Abstract
NMDA receptors (NMDARs) are glutamate-gated ion channels present at most excitatory mammalian synapses. The four GluN2 subunits (GluN2A–D) contribute to four diheteromeric NMDAR subtypes that play divergent physiological and pathological roles. Channel properties fundamental to NMDAR function vary among subtypes. We investigated the amino acid residues responsible for variations in channel properties by creating and examining NMDARs containing mutant GluN2 subunits. Unexpectedly, we found that the NMDAR subtype specificity of three crucial channel properties, Mg2+ block, selective permeability to Ca2+, and single-channel conductance, all are controlled primarily by the residue at a single GluN2 site in the M3 transmembrane region. Mutant cycle analysis guided by molecular modeling revealed that a GluN2-GluN1 subunit interaction mediates the site’s effects. We conclude that a single GluN2 subunit residue couples with the pore-forming loop of the GluN1 subunit to create naturally-occurring variations in NMDAR properties that are critical to synaptic plasticity and learning.
INTRODUCTION
Glutamate mediates the majority of fast excitatory neurotransmission in the vertebrate brain. Glutamate receptors (GluRs) transduce signals in two ways: metabotropic GluRs signal via intracellular G proteins, whereas ionotropic GluRs (iGluRs) open intrinsic ion channels in response to agonist binding. NMDARs are glutamate- and glycine-gated iGluRs that play critical roles in spatial learning, contextual fear memory acquisition, and synaptogenesis1,2. Particularly high Ca2+ permeability and strongly voltage-dependent channel block by external Mg2+ distinguish NMDARs from other iGluRs2. Mg2+ channel block of NMDARs inhibits current influx through the majority of agonist-bound, open NMDARs at resting membrane potentials, but this block is relieved by depolarization. Thus, substantial current flow through NMDARs requires coincident presynaptic activity (glutamate release) and postsynaptic activity (depolarization to relieve Mg2+ channel block), conferring on NMDARs a coincidence detection capability critical to, for example, NMDAR-dependent long-term potentiation (LTP). LTP strengthens synapses following coincident pre- and postsynaptic activity and is necessary for many types of learning and memory1. To mediate this and other important functions, NMDARs require tight regulation of the voltage-dependent Mg2+ block that controls current flow and Ca2+ influx.
Most NMDARs are tetramers thought to be composed of two GluN1 and two GluN2 subunits2. Each GluN1 and GluN2 subunit contains an N-terminal domain, an extracellular agonist binding domain, three transmembrane regions (M1, M3, and M4), a reentrant loop (M2/p-loop) with a pore-lining segment, and an intracellular C-terminal domain2 (Fig. 1a). The p-loop, which forms the narrowest part of the pore toward the intracellular aspect of the channel, creates the selectivity filter2. M1, M3 and M4 residues participate in forming the large extracellular vestibule just external to the selectivity filter3,4.
Figure 1.
Transmembrane topology and sequence alignment of NMDARs. (a) NMDAR transmembrane topology. Each NMDAR subunit contains extracellular N-terminal and agonist binding domains, three transmembrane regions (M1, M3, M4), a re-entrant loop (M2/p-loop) and an intracellular C-terminal domain. For clarity, only two of the four subunits are shown. This depiction does not indicate subunit arrangement around the pore. (b) Amino acid residue sequence alignment of the M1–M4 regions of GluN2A–D subunits, with membrane regions labeled. Slashes indicate sequence discontinuity. Asterisks mark residues examined in this study. Large asterisk marks the GluN2 S/L site.
Both GluN1 and GluN2 subunits are necessary to form functional glutamate-gated NMDARs in mammalian systems2. Expression of the four principal NMDAR subtypes, defined by the GluN2 subunit that is coassembled with GluN1 (GluN1/2A –GluN1/2D receptors), is highly regulated and varies by brain region, developmental stage, experience, and disease state5–7, suggesting that NMDAR subtypes play distinct physiological roles. Triheteromeric receptors composed of GluN1 and two different types of GluN2 subunits also are widely expressed2, although their functional properties are not well characterized.
The GluN2 subunit present in a receptor shapes numerous NMDAR properties, which therefore vary among NMDAR subtypes. The NMDAR subtype-dependence of several properties, including channel open probability and agonist potency, are largely conferred by the N-terminal domain and the linker to the agonist binding domain8,9. Less understood are the structural bases of the NMDAR subtype-dependence of open channel properties, including single-channel conductance, Ca2+ permeability, and Mg2+ block. The NMDAR subtype-dependent variation in Mg2+ block has been found to depend on three portions of the M1–M4 regions, as well as the agonist binding domain10,11. To our knowledge, there is no information on the origin of the NMDAR subtype dependence of Ca2+ permeability or single-channel conductance.
RESULTS
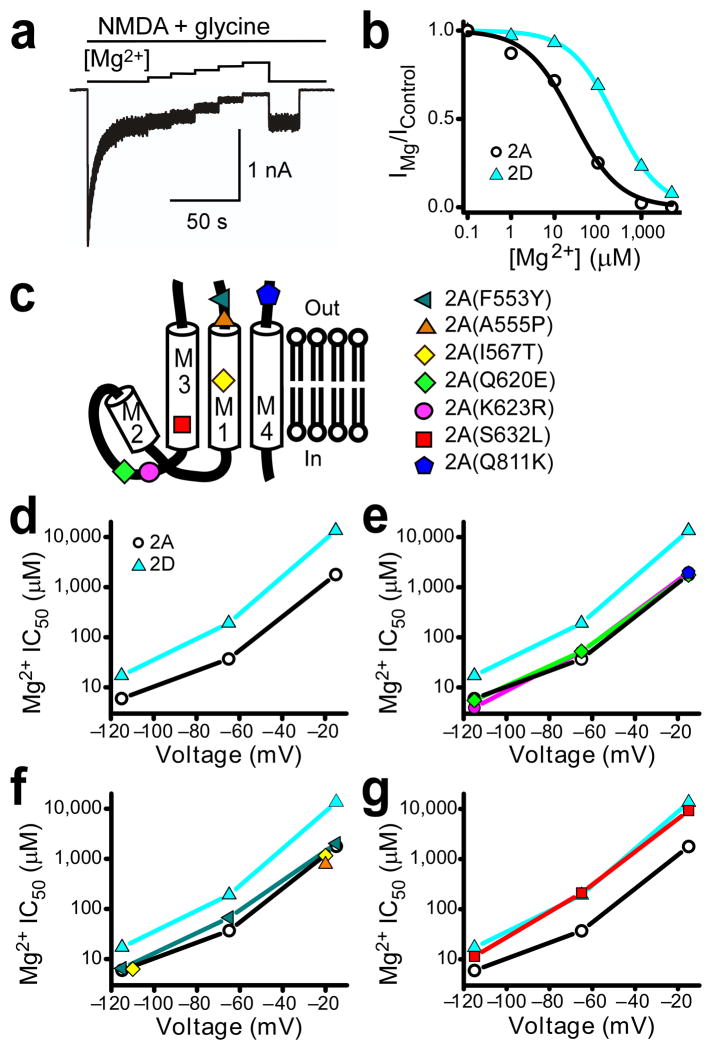
To identify a structural basis for the NMDAR subtype dependence of Mg2+ block, we took advantage of clustering of open channel properties that divide NMDARs into two groups: GluN1/2A and GluN1/2B receptors versus GluN1/2C and GluN1/2D receptors. GluN1/2A and GluN1/2B receptors exhibit similarly high Mg2+ affinities and single-channel conductances; GluN1/2C and GluN1/2D receptors exhibit similarly low Mg2+ affinities and single-channel conductances6. Likewise, GluN1/2A and GluN1/2B receptors have higher Ca2+ permeabilities than do GluN1/2C receptors12,13 (the Ca2+ permeability of GluN1/2D receptors has not been quantified). Furthermore, in the M1–M4 regions, GluN2A and GluN2B subunits have more sequence identity with each other than with GluN2C or GluN2D subunits; GluN2C and GluN2D subunits also share more sequence identity with each other than with the other GluN2 subunits (Fig. 1b). We targeted sites in the M1–M4 regions for mutagenesis based on this pattern (Fig. 1b and 2c): sites at which GluN2A and GluN2B subunits contain the same residue, but at which GluN2C and GluN2D subunits share a different residue. In GluN2A subunits, the wild type residue was substituted with the residue found in GluN2C and GluN2D subunits.
