are using an alternative approach which consists in identifying genes that are highly expressed in one specific cell type. Using a compendium of public microarray data, we identified the gene signatures of most peripheral blood mononuclear cells (i.e., Monocytes, myeloid dendritic cells, plasmacytoid dendritic cells, B cells, T cells, and Natural killer cells). We then compared these immune signatures to the genes identified in our blood transcriptomics analysis in order to find significant enrichment of specific cell types.26
Prediction of Vaccine Immunogenicity
The identification of well-defined correlates of protection is one of the biggest challenges of vaccine development. For most vaccines, the humoral immunity is considered to be the correlate of protection71; however, for other vaccines the magnitude of antibody titers may not be the only, or even the best, correlate of protection.5 In such cases, one or more components of the immune response (e.g., innate or antigen-specific T cell responses) may represent correlates of protection or immunogenicity.
In our previous reviews, we proposed the development of a ‘vaccine chip’ for evaluating vaccine efficacy.5,16 We postulated that vaccines with similar correlates of protection may share similar molecular signatures. To be broadly useful, the vaccine chip should accommodate predictors for different molecular signatures, each reflecting the different arms of vaccine-induced immune responses (e.g., Th2 T cell responses or macrophage activation).5 We envision that such a chip would be useful to rapid evaluate the strength, type, duration, and quality of protective immune responses stimulated by many vaccines.5
Prediction of vaccine immunogenicity or protection is a promising application of systems vaccinology. However, identification of reliable predictors will probably require extensive validation in many clinical trials. Cancer genomics is a good example of our future challenge. Hundreds of studies have used microarray data to predict cancer diagnosis or progression; however, there is only one microarray-based test, approved by FDA, for diagnosis and prognosis of breast cancer.72 Since experimental measurements are usually taken at very specific and limited conditions, the robustness of predictors derived therein is a primary concern.73
CONCLUSION
The application of systems biological methods to vaccinology is beginning to offer insights about the mechanisms by which vaccines stimulate immune responses, and provide strategies for predicting the immunogenicity of vaccines. However, major computational and biological challenges remain to be addressed for the fruitful integration of systems approaches to vaccinology. We hope that the coming decade will provide solutions to these challenges.
References
- 1.Plotkin SA. Vaccines: the fourth century. Clin Vaccine Immunol. 2009;16:1709–1719. doi: 10.1128/CVI.00290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: implications for vaccine development. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 3.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 4.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 5.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J ExpMed. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vahey MT, Wang Z, Kester KE, Cummings J, Heppner DG, Jr, Nau ME, Ofori-Anyinam O, Cohen J, Coche T, Ballou WR, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS, S malaria vaccine. J Infect Dis. 2010;201:580–589. doi: 10.1086/650310. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Pineres AJ, Hildesheim A, Dodd L, Kemp TJ, Yang J, Fullmer B, Harro C, Lowy DR, Lempicki RA, Pinto LA. Gene expression patterns induced by HPV-16 L1 virus-like particles in leukocytes from vaccine recipients. J Immunol. 2009;182:1706–1729. doi: 10.4049/jimmunol.182.3.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chahroudi A, Garber DA, Reeves P, Liu L, Kalman D, Feinberg MB. Differences and similarities in viral life cycle progression and host cell physiology after infection of human dendritic cells with modified vaccinia virus Ankara and vaccinia virus. J Virol. 2006;80:8469–8481. doi: 10.1128/JVI.02749-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams JN, Skipp PJ, O’Connor CD, Christodoulides M, Heckels JE. Immunoproteomic analysis of the development of natural immunity in subjects colonized by Neisseria meningitidis reveals potential vaccine candidates. Infect Immun. 2009;77:5080–5089. doi: 10.1128/IAI.00701-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vytvytska O, Nagy E, Bluggel M, Meyer HE, Kurzbauer R, Huber LA, Klade CS. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics. 2002;2:580–590. doi: 10.1002/1615-9861(200205)2:5<580::AID-PROT580>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 13.Reif DM, Motsinger-Reif AA, McKinney BA, Rock MT, Crowe JE, Jr, Moore JH. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun. 2009;10:112–119. doi: 10.1038/gene.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies DH, Molina DM, Wrammert J, Miller J, Hirst S, Mu Y, Pablo J, Unal B, Nakajima-Sasaki R, Liang X, et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics. 2007;7:1678–1686. doi: 10.1002/pmic.200600926. [DOI] [PubMed] [Google Scholar]
- 16.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 17.Reinhardt B, Jaspert R, Niedrig M, Kostner C, L’Age-Stehr J. Development of viremia and humoral and cellular parameters of immune activation after vaccination with yellow fever virus strain 17D: a model of human flavivirus infection. J Med Virol. 1998;56:159–167. doi: 10.1002/(sici)1096-9071(199810)56:2<159::aid-jmv10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 18.Miller JD, van der Most RG, Akondy RS, Glidewell JT, Albott S, Masopust D, Murali-Krishna K, Mahar PL, Edupuganti S, Lalor S, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J ExpMed. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dabney AR. Classification of microarrays to nearest centroids. Bioinformatics. 2005;21:4148–4154. doi: 10.1093/bioinformatics/bti681. [DOI] [PubMed] [Google Scholar]