Figure 2.
Search for GluN2 residues that affect the NMDAR subtype specificity of Mg2+ block. (a) Example trace of GluN1/2A receptor-mediated currents at −65 mV and current inhibition by addition of Mg2+ at concentrations from 1 μM to 5 mM. (b) [Mg2+]-inhibition curves constructed from the GluN1/2A trace in (a) (black circles) and similar data from GluN1/2D receptors (blue triangles). (c) Transmembrane topology of GluN2 subunits and approximate locations of targeted residues. (d–g) Plots of average Mg2+ IC50s at −115, −65, and −15 mV in wild-type receptors and receptors containing amino acid residue substitutions (point mutants in which a residue in the GluN2A subunit was replaced with the homologous residue found in GluN2C and GluN2D subunits). The mutant subunit represented by each symbol is defined in (c) (right). (d) Mg2+ IC50s of wild-type GluN1/2A and GluN1/2D receptors (these data are replotted in e–g). (e) Mg2+ IC50s of GluN1/2A(Q620E), GluN1/2A(K623R), GluN1/2A(Q811K) and wild-type receptors. (f) Mg2+ IC50s of GluN1/2A(F553Y), GluN1/2A(A555P), GluN1/2A(I567T) and wild-type receptors. (g) Mg2+ IC50s of GluN1/2A(S632L) and wild-type receptors. n = 2 – 24 cells per point.
We compared the Mg2+ block properties of receptors composed of GluN1 combined with wild type GluN2A, wild type GluN2D, or mutant GluN2A subunits. For each NMDAR subunit combination, the Mg2+ IC50 (the concentration of Mg2+ ([Mg2+]) necessary to inhibit 50% of NMDAR-mediated current) was quantified (Fig. 2a,b). Data typically were gathered at several voltages because of the strong voltage dependence of Mg2+ block of NMDARs. Consistent with previous data5,10,14,15, GluN1/2A receptors had lower Mg2+ IC50s than did GluN1/2D receptors across voltages (Fig. 2b,d).
One residue controls NMDAR subtype dependence of Mg2+ block
Mg2+ IC50s of NMDARs with the GluN2C and GluN2D subunit residue substituted at the corresponding GluN2A subunit site were measured. Six mutant GluN1/2A receptors, each containing a single substitution, displayed Mg2+ IC50s comparable to wild type GluN1/2A receptors (Figs. 1b and 2c,e,f). NMDARS in which the GluN2C and GluN2D leucine (L) was substituted in place of the GluN2A and GluN2B serine (S) at GluN2A(S632) in M3, however, resulted in Mg2+ IC50s that were strikingly similar to GluN1/2D receptor Mg2+ IC50s (Fig. 2g).
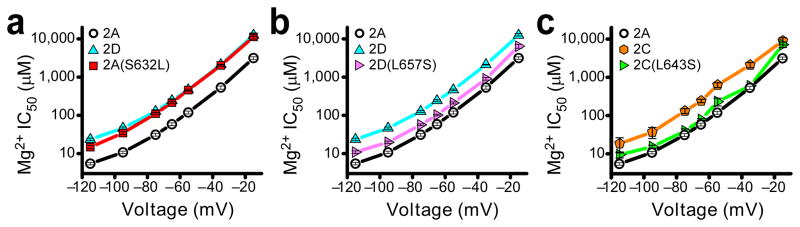
Investigation of the influence of GluN2A(S632) on the interaction of Mg2+ with NMDARs required careful measurement of the voltage dependence of Mg2+ IC50 from numerous wild type and mutant receptors (e.g., Figs. 3,6,8). Characterization of the voltage dependence of Mg2+ IC50 using the protocol shown in Figure 2a was relatively inefficient. We therefore developed a method to rapidly determine NMDAR Mg2+ IC50 at seven voltages from −115 mV to −15 mV by measuring I-V curves near the end of 5-second applications of each Mg2+-containing solution (Supplementary Fig. 1a,b). This protocol yielded results similar to those made with the slower protocol (Fig. 2a; Supplementary Fig. 1c–e). Both protocols revealed that GluN1/2A(S632L) receptor Mg2+ IC50s were very close to those of GluN1/2D receptors (Figs. 2g and 3a). Substitution of leucine for serine at this site (the “GluN2 S/L site”) accounted for 88% of the difference between GluN1/2A and GluN1/2D receptor Mg2+ IC50s (averaged across voltages; Online Methods). We then performed the reverse mutation by substituting serine for the homologous leucine at the GluN2 S/L site in GluN2C and GluN2D subunits. The GluN1/2D(L657S) and GluN1/2C(L643S) receptor Mg2+ IC50s were decreased toward those of GluN1/2A receptors by an average of 57% and 67%, respectively (Fig. 3b,c). GluN2 subunit regions that may account for the remaining 33–43% of the NMDAR subtype dependence of Mg2+ IC50 are considered in the Discussion.
Figure 3.
The GluN2 S/L site regulates the NMDAR subtype specificity of Mg2+ block. (a) GluN1/2A(S632L) receptor Mg2+ IC50s are similar to GluN1/2D receptor IC50s across a wide range of voltages. Plot of the voltage-dependence of Mg2+ IC50s of GluN1/2A, GluN1/2D, and GluN1/2A(S632L) receptors. IC50s in this figure and in Figures 6 and 8 were determined using a rapid measurement protocol (Supplementary Fig. 1). (b) GluN1/2D(L657S) receptor Mg2+ IC50s approach those of GluN1/2A receptors across a wide range of voltages. (c) GluN1/2C(L643S) receptor Mg2+ IC50s approach GluN1/2A receptor Mg2+ IC50s across a wide range of voltages.
Figure 6.
Influence on Mg2+ inhibition of mutations at the GluN2 S/L site. (a) Voltage-dependence of Mg2+ IC50s of NMDARs containing wild-type GluN2A subunits, wild-type GluN2D subunits, or GluN2A subunits mutated at the GluN2 S/L site. (b) Mg2+ IC50s of wild-type and mutant NMDARs at −75 mV. Examination of correlations between amino acid properties and Mg2+ IC50s can be found in Supplementary Figure 3.
Figure 8.
Mutant cycle examination of intersubunit interactions involved in NMDAR subtype specificity of Mg2+ block. (a,b) Mutant cycle results and schematics. Left, plot of the voltage-dependence of wild-type and mutant NMDAR Mg2+ IC50s (n = 4 cells for each NMDAR subunit combination). *p < 0.05; ** p < 0.01; ***p < 0.001. Right, mutant cycle schematic and representative mutant cycle results at −65 mV. GluN1m and 2Am represent mutant subunits. Above each NMDAR subunit combination name is the Mg2+ IC50. Adjacent to each arrow is the fold-change of Mg2+ IC50s presented as the ratio (mean Mg2+ IC50 of NMDARs containing a wild-type subunit)/(mean Mg2+ IC50 of NMDARs containing a mutant subunit). If a GluN1 residue is not coupled to the GluN2 S/L site, then the fold-change of Mg2+ IC50 should be the same for the left and right vertical arrows, and the same for the upper and lower horizontal arrows. The coupling coefficient Ω at −65 mV is in the center of the schematic. (a) GluN1(W611)-GluN2A(S632) mutant cycle. Two-way ANOVA results: −115 mV, p = 0.51; −95 mV, p = 0.38; −75 mV, p = 0.53; −65 mV, p = 0.09; −55 mV, p = 0.26; −35 mV, p = 0.06. Ω = 1.6, averaged across voltages. (b) GluN1(W608)-GluN2A(S632) mutant cycle. Two-way ANOVA results: −115 mV, p = 0.03; −95 mV, p = 0.01; −75 mV, p = 0.00001; −65 mV, p < 0.00001; −55 mV, p = 0.00004; −35 mV, p = 0.001. Ω = 3.5, averaged across voltages.
Effect of the GluN2 S/L site on relative Ca2+ permeability
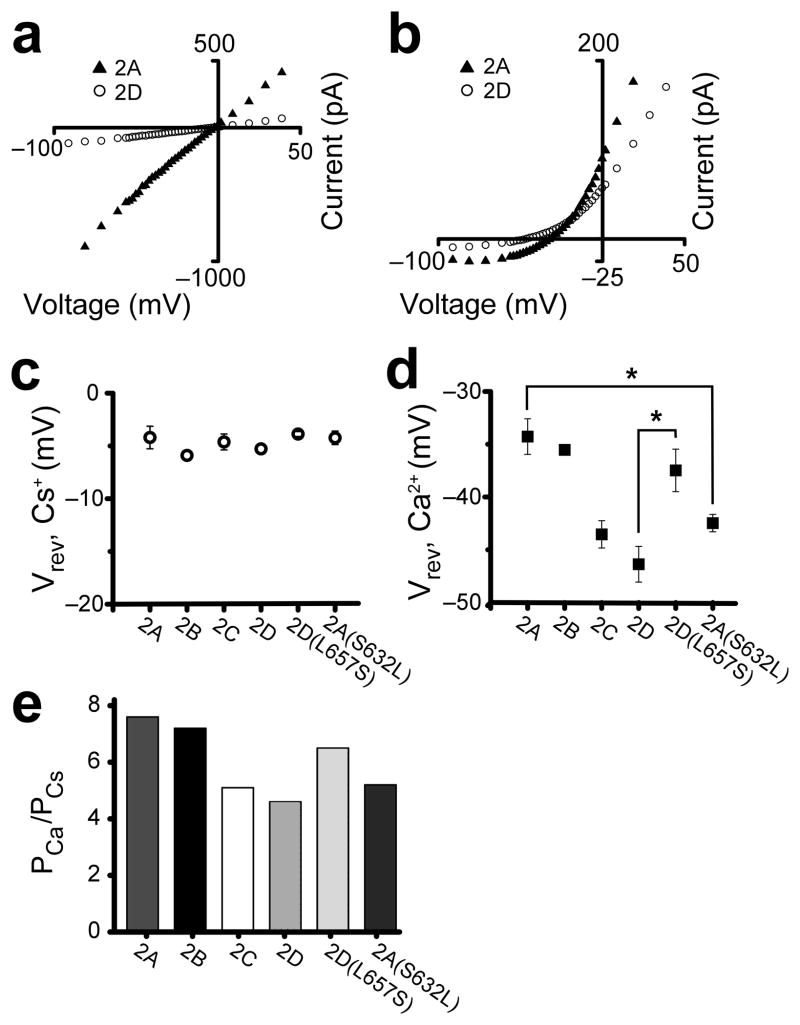
To determine whether the effects of the GluN2 S/L site are specific to Mg2+ IC50, we examined selective permeability to Ca2+, another NMDAR subtype-dependent channel property of central physiological importance. GluN1/2A and GluN1/2B receptors display similarly high Ca2+ permeabilities13, whereas GluN1/2C receptors display moderately lower Ca2+ permeability12. To our knowledge, the relative Ca2+ permeability ofGluN1/2D receptors has not been quantified. We used reversal potential (Vrev) measurements to quantify Ca2+ permeability relative to Cs+ permeability (PCa/PCs), and first compared the PCa/PCs ratios of all four wild-type NMDAR subtype. Vrevs were determined with a voltage step protocol in biionic conditions in which the sole permeant intracellular ion was Cs+ (146 mM)and the sole permeant extracellular ion was either 143 mM Cs+ (Fig. 4a) or 1.8 mM Ca2+ (Fig. 4b; Online Methods). We observed no significant difference among NMDAR subtype Vrevs in 143 mM extracellular Cs+ (Fig. 4c; see Supplementary Table 1 for Vrev and PCa/PCs values). We therefore performed model-independent statistical comparison of Ca2+ permeability based on Vrev values in 1.8 mM extracellular Ca2+, and found: GluN1/2A and GluN1/2B receptor values were not significantly different; GluN1/C and GluN1/2D receptor values were not significantly different; GluN1/2A and GluN1/2B receptor values were significantly different from GluN1/C and GluN1/2D receptor values (Fig. 4d). We also calculated PCa/PCs ratios by inserting average Vrevs into a modified version of the Lewis equation (Online Methods, Equation 2)16,17. In agreement with previously reported data, GluN1/2A and GluN1/2B receptor PCa/PCss were similar, and each was higher than the PCa/PCs of GluN1/2C receptors (Fig. 4e). The GluN1/2D receptor PCa/PCs was similar to the PCa/PCs of GluN1/2C receptors, and lower than the PCa/PCss of GluN1/2A and GluN1/2B receptors (Fig. 4e). Hence, the subtype-dependent clustering of open-channel properties includes selective permeability to Ca2+ as well as Mg2+ IC50 and single-channel conductance.
Figure 4.
The GluN2 S/L site contributes to NMDAR subtype specificity of relative Ca2+ permeability. (a,b) Representative I–V curves in 143 mM extracellular Cs+ (a) and 1.8 mM extracellular Ca2+ (b). (c,d) Average Vrevs, ± s.e.m., of wild-type and mutant NMDARs in 143 mM extracellular Cs+ (c) and 1.8 mM extracellular Ca2+ (d). (c) No significant differences were detected among wild-type and mutant receptor Vrevs in 143 mM extracellular Cs+ (one-way ANOVA, p = 0.63). (d) Significantly different Vrevs (one-way ANOVA, p < 0.05) between wild-type and mutant receptors are marked with an asterisk. (e) PCa/PCs values of wild-type and mutant NMDARs.
We performed similar measurements to evaluate the Ca2+ permeability of GluN1/2A(S632L) and GluN1/2D(L657S) receptors. No significant differences were detected among wild-type and mutant receptor Vrevs in 143 mM extracellular Cs+ (Fig. 4c). In 1.8 mM Ca2+, GluN1/2A(S632L) receptor Vrevs were significantly different from those of wild-type GluN1/2A receptors, but not from GluN1/2C or GluN1/2D receptor Vrevs (Fig. 4d). GluN1/2D(L657S) receptor Vrevs in 1.8 mM Ca2+ were significantly different from GluN1/2D receptor Vrevs, but not from GluN1/2A or GluN1/2B receptor Vrevs (Fig. 4d). The PCa/PCs ratio for GluN1/2A(S632L) receptors was lower than the ratio for GluN1/2A receptors, and the PCa/PCs ratio for GluN1/2D(L657S) receptors was higher than the ratio for GluN1/2D receptors (Fig. 4e). Thus, the GluN2 S/L site regulates the NMDAR subtype dependence of selective permeability to Ca2+ as well as Mg2+ IC50s.
Effects of GluN2 S/L site on single-channel conductance
We also examined the effect of the GluN2 S/L site on the NMDAR subtype dependence of single-channel conductance. Single-channel conductance, along with reversal potential, defines how much current passes through an open channel at any voltage. It is a hallmark characteristic that distinguishes NMDAR subtypes, and determination of single-channel conductance is an important tool for identifying NMDAR subunit composition of native channels in the nervous system18,19.
We compared single-channel currents (Fig. 5a), current amplitudes (Fig. 5b), and conductances (Fig. 5c) from GluN1/2A, GluN1/2A(S632L), GluN1/2D, and GluN1/2D(L657S) receptors. The main state conductance of wild-type GluN1/2A receptors was 54.6 ± 2.1 pS, similar to other reported values20. The main state conductance of GluN1/2A(S632L) receptors was 34.8 ± 1.4 pS, significantly different from the GluN1/2A receptor main state conductance (t-test for heterogeneity of slopes, p < 0.0001), but not significantly different from that of GluN1/2D receptors (37.4 ± 1.3 pS, measured previously in our lab15, p = 0.252, and consistent with other reported values21). The GluN1/2D(L657S) receptor main state conductance was 54.9 ± 1.5 pS, significantly different from the GluN1/2D receptor main state conductance15 (p<0.0001), but not significantly different from that of GluN1/2A receptors (p = 0.233).
Figure 5.
The GluN2 S/L site controls the NMDAR subtype specificity of single-channel conductance. (a,b) Representative single-channel current traces (a) and amplitude histograms (b) of GluN1/2A (left), GluN1/2A(S632L) (center left), GluN1/2D (center right), and GluN1/2D(L657S) (right) receptors recorded in the outside-out patch configuration at −75 mV. (c) i-V plots of single-channel currents and linear regression fits. The slope of the linear fit to each i-V plot (single-channel conductance) is shown next to each fit. Left, GluN1/2A receptors, (n = 17 single-channel current recordings); center left, GluN1/2A(S632L) receptors ( n = 17); center right, GluN1/2D receptors (n = 24; GluN1/2D data from Qian et al. (2005)15); right GluN1/2D(L657S) receptors (n = 16).