- 21.Lee EK. Large-scale optimization-based classification models inmedicine and biology. AnnBiomedEng. 2007;35:1095–1109. doi: 10.1007/s10439-007-9317-7. [DOI] [PubMed] [Google Scholar]
- 22.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 23.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 24.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 25.Woodland RT, Schmidt MR, Thompson CB. BLyS and B cell homeostasis. Semin Immunol. 2006;18:318–326. doi: 10.1016/j.smim.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–795. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhiman N, Smith DI, Poland GA. Next-generation sequencing: a transformative tool for vaccinology. Expert Rev Vaccines. 2009;8:963–967. doi: 10.1586/erv.09.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haining WN, Wherry EJ. Integrating genomic signatures for immunologic discovery. Immunity. 2010;32:152–161. doi: 10.1016/j.immuni.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Han JDJ. Understanding biological functions through molecular networks. Cell Res. 2008;18:224–237. doi: 10.1038/cr.2008.16. [DOI] [PubMed] [Google Scholar]
- 31.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 32.Hyduke DR, Palsson BO. Towards genome-scale signalling-network reconstructions. Nat Rev Genet. 2010;11:297–307. doi: 10.1038/nrg2750. [DOI] [PubMed] [Google Scholar]
- 33.Russell RB, Aloy P. Targeting and tinkering with interaction networks. Nat Chem Biol. 2008;4:666–673. doi: 10.1038/nchembio.119. [DOI] [PubMed] [Google Scholar]
- 34.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 36.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 40.Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y. Gene-set analysis and reduction. Brief Bioinform. 2009;10:24–34. doi: 10.1093/bib/bbn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genomewide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y. Improving gene set analysis of microarray data by SAM-GS. BMC Bioinform. 2007;8:242. doi: 10.1186/1471-2105-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 47.Shingai M, Ebihara T, Begum NA, Kato A, Honma T, Matsumoto K, Saito H, Ogura H, Matsumoto M, Seya T. Differential type I IFN-inducing abilities of wildtype versus vaccine strains of measles virus. J Immunol. 2007;179:6123–6133. doi: 10.4049/jimmunol.179.9.6123. [DOI] [PubMed] [Google Scholar]
- 48.Scherer CA, Magness CL, Steiger KV, Poitinger ND, Caputo CM, Miner DG, Winokur PL, Klinzman D, McKee J, Pilar C, et al. Distinct gene expression profiles in peripheral blood mononuclear cells from patients infected with vaccinia virus, yellow fever 17D virus, or upper respiratory infections. Vaccine. 2007;25:6458–6473. doi: 10.1016/j.vaccine.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JW, Lee JB, Park M, Song SH. An extensive comparison of recent classification tools applied to microarray data. Comp Stat Data Anal. 2005;48:869–885. [Google Scholar]
- 53.Brown MPS, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci U S A. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diaz-Uriarte R, de Andres SA. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim HD, Shay T, O’Shea EK, Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325:429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBP delta in a regulatory circuit that discriminates between transient and persistent TLR4- induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manicassamy S, Pulendran B. Modulation of adaptive immunity with Toll-like receptors. Semin Immunol. 2009;21:185–193. doi: 10.1016/j.smim.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 2010;6:337. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 61.Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, Rajbhandari P, Shen Q, Nemenman I, Basso K, Margolin AA, et al. Genome-wide identification of posttranslational modulators of transcription factor activity in human B cells. Nat Biotechnol. 2009;27:829–839. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stumpf MPH, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jensen ON. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 64.Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W, Li F, Nie L. Integrating multiple ‘omics’ analysis for microbial biology: application and methodologies. Microbiology. 2010;156:287–301. doi: 10.1099/mic.0.034793-0. [DOI] [PubMed] [Google Scholar]
- 66.Chaussabel D, Pascual V, Banchereau J. Assessing the human immune system through blood transcriptomics. BMC Biol. 2010;8:84. doi: 10.1186/1741-7007-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, Hastie T, Sarwal MM, Davis MM, Butte AJ. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7:287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hyatt G, Melamed R, Park R, Seguritan R, Laplace C, Poirot L, Zucchelli S, Obst R, Matos M, Venanzi E, et al. Gene expression microarrays: glimpses of the immunological genome. Nat Immunol. 2006;7:686–691. doi: 10.1038/ni0706-686. [DOI] [PubMed] [Google Scholar]
- 70.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 72.Glas AM, Floore A, Delahaye LJ, Witteveen AT, Pover RC, Bakx N, Lahti-Domenici JS, Bruinsma TJ, Warmoes MO, Bernards R, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genom. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
FURTHER READING
- Hughey JJ, Lee TK, Covert MW. Computational modeling of mammalian signaling networks. Wiley Interdiscip Rev Syst Biol Med. 2010;2:194–209. doi: 10.1002/wsbm.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzer M, Maynard ND, Covert MW, Stelling J. Genome-scale metabolic networks. Wiley Interdiscip Rev Syst Biol Med. 2009;1:285–297. doi: 10.1002/wsbm.37. [DOI] [PubMed] [Google Scholar]