Another single-channel property that distinguishes NMDAR subtypes is the prominent subconductance state of ~20 pS that is exhibited by GluN1/2C and GluN1/2D receptors15,21; GluN1/2A or GluN1/2B receptors, in contrast, exhibit a larger subconductance state (~40 pS) 20 that is less commonly occupied. GluN1/2A(S632L) receptors exhibited a subconductance state with a conductance (17.0 ± 0.7 pS) not significantly different from the subconductance state of GluN1/2D receptors (20.2 ± 1.3 pS; p = 0.052; Fig. 5c). In contrast, the GluN1/2D(L657S) receptor subconductance state was 49.6 ± 5.6 pS, significantly higher than that of GluN1/2D receptors (p < 0.0001). We could not consistently resolve a subconductance state in GluN1/2A receptors, although in some patches a subconductance state of ~40 ps was occupied infrequently. Our ability to resolve consistently the subconductance state of GluN1/2D and GluN1/2D(L657S) receptors, but not of GluN1/2A receptors, suggests that subconductance state occupancy by GluN1/2D receptors was not dramatically reduced by the mutation. Thus, mutation of the GluN2 S/L site in GluN2D receptors has a powerful effect on the conductance of the subconductance state, but appears to have a weaker effect on subconductance state occupancy.
To determine whether single-channel kinetics are influenced by the GluN2 S/L site, we constructed open period and shut time histograms (Supplementary Fig. 2 and Supplementary Table 2) and compared statistically the values of weighted mean open periods. The weighted mean open periods of GluN1/2A and GluN1/2A(S632L) receptors were not significantly different, nor were the weighted mean open periods of GluN1/2D and GluN1/2D(L657S) receptors (see Supplementary Table 2). Hence, consistent with previous studies showing that NMDAR subtype-dependence of channel gating depends on the N-terminal domain8,9, the GluN2 S/L site does not appear to have a substantial effect on channel kinetics.
We conclude that the naturally-occurring residue replacement in GluN2 subunits that we mimicked by creating GluN2A(S632L), GluN2C(L643S), and GluN2D(L657S) subunits underlies fundamental NMDAR subtype-dependent variations in multiple channel characteristics: Mg2+ block, relative Ca2+ permeability, and single-channel conductance of both main and subconductance states. However, the way in which the GluN2 S/L site affects pore properties is unclear based on data presented thus far.
Mechanism of GluN2 S/L site influence on Mg2+ block
In search of mechanisms by which the GluN2 S/L site transmits its effects to the pore, we created additional GluN2A subunits with mutations at the GluN2 S/L site and tested whether the Mg2+ IC50s of NMDARs containing these mutations correlated with chemical or physical properties of the substituted amino acid residues. We mutated GluN2A(S632) to asparagine (N), isoleucine (I), lysine (K), phenylalanine (F), aspartate (D), alanine (A), and tryptophan (W), and compared Mg2+ IC50s of NMDARs containing GluN2 S/L site point mutant or wild-type subunits (Fig. 6 and Supplementary Table 3). Linear regression analyses to test the dependence of mutant and wild type NMDAR Mg2+ IC50s at −75 mV on amino acid polarity, hydrophobicity, hydropathy, and volume revealed no significant correlation if corrected for multiple comparisons (Supplementary Fig. 3). We next tried an alternative approach to developing a hypothesis to explain how the GluN2 S/L site regulates channel properties.
The GluN2 S/L site is unlikely to interact with permeant and blocking ions directly due to its location at the base of the M3 region (Fig. 2c). We therefore hypothesized that the GluN2 S/L site exerts its influence over Mg2+ block, Ca2+ permeability, and single-channel conductance through interactions with amino acid residues that are closer to the pore. To predict which residues might interact with the GluN2 S/L site, the molecular homology modeling software Modeller22 was employed to create a structural model of the NMDAR M2–M3 region. Homology modeling based on crystallized K+ channels, especially the KcsA channel, has been used and validated extensively to predict NMDAR structure and its functional implications23–25. The model presented here is based on the crystal structure of the Bacillus cereus NaK channel26. The NaK channel is a member of the cyclic nucleotide-gated channel family of non-selective tetrameric cation channels, shares high sequence homology with K+ channels (Supplementary Fig. 4), and is probably evolutionarily related to K+ channels and NMDARs24,26. We chose to base the NMDAR homology model on the NaK rather than KcsA channel structure because, like NMDARs, the NaK channel is permeable to Na+ and Ca2+ as well as K+ 26,27. We also produced and examined homology models based on the crystal structure of the KcsA channel28,29 (Supplementary Fig. 5). Homology models based on the GluA2 glutamate receptor structure30 were not initially considered because it was published after we had tested homology model predictions. We subsequently created and examined a homology model based on the GluA2 receptor crystal structure (Supplementary Discussion and Supplementary Fig. 6).
The NaK channel contains two transmembrane regions (M1 and M2) separated by a reentrant p-loop. Like an upside-down NMDAR channel, the NaK channel p-loop extends from near the extracellular surface of the membrane to approximately halfway through the channel. Several studies have demonstrated the structural conservation of glutamate receptor M2–M3 regions and the homologous regions of proteins related to the NaK channel23–25,31; we therefore limited the NaK channel-based homology model to the M2–M3 regions (Fig. 7a). To ensure sequence alignment validity, we generated a ClustalX multiple sequence alignment of NMDAR subunits, the NaK channel, and nine K+ channels for which crystal structures of the pore region are available (Supplementary Fig. 4). This alignment, which involved no residue insertions or deletions, agreed with previously published alignments based on both experimental and computational methods4,23,25,32.
Figure 7.
Homology model of the GluN1/2A receptor M2–M3 regions based on the NaK channel structure. (a) Sequence alignment used for the NMDAR homology model, showing the M2–M3 region of GluN1 and GluN2A subunits and homologous region of the NaK channel subunit. Yellow highlight, selectivity filter residues; red font, GluN2 S/L site, GluN1(W608), and GluN1(W611); asterisks, residues that are identical in NaK channel, GluN1 and GluN2A subunits (black), identical in NaK channel and GluN1 subunits (green), or identical in NaK channel and the GluN2A subunit (blue). (b) Ribbon diagram of the NMDAR homology model, viewed from the extracellular surface. Green ribbons, GluN1 subunits; blue ribbons, GluN2A subunits. (c) Ribbon diagram of adjacent GluN1 and GluN2A M2–M3 regions in the NMDAR model, viewed from the membrane outside the channel, overlaid onto a schematic of membrane regions. The α-carbons of GluN2A(S632) (red), GluN1(W608) (orange) and GluN1(W611) (cyan) are displayed as spheres. GluN1 and GluN2A subunits on the far side of the channel, and schematic of M1 and M4 regions of GluN2A, were removed for clarity. (d) Ribbon diagram of the NMDAR model, viewed from outside the channel. Spheres, α-carbons of three residues as in c. The GluN1 and GluN2A subunits on the far side of the channel were removed for clarity. Box, area enlarged in e. (e) Stereo view of intersubunit residue-residue interactions in the NMDAR model. Theα-helical sections of GluN1 subunit M2 and GluN2A subunit M3 regions are shown as ribbons. Space-filling residues are colored as in c.
Our GluN1/2A receptor model based on the NaK channel has a GluN1-2A-1-2A subunit arrangement30 (Fig. 7b). The GluN2 S/L site in the model interfaces with the p-loop of GluN1 subunits (Fig. 7c–e). When viewed as a space-filling residue, GluN2A(S632) appears to be very close to two GluN1 tryptophans in the p-loop helix: GluN1(W608) and GluN1(W611), which are separated by approximately one α-helical turn (Fig. 7e). Our homology model based on the KcsA channel yielded very similar predictions (Supplementary Fig. 5). To test experimentally whether GluN1(W608) or GluN1(W611) couples with the GluN2 S/L site to influence GluN2 subunit-dependent channel properties, we performed mutant cycle analyses33,34. In a mutant cycle analysis, the effects of two or more point mutations are evaluated to probe for functional evidence of coupling between residues at these sites.
For each mutant cycle, we measured the Mg2+ IC50s of four NMDAR subunit combinations: wild-type GluN1 and wild-type GluN2A subunits (wt/wt receptors); wt GluN1 and mutant GluN2A subunits (wt/mut receptors); mut GluN1 and wt GluN2A subunits (mut/wt receptors); and mut GluN1 and mut GluN2A subunits (mut/mut receptors). If there is no coupling between the GluN1 and GluN2 subunit residues, then inclusion of a mutant GluN1 subunit in an NMDAR should result in the same degree of functional change, regardless of whether that mutant GluN1 subunit is expressed with a wild-type or mutant GluN2A subunit. That is, if the mutated residues do not interact, then the magnitude of change in Mg2+ IC50 from wt/wt to mut/wt should equal the magnitude of change from wt/mut to mut/mut. Likewise, the magnitude of change in Mg2+ IC50 from wt/wt to wt/mut should be the same as that from mut/wt to mut/mut. Mutant cycle analyses are commonly quantified by calculation of a coupling coefficient Ω. In our experiments, putative coupling between residues was assessed indirectly by measuring the Mg2+ IC50s of each subunit combination, resulting in the following equation for Ω:
When Ω = 1, there is no evidence for coupling between two residues; Ω > 1 or Ω < 1 argues for coupling between the residues of interest. Ω values do not provide information on the statistical significance of our data; therefore, we performed two-way ANOVA interaction tests at each voltage tested (Online Methods) to determine whether the mutant cycles provided statistically significant evidence for residue-residue coupling.
We first probed for coupling between GluN1(W611) and GluN2A(S632). Several attempted mutant cycles were unsuccessful because of mutant receptors that expressed currents too small to permit accurate measurement of Mg2+ IC50s (Online Methods). We eventually found that GluN1(W611A) produced currents large enough to permit Mg2+ IC50 measurements when coexpressed with GluN2A and when coexpressed with GluN2A(S632L). Mutating GluN1(W611) to alanine had little effect on Mg2+ IC50s when coexpressed with GluN2A, and little effect when coexpressed with GluN2A(S632L) (Fig. 8a); at no voltage was there statistically significant coupling. Thus, our data do not provide evidence for coupling between GluN1(W611) and GluN2A(S632).
To determine whether GluN1(W608) interacts with the GluN2 S/L site, we replaced GluN1(W608) with serine, creating GluN1(W608S), and replaced the GluN2A serine with tryptophan, creating GluN2A(S632W). We predicted that mutating the GluN1 tryptophan to serine might partially compensate for the effects of mutating the GluN2A serine to tryptophan because the tryptophan and serine would be exchanged in the mut/mut receptor. Expression of GluN1(W608S) with wild-type GluN2A slightly increased the Mg2+ IC50 values compared to GluN1/2A receptor values (Fig. 8b). If there is no coupling between GluN1(W608) and GluN2A(S632), then expression of GluN1(W608S) with GluN2A(S632W) similarly should slightly increase the Mg2+ IC50 values compared to GluN1/2A(S632W) receptor values. Mutating GluN2A(S632) to tryptophan, however, markedly decreased Mg2+ IC50s of NMDARs when coexpressed with GluN1(W608S) subunits (Fig. 8b). Statistically significant coupling was found at every voltage. Thus, mutating GluN1(W608) to serine partially compensates for the effects of mutating GluN2A(S632) to tryptophan. We subsequently performed another mutant cycle analysis to test for coupling between GluN1(W608) and GluN2A(S632), but using the mutants GluN1(W608S) and GluN2A(S632L) (Supplementary Fig. 7). Evidence for coupling between the residues was still observed, although of smaller magnitude and significance than for the GluN1(W608S) and GluN2A(S632W) cycle. The coupling may be weaker because a tryptophan-serine residue exchange was not performed, and/or because mutating GluN2A(S632) to leucine had a smaller effect on Mg2+ IC50 than mutation to tryptophan.
These mutant cycle data indicate that the NMDAR subtype dependence of Mg2+ inhibition depends on coupling between the GluN2 S/L site and GluN1(W608), while leaving open the possible involvement of additional pairs of residues. Although residue-residue coupling usually implies close physical proximity of residues34, it is possible that coupling between residues may result from indirect interactions. Of particular importance, the data unveil an intersubunit interaction between GluN1 and GluN2 that is critical to an NMDAR subtype-dependent channel property.
DISCUSSION
We have identified a naturally-occurring residue substitution in GluN2 subunits that is predominantly responsible for the NMDAR subtype dependence of Mg2+ inhibition, Ca2+ permeability, and single-channel conductance. Using structural homology models, we predicted that the GluN2 S/L site, which is at the base of the M3 region, conveys its effects on the pore by interacting with the p-loop of the adjacent GluN1 subunit. Results from mutant cycle experiments support the model’s prediction, identifying GluN1(W608) as a p-loop residue that is coupled to the GluN2 S/L site.
Based on the results of this study and recent previous work, the spectrum of subtype-dependent NMDAR properties can be divided into two clusters. One cluster of properties is related to channel gating and binding of ligands that influence gating, and includes maximal channel open probability, pH sensitivity, Zn2+ inhibition, agonist potency, and deactivation kinetics. The NMDAR subtype dependence of this gating property cluster depends principally on the large N-terminal domain and its linker to the agonist binding domain8,9, which are distant from the GluN2 S/L site. We demonstrate here that the NMDAR subtype dependence of a second cluster, properties of the open channel, depends, in contrast, on a single amino acid difference in the M3 transmembrane region. Together, differences in and near the N-terminal domain and at the GluN2 S/L site underlie the great majority of functional variation among NMDAR subtypes.
The GluN2 S/L site regulates ion permeation of the NMDAR channel, as demonstrated by its effects on relative Ca2+ permeability and single-channel conductance. The influence of the GluN2 S/L site on Mg2+ block also may be partly mediated by an effect on ion permeation; the single greatest difference between the kinetics of Mg2+ block of GluN1/2A and GluN1/2D receptors is an enhanced rate of Mg2+ permeation through GluN1/2D receptors15,35. The GluN2 S/L site and GluN1(W608), situated near the intracellular end of the pore, appear to be well-located to affect ion permeation.
The subtype dependence of channel block by Mg2+ is not solely determined by the GluN2 S/L site. This was particularly clear from measurements showing that NR2 S/L site leucine to serine mutations accounted for 67% of the Mg2+ IC50 difference between GluN1/2C and GluN1/2A receptors, and 57% of the difference between GluN1/2D and GluN1/2A receptors (Fig. 5b,c). Based on previous studies, it appears likely that the remaining NMDAR subtype dependence of Mg2+ IC50 may be accounted for by multiple subunit regions, including the M1 and M4 transmembrane regions, the M2–M3 linker10, and the agonist binding domain11. Hence, the majority of the NMDAR subtype dependence of Mg2+ IC50 depends on the single residue difference at the GluN2 S/L site; the remaining NMDAR subtype dependence of Mg2+ IC50 is likely to depend on a number of the >700 residue differences between GluN2A or GluN2B and GluN2C or GluN2D subunits, spread across several regions of the subunits.
In a series of experiments related to those presented here, Mg2+ block of GluN1/2C receptors was examined using GluN2C subunits modified to contain parts of the GluN2B subunit10. As noted above, multiple sections of GluN2 subunits were found to influence the NMDAR subtype dependence of Mg2+ IC50. The authors found that receptors containing a GluN2C subunit with a leucine-to-serine substitution at the GluN2 S/L site exhibited increased voltage dependence of Mg2+ block, but no change in Mg2+ IC50 at −100 mV. The apparent discord with our findings may stem from the use of different preparations: Xenopus oocytes were used in the previous study10 whereas HEK cells were used here, resulting in multiple experimental differences, including permeant ion concentrations, which strongly affect Mg2+ block36.
Insight into why NMDAR channels exhibit weak monovalent cation selectivity is suggested by comparing selectivity filters of homologous channels with known structures. Although the GluA2 AMPA receptor cystal structure30 cannot be used because its selectivity filter was not resolved (see Supplementary Discussion), previous studies have demonstrated structural similarities between NMDARs and K+ channels24,37. The NaK channel is structurally similar to the NMDAR channel as well, but unlike K+ channels is cation-nonselective26,27. Whereas in K+ channels, four K+ binding sites are formed along the selectivity filter by four rings of carbonyl oxygens projecting into the channel28,38,39, the NaK channel selectivity filter contains only two rings of carbonyl oxygens26. NaK channel residues along the pore-lining section of the p-loop that do not contribute to these carbonyl oxygen rings form an external vestibule that is wider than the selectivity filter; this vestibule appears to contribute to non-specific cation permeability27. The monovalent cation nonselectivity of NMDARs similarly may depend on a relatively short selectivity filter and wide vestibule. The NMDAR selectivity filter nevertheless must exhibit powerful selectivity among divalent cations.
The mutant cycle results presented here suggest that the GluN2 S/L site regulates NMDAR subtype specificity of Mg2+ block at least in part through coupling with GluN1(W608). The mechanism by which GluN1(W608) mediates communication between the GluN2 S/L site and the pore, however, is unknown. A substituted-cysteine accessibility study showed that GluN1(W608) is solvent-accessible from the intracellular, but not the extracellular, aspect of the membrane40. Mutations of the homologous residue in GluN2B subunits, also a tryptophan, strongly affects Mg2+ block and permeation41. Intriguingly, the tryptophan at GluN1(W608) is highly conserved. All known mammalian iGluR subunits contain a tryptophan at homologous positions, with the exception of the GluN3A and GluN3B subunits. In fact, this tryptophan is present at the homologous sites of nematode (Caenorhabditis elegans) and insect (Drosophila melanogaster) iGluRs, as well as in predicted iGluR subunits and glutamate binding proteins of the plant Arabidopsis42,43. Furthermore, the tryptophan at this site is conserved in the bacterial GluR0 channel and in some potassium channels43. Thus, GluN1(W608) and the tryptophans at equivalent sites in NMDAR subunit homologs are likely to play a critical role in channel function.
METHODS
Cell culture and transfection
HEK293T cell (whole-cell experiments; source, ATCC) and HEK293 cells (single-channel experiments; source, ATCC) were maintained as previously described15. Calcium phosphate transfections44 or FuGENE transfections (Roche) were performed with a cDNA ratio of 1 eGFP:1 GluN1:2 GluN2 or 1 eGFP:3 GluN1:6 GluN2.
Mutagenesis and cDNA preparation
cDNAs encoding rattus GluN1-1a (GenBank accession number (ACCN) X63255), GluN2A (ACCN M91561), GluN2C (ACCN M91562), and GluN2D (ACCN L31611) subunit genes in ampicillan resistance-encoding plasmids (pcDNA 3.1 or pcDNA1) were mutagenized with the Stratagene Quik-Change XL sited-directed mutagenesis kit. Mutagenized NMDAR subunit cDNAs from isolated colonies were sequenced from 100–200 bases upstream to 100–200 bases downstream of each mutation (University of Pittsburgh Genomics and Proteomics Core Laboratories). In most cases, mutant-containing plasmids from at least two colonies were expressed for electrophysiological experiments to ensure that results did not arise from unintended mutations. Furthermore, the entire coding sequences of the mutant subunits GluN2A(S632L) and GluN2D(L657S) were sequenced (GeneWIZ). The amino acid residue sequence encoded by the GluN2A(S632L) mutant had two differences from the source sequence: the intended mutation (S632L) and S758T. The latter residue (GluN2A(T758)) is present in another GluN2A sequence (ACCN D13211), suggesting it was present in the wild-type sequence. The amino acid residue sequence encoded by the GluN2D(L657S) mutant had three differences from the source sequence: the intended mutation (L657S), P94R, and A305R. The latter two residues (GluN2D(R94) and GluN2D(R305)) are present in another GluN2D sequence (ACCN U08260), suggesting they were present in the wild-type sequence.
Electrophysiology
Experiments was performed 12–72 hours after initiation of transfection. Borosilicate micropipettes of 2–5 MΩ (whole-cell recordings) or 5–11 MΩ (single-channel recordings) resistance were pulled on a Sutter Instruments P-97 electrode puller. Micropipettes used in single-channel recordings were coated with Sylgard 184. In all recordings except Ca2+ permeabilitiy experiments (see below), bath solutions were grounded with a silver chloride pellet. Intracellular solution for whole-cell experiments consisted of (in mM): CsCl (125), HEPES (10), EGTA (10); intracellular solution composition for single-channel experiments was (in mM): CsFl (115), CsCl (10), HEPES (10), EGTA (10). CsOH (~30 mM) was added to both intracellular solutions to arrive at pH 7.2 ± .05 and 275 ± 5 mOsmol/kg. The normal extracellular solution, which was used in all experiments except Ca2+ permeability measurements, consisted of (in mM): NaCl (140), KCl (2.8), CaCl2 (1.0), HEPES (10). 1.8 mM extracellular Ca2+ solution consisted of (in mM): 140 NMDG, 10 Hepes, 1.8 Ca2+, pH adjusted to 7.2± 0.05 with HCl. 143 mM extracellular Cs+ solution contained (in mM): 140 CsCl, 10 HEPES, pH adjusted to 7.2± 0.05 by addition of ~3 mM CsOH. When necessary, sucrose was added to reach 290± 10 mOsmol/kg. NMDAR responses were elicited by 30 μM glycine plus 10 μM NMDA (Mg2+ IC50 experiments), 50 μM glycine plus 1 mM glutamate (Ca2+ permeability experiments), or 30 μM glycine plus 30 μM NMDA (single-channel experiments), referred to as “agonists”. NMDA was used as an agonist in some experiments to facilitate comparison with previous data15. Magnesium-containing solutions were composed of the normal extracellular solution to which agonists and MgCl2 were added.
Outside-out patch and whole-cell currents were amplified using an Axopatch 200B or 200 patch-clamp amplifier (Molecular Devices) and recorded on video tape and/or computer in pClamp version 9 or 10 format (Molecular Devices). Currents were filtered at 1 kHz (whole-cell recordings) or 2.5 kHz (single-channel recordings) with a Warner Instruments LPF-8 and sampled at 20 kHz (whole-cell recordings) or 50 kHz (single-channel recordings). Correction for liquid junction potentials (Vps) between the intracellular and extracellular solutions was applied in all experiments (whole-cell experiments: Vp = −6.3 mV in normal extracellular solution; Vp = −13.0 mV in 1.8 mM Ca2+ extracellular solution; single-channel experiments: Vp = −9.7 mV in normal extracellular solution).
We developed a method for rapid measurement of Mg2+ IC50s at seven voltages (Supplementary Fig. 1). Normal (Mg2+-free) extracellular solution plus agonists was applied until a steady-state current was reached. Successively higher [Mg2+]s (0–10 mM in normal extracellular solution plus agonists) were applied to the cell, using a 5-s application of each [Mg2+], via a gravity-fed 10-barrel fast perfusion system (Supplementary Fig. 1). During the last 1.5 s of each solution application, the command voltage was stepped from a holding potential of −65 mV through a series of voltages ranging from −115 mV to −15 mV, in 20 mV increments. Each stepped command voltage was maintained for 210 ms. The current at each command voltage was averaged over the last 150 ms of the voltage step. Before and after application of agonists, baseline current (0 agonists) at each command voltage was measured using the same sequence of voltage steps; baseline currents were averaged and subtracted from the agonist-activated currents in each [Mg2+] at each command voltage to calculate the NMDAR-mediated current. Using this approach, current measurements in ten [Mg2+]s at seven voltages each were made in ~1.5 minutes. The ten [Mg2+]s covered a wide concentration range so that an accurate estimate of Mg2+ IC50 could be made at each of the voltages used. A complete data set for one cell was gathered by repeating this protocol at least 3 times and averaging the NMDAR-mediated current at each [Mg2+] and command voltage across trials.
For each cell, at each voltage, fractional current (IMg/Icontrol) measured in each [Mg2+] was plotted on a [Mg2+]-inhibition curve and fit with Equation (1):
| (1) |
where n is the Hill coefficient, IMg is the amplitude of agonist-induced current in [Mg2+], and Icontrol is the amplitude of NMDAR-mediated current in 0 Mg2+ (Fig. 2b and Supplementary Fig. 1b).
To asses relative Ca2+ permeability (PCa/PCs), Vrevs of mutant and wild-type NMDARs were measured with whole-cell recordings in biionic conditions. Extracellular solution containing a single permeant ion (Cs+ or Ca2+) was bath-applied via a gravity-fed perfusion system for several minutes to ensure complete solution exchange. Agonists were then bath-applied for 30–60 s, until desensitization was complete. In the continued presence of agonists, the command voltage was stepped from a holding potential of −65 mV (before correction for junction potential) through a series of voltages ranging from −95 mV to −55 mV in 10 mV increments, then stepped from −50 mV to 10 mV in 2 mV increments, and finally from 15 mV to −55 mV in 10 mV increments. Each command voltage was maintained for 210 ms. Subtraction of baseline current was performed as described for Mg2+ IC50 experiments. We used two approaches to minimize errors due to voltage changes at the interface between the bath solution and the AgCl ground pellet that could result from changing bath solution. First, in some experiments, we grounded the bath through a 3 M KCl agarose bridge to minimize voltage changes at the agarose-bath solution interface and contamination of the bath solution, which was constantly perfused (3–6 ml/min; bath volume 250–300 μl). Second, in most experiments we grounded the bath with a gravity-fed flowing KCl bridge. This bridge consisted of a 1 ml syringe filled with 3M KCl into which an AgCl ground pellet was placed, attached to a 32 gauge needle (0.108 mm inner diameter) (Hamilton Company). The needle was inserted into the bath efflux tubing,, minimizing the possibility of bath solution contamination. Data from experiments in which voltage drift exceeded 2 mV were excluded from analysis. Relative Ca2+ permeability was calculated with a modified Lewis equation16,17:
| (2) |
where [Cs+]o and [Ca2+]o are the extracellular concentrations of Cs+ and Ca2+, and PCa/PCs is the permeability ratio of Ca2+ to Cs+.
Single-channel data analysis
Idealization of single-channel currents was performed using the time-course fitting method45. To determine single-channel current amplitudes, Gaussian functions were fit to single-channel current amplitude event histograms using the maximum likelihood method45. Conductances were calculated as the slope of the best-fit linear regression to an i-V graph containing data collected at four or more voltages. Open period and shut time histograms (Supplementary Fig. 2 and Supplementary Table 2) were constructed from data collected at −75 mV. Only patches in which less than 5% of the openings were double openings were used. Fits comprised of two to four components were generated using the maximum likelihood method. Time-course fitting and maximum likelihood fitting were performed using the DC Analysis programs (http://www.ucl.ac.uk/Pharmacology/dcpr95.html) SCAN and EKDIST, respectively.
Structural modeling
The structural homology modeling program Modeller (version 9.7) was used to create energetically-plausible models of the NMDAR M2–M3 region, based on homologous regions of the crystalized NaK channel (PBD ID 2AHZ)26 and KcsA channel (PBD IDs 1BL8 and 3FB5)28,29, or of the entire NMDAR minus the C-terminus and a portion of the p-loop, based on the crystalized GluA2 channel (PDB ID 3KG2). The molecular graphics program VMD46 was used to visualize NMDAR models and to generate Figure 7b–e and Supplementary Figures 5b–e and 6.
Mutant cycle analysis
Several point mutants at each GluN1 and GluN2 subunit site of interest were created. Mutant cycle measurements were performed if whole-cell recordings from HEK293T cells expressing each relevant mutant subunit combination typically yielded NMDAR current amplitudes more negative than −100 pA at −65 mV in 0 Mg2+. Cells with smaller current amplitudes did not yield reliable [Mg2+]-inhibition curves, especially at less negative voltages. We attempted to perform experiments with GluN1(W611S) and GluN2A(S632W) subunits based on the idea that the tryptophan-serine exchange in the mut/mut receptor might partially compensate for the effects of the GluN2A serine to tryptophan mutation. Currents through GluN1(W611S)/2A(S632W) receptors, however, were too small to permit accurate measurement of Mg2+ IC50s. We constructed several more GluN1(W611) point mutants, but most appeared to form nonfunctional channels when co-expressed with wild-type or mutant GluN2A subunits. Thus, we performed a mutant cycle using GluN1(W611A) and GluN2A(S632L) subunits (Fig. 8a). We attempted to supplement the mutant cycle experiment in which mutant subunits GluN1(W608S) and GluN2A(S632W) were used (Fig. 8) with a mutant cycle experiment that paralleled the GluN1(W611A)-GluN2A(S632L) experiments. We observed no NMDAR-mediated current, however, in GluN1(W608A)/2A(S632L) receptors. In each mutant cycle presented, data at −15 mV were excluded because some mutants did not pass enough current to accurately estimate Mg2+ IC50s at that voltage.
Statistical analysis
Most statistical tests were performed using Origin 7 (OriginLab Corporation). T-tests for heterogeneity of slopes were performed in the R software environment47 (http://www.R-project.org) with Package HH48. In Figures 3, 6, and 8, and Supplementary Figures 3 and 7, Mg2+ IC50 data are presented as mean Mg2+ IC50 value (averaged across cells) ± s.e.m. For each mutant cycle, six two-way ANOVA interaction tests of log-transformed Mg2+ IC50 data were performed (one test per voltage) (Fig. 8). Whereas untransformed IC50 measurement data do not follow a normal distribution, log-transformed IC50 data are normally distributed49. Because ANOVA tests assume normal distributions of data, the log transformation was used. The factors in each two-way ANOVA were GluN1 subunit identity (wild type or mutant) and GluN2A subunit identity (wild type or mutant). A significant interaction effect implies that the magnitude of the effect of one factor (e.g., mutation of GluN1 subunit) depended on the other factor (e.g., the identity of the coexpressed GluN2A subunit), which would reflect coupling between the mutated GluN1 and GluN2A residues. Interactions were considered significant at p < 0.05 (significance level α = 0.05).
The percent of the difference between GluN1/2A and GluN1/2D receptor Mg2+ IC50s that was accounted for by mutation of the GluN2 S/L site was calculated for the GluN2A(S632L) mutation (88%; see Results) using the equation:
| (3) |
A related equation was used to calculate percent of the difference between Mg2+ IC50 of GluN1/2A and of GluN1/2C or GluN1/2D receptor accounted for by the GluN2C(L643S) mutation (67%) and the GluN2D(L657S) mutation (57%). Although use of log-transformed IC50 data is the more appropriate approach50, we performed similar calculations without log-transformation, yielding 81% for the GluN2A(S632L) mutation, 73% for the GluN2D(L657S) mutation, and 77% for the GluN2C(L643S) mutation.
Supplementary Material
Supplementary Figure 1. A rapid approach for measurement of Mg2+ IC50s at 7 voltages
Supplementary Figure 2. Single-channel kinetics of wild-type and mutant NMDARs
Supplementary Figure 3. Correlation between NMDAR Mg2+ IC50 and properties of the amino acid at the GluN2 S/L site
Supplementary Figure 4. Multiple sequence alignment of K+ channel and NMDAR subunits
Supplementary Figure 5. Homology model of a GluN1/2A receptor based on the crystal structure of the KcsA channel
Supplementary Figure 6. Homology model of the GluN1/2A receptor based on the crystal structure of the GluA2 homomer
Supplementary Figure 7. GluN1(W608S)-GluN2A(S632L) mutant cycle
Supplementary Table 1. Vrevs of wild-type and mutant NMDARs and calculated PCa/PCs values
Supplementary Table 2. Single-channel kinetics of wild-type and mutant NMDARs
Supplementary Table 3. Mg2+ IC50s of wild-type NMDARs and GluN1/2A(S632) mutant receptors
Supplementary Discussion
Acknowledgments
The authors thank A. Qian for valuable contributions to and advice on initial experiments, K. Bouch and C. Shiber for excellent technical assistance, M. Pellegrino for generous help with statistical analyses, M. Casio and A. Retchless for helpful discussions and comments on the manuscript, and S. Meriney, M. Grabe, D. Wood, and Johnson lab members for helpful discussions, and D. Colquhoun for making available the DC Analysis programs for single-channel analysis. This work was supported by National Institute of Health grants R01 MH045817 (J.W.J.) and F31 MH079755 (B.S.R.)
Footnotes
Note: Supplementary Information is available online.
AUTHOR CONTRIBUTIONS
All authors participated in experimental design and analysis, and in revision of the manuscript; BSR and WG performed experiments; BSR and JWJ wrote the manuscript.
References
- 1.Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- 2.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobolevsky AI, Beck C, Wollmuth LP. Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron. 2002;33:75–85. doi: 10.1016/s0896-6273(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 4.Beck C, Wollmuth LP, Seeburg PH, Sakmann B, Kuner T. NMDAR channel segments forming the extracellular vestibule inferred from the accessibility of substituted cysteines. Neuron. 1999;22:559–570. doi: 10.1016/s0896-6273(00)80710-2. [DOI] [PubMed] [Google Scholar]
- 5.Ishii T, et al. Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J Biol Chem. 1993;268:2836–2843. [PubMed] [Google Scholar]
- 6.Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004;2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 7.Clinton SM, Meador-Woodruff JH. Abnormalities of the NMDA receptor and associated intracellular molecules in the thalamus in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2004;29:1353–1362. doi: 10.1038/sj.npp.1300451. [DOI] [PubMed] [Google Scholar]
- 8.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuner T, Schoepfer R. Multiple structural elements determine subunit specificity of Mg2+ block in NMDA receptor channels. J Neurosci. 1996;16:3549–3558. doi: 10.1523/JNEUROSCI.16-11-03549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrighton DC, Baker EJ, Chen PE, Wyllie DJ. Mg2+ and memantine block of rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J Physiol. 2008;586:211–225. doi: 10.1113/jphysiol.2007.143164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnashev N, Zhou Z, Neher E, Sakmann B. Fractional calcium currents through recombinant GluR channels of the NMDA, AMPA and kainate receptor subtypes. J Physiol. 1995;485 ( Pt 2):403–418. doi: 10.1113/jphysiol.1995.sp020738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneggenburger R. Simultaneous measurement of Ca2+ influx and reversal potentials in recombinant N-methyl-D-aspartate receptor channels. Biophys J. 1996;70:2165–2174. doi: 10.1016/S0006-3495(96)79782-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monyer H, et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 15.Qian A, Buller AL, Johnson JW. NR2 subunit-dependence of NMDA receptor channel block by external Mg2+ J Physiol. 2005;562:319–331. doi: 10.1113/jphysiol.2004.076737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis CA. Ion-concentration dependence of the reversal potential and the single channel conductance of ion channels at the frog neuromuscular junction. J Physiol. 1979;286:417–445. doi: 10.1113/jphysiol.1979.sp012629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wollmuth LP, Sakmann B. Different mechanisms of Ca2+ transport in NMDA and Ca2+-permeable AMPA glutamate receptor channels. J Gen Physiol. 1998;112:623–636. doi: 10.1085/jgp.112.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrant M, Feldmeyer D, Takahashi T, Cull-Candy SG. NMDA-receptor channel diversity in the developing cerebellum. Nature. 1994;368:335–339. doi: 10.1038/368335a0. [DOI] [PubMed] [Google Scholar]
- 19.Cull-Candy SG, et al. NMDA receptor diversity in the cerebellum: identification of subunits contributing to functional receptors. Neuropharmacology. 1998;37:1369–1380. doi: 10.1016/s0028-3908(98)00119-1. [DOI] [PubMed] [Google Scholar]
- 20.Stern P, Cik M, Colquhoun D, Stephenson FA. Single channel properties of cloned NMDA receptors in a human cell line: comparison with results from Xenopus oocytes. J Physiol. 1994;476:391–397. doi: 10.1113/jphysiol.1994.sp020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyllie DJ, Behe P, Nassar M, Schoepfer R, Colquhoun D. Single-channel currents from recombinant NMDA NR1a/NR2D receptors expressed in Xenopus oocytes. Proc Biol Sci. 1996;263:1079–1086. doi: 10.1098/rspb.1996.0159. [DOI] [PubMed] [Google Scholar]
- 22.Eswar N, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;Chapter 2(Unit 2):9. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 23.Panchenko VA, Glasser CR, Mayer ML. Structural similarities between glutamate receptor channels and K+ channels examined by scanning mutagenesis. J Gen Physiol. 2001;117:345–360. doi: 10.1085/jgp.117.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood MW, VanDongen HM, VanDongen AM. Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proc Natl Acad Sci U S A. 1995;92:4882–4886. doi: 10.1073/pnas.92.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tikhonov DB. Ion channels of glutamate receptors: structural modeling. Mol Membr Biol. 2007;24:135–147. doi: 10.1080/09687860601008806. [DOI] [PubMed] [Google Scholar]
- 26.Shi N, Ye S, Alam A, Chen L, Jiang Y. Atomic structure of a Na+- and K+-conducting channel. Nature. 2006;440:570–574. doi: 10.1038/nature04508. [DOI] [PubMed] [Google Scholar]
- 27.Alam A, Shi N, Jiang Y. Structural insight into Ca2+ specificity in tetrameric cation channels. Proc Natl Acad Sci U S A. 2007;104:15334–15339. doi: 10.1073/pnas.0707324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 29.Cuello LG, Jogini V, Cortes DM, Perozo E. Structural mechanism of C-type inactivation in K+ channels. Nature. 2010;466:203–208. doi: 10.1038/nature09153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999;402:817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- 32.Tikhonov DB, Mellor JR, Usherwood PN, Magazanik LG. Modeling of the pore domain of the GLUR1 channel: homology with K+ channel and binding of channel blockers. Biophys J. 2002;82:1884–1893. doi: 10.1016/S0006-3495(02)75538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hidalgo P, MacKinnon R. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 1995;268:307–310. doi: 10.1126/science.7716527. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. Journal of molecular biology. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 35.Qian A, Johnson JW. Permeant ion effects on external Mg2+ block of NR1/2D NMDA receptors. J Neurosci. 2006;26:10899–10910. doi: 10.1523/JNEUROSCI.3453-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonov SM, Johnson JW. Permeant ion regulation of N-methyl-D-aspartate receptor channel block by Mg2+ Proc Natl Acad Sci U S A. 1999;96:14571–14576. doi: 10.1073/pnas.96.25.14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones KS, VanDongen HM, VanDongen AM. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J Neurosci. 2002;22:2044–2053. doi: 10.1523/JNEUROSCI.22-06-02044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, et al. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 39.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 40.Kuner T, Wollmuth LP, Karlin A, Seeburg PH, Sakmann B. Structure of the NMDA receptor channel M2 segment inferred from the accessibility of substituted cysteines. Neuron. 1996;17:343–352. doi: 10.1016/s0896-6273(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 41.Williams K, et al. The selectivity filter of the N-methyl-D-aspartate receptor: a tryptophan residue controls block and permeation of Mg2+ Mol Pharmacol. 1998;53:933–941. [PubMed] [Google Scholar]
- 42.Chiu JC, et al. Phylogenetic and expression analysis of the glutamate-receptor-like gene family in Arabidopsis thaliana. Mol Biol Evol. 2002;19:1066–1082. doi: 10.1093/oxfordjournals.molbev.a004165. [DOI] [PubMed] [Google Scholar]
- 43.Sprengel R, et al. Glutamate receptor channel signatures. Trends Pharmacol Sci. 2001;22:7–10. doi: 10.1016/s0165-6147(00)01588-1. [DOI] [PubMed] [Google Scholar]
- 44.Clarke R, Johnson J. NMDA receptor NR2 subunit dependence of the slow component of magnesium unblock. Journal of Neuroscience. 2006;26:5825–5834. doi: 10.1523/JNEUROSCI.0577-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colquhoun D, Sigworth FJ. Analysis of single ion channel data. In: Sakmann B, Neher E, editors. Single-channel recording. Plenum Press; New York: 1995. pp. 483–587. [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- 48.Heiberger RM, Holland B. Statistical analysis and data display: an intermediate course with examples in S-plus, R, and SAS. Springer; New York: 2004. [Google Scholar]
- 49.Fleming WW, Westfall DP, De la Lande IS, Jellett LB. Log-normal distribution of equieffective doses of norepinephrine and acetylcholine in several tissues. J Pharmacol Exp Ther. 1972;181:339–345. [PubMed] [Google Scholar]
- 50.Colquhoun D. Binding, gating, affinity and efficacy: the interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. British Journal of Pharmacology. 1998;125:924–947. doi: 10.1038/sj.bjp.0702164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. A rapid approach for measurement of Mg2+ IC50s at 7 voltages
Supplementary Figure 2. Single-channel kinetics of wild-type and mutant NMDARs
Supplementary Figure 3. Correlation between NMDAR Mg2+ IC50 and properties of the amino acid at the GluN2 S/L site
Supplementary Figure 4. Multiple sequence alignment of K+ channel and NMDAR subunits
Supplementary Figure 5. Homology model of a GluN1/2A receptor based on the crystal structure of the KcsA channel
Supplementary Figure 6. Homology model of the GluN1/2A receptor based on the crystal structure of the GluA2 homomer
Supplementary Figure 7. GluN1(W608S)-GluN2A(S632L) mutant cycle
Supplementary Table 1. Vrevs of wild-type and mutant NMDARs and calculated PCa/PCs values
Supplementary Table 2. Single-channel kinetics of wild-type and mutant NMDARs
Supplementary Table 3. Mg2+ IC50s of wild-type NMDARs and GluN1/2A(S632) mutant receptors
Supplementary Discussion