Summary
The eukaryotic bZIP transcription factors are critical players in organismal response to environmental challenges. In fungi, the production of secondary metabolites (SMs) is hypothesized as one of the responses to environmental insults, e.g. attack by fungivorous insects, yet little data to support this hypothesis exists. Here we establish a mechanism of bZIP regulation of SMs through RsmA, a recently discovered YAP-like bZIP protein. RsmA greatly increases SM production by binding to two sites in the A. nidulans AflR promoter region, a C6 transcription factor known for activating production of the carcinogenic and anti-predation SM, sterigmatocystin (ST). Deletion of aflR in an overexpression rsmA (OE::rsmA) background not only eliminates ST production but also significantly reduces asperthecin synthesis. Furthermore, the fungivore, Folsomia candida, exhibited a distinct preference for feeding on wild type rather than an OE::rsmA strain. RsmA may thus have a critical function in mediating direct chemical resistance against predation. Taken together, these results suggest RsmA represents a bZIP pathway hardwired for defensive SM production.
Keywords: sterigmatocystin, asperthecin, fungivory, Yap, rsmA
Introduction
The fungal kingdom is remarkable for the production of bioactive molecules commonly referred to as secondary metabolites (SMs). In general these metabolites are considered to provide fitness attributes to the producing organism, often as a defense against environmental insult. For example, stress responses have been linked to increased aflatoxin level in A. parasiticus (Roze et al. 2011; Chang et al. 2008) and a series of recent works have definitively shown that SM production is associated with increased fungal fitness in confrontations with insects where both LaeA (a global regulator of fungal secondary metabolism, Bok et al. 2004) and AflR (the C6 transcription factor responsible for aflatoxin and ST synthesis in Aspergilli, Fernandes et al. 1998; Chang et al. 1995) are required for protection from fungivores (Rohlfs et al. 2007; Trienens et al. 2010, Staaden et al. 2010).
In fungi, developmental processes in response to various abiotic or biotic external triggers are commonly associated with secondary metabolism (Calvo et al. 2002; Braus et al. 2010). A breakthrough in fungal biology was the discovery of heterotrimeric VelB/VeA/LaeA transcriptional complex (known as the Velvet complex) which connects SM with light signal (Bayram et al. 2008). Different members of the velvet protein family partner with each other, coordinating SM biosynthesis with fungal development through various signal transduction pathways (reviewed in Bayram and Braus, 2011). However, how Velvet controls SM production is unknown. Because many genes for the synthesis of secondary metabolites are arranged in gene clusters (Keller and Hohn, 1997), the considerable evidence for SM gene regulation can be in part explained by transcriptional control through hierarchical levels of transcriptional regulatory elements including cluster specific regulatory elements, global regulators as well as transcriptional complexes like Velvet (reviewed in Yin and Keller, 2011).
Identification of SM regulatory elements could potentially provide a means of increasing production of beneficial metabolites, aid in the identification of “silent” natural products, and, importantly, also contribute to a broader understanding of the molecular mechanisms by which SM are produced. The case for a fungal SM stress response pathway was recently strengthened by the finding of RsmA (restorer of secondary metabolism A), a putative YAP-like bZIP protein identified in a multicopy suppressor screen for restoration of sterigmatocystin (a carcinogenic and anti-predation SM), in Velvet complex mutants (Shaaban et al. 2010). Overexpression of RsmA conferred a remarkable ability to greatly increase ST in multiple genetic backgrounds of A. nidulans. YAP proteins are well known mediators of stress response pathways throughout the spectrum of life from yeast to humankind (Rodrigues-Pousada et al. 2010). Several bZIP proteins have been characterized in Aspergillus spp. as responding to oxidative, osmotic, drug, nutrient and iron stress (Roze et al. 2011; Asano et al. 2007; Hagiwara et al. 2008; Balazs et al. 2010; Qiao et al. 2008). Here we uncover the mechanism by which RsmA regulates SM production. We find that RsmA operates through activation of aflR, encoding the C6 transcription factor embedded in the ST gene cluster. RsmA binds to two sites in the aflR promoter and elimination of either site or aflR eliminates not only enhanced ST synthesis by overexpression of rsmA but also reduces asperthecin production, a metabolite produced by a SM cluster containing no transcription factor. Furthermore, when given the choice between wild type (WT) and a rsmA overexpression strain, fungivorous insects overwhelmingly refuse to feed on the latter strain in accordance with the role of ST as a metabolite mediating resistance against predation.
Results
RsmA is involved in SM regulation
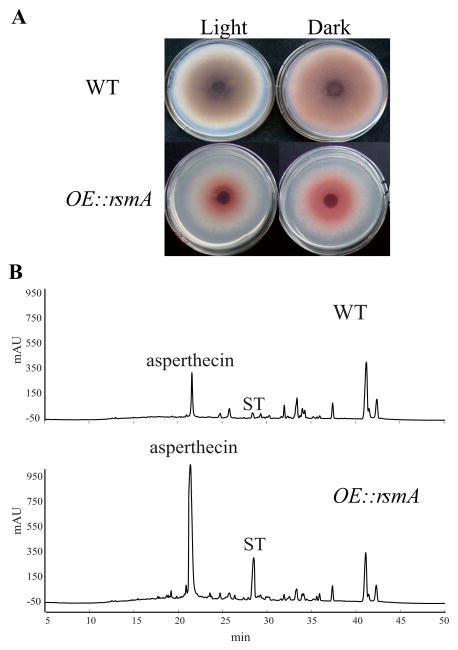
The initial assessment of RsmA as a multicopy suppressor of loss of laeA and veA expression focused on the enhanced sterigmatocystin synthesis in the overexpression RsmA (OE::rsmA) strain (Shaaban et al., 2010). To examine impact of OE::rsmA on other aspects of Aspergillus biology, we first made a new version of the overexpression allele. To minimize impact of possible ectopic affects on RsmA function, we replaced the native promoter with the constitutive glyceraldehyde-3-phosphate dehydrogenase gene (gpdA) promoter, rather than placing the allele ectopically as described in Shaaban et al. (2010). Assessment of this new strain showed that it looked identical to the original strain and also produced enhanced quantities of ST (Fig. 1A and B). LC-MS analysis of this strain grown on solid medium revealed greatly increased production of not only ST but also asperthecin (Fig. 1B), a recently characterized anthraquinone (Szewczyk et al. 2009).
Figure 1. RsmA overexpression increases secondary metabolite production.
Panel A Bottom side of plates of WT (RDIT9.32) and RsmA overexpression (OE::rsmA, RWY2.12) strains grown on solid GMM media under light and dark conditions. These strains were incubated with point inoculation at 37 °C for 4 days.
Panel B. LC-MS analysis of secondary metabolite production by WT and OE::rsmA respectively.
Role for RsmA in fungivore resistance but not canonical stress response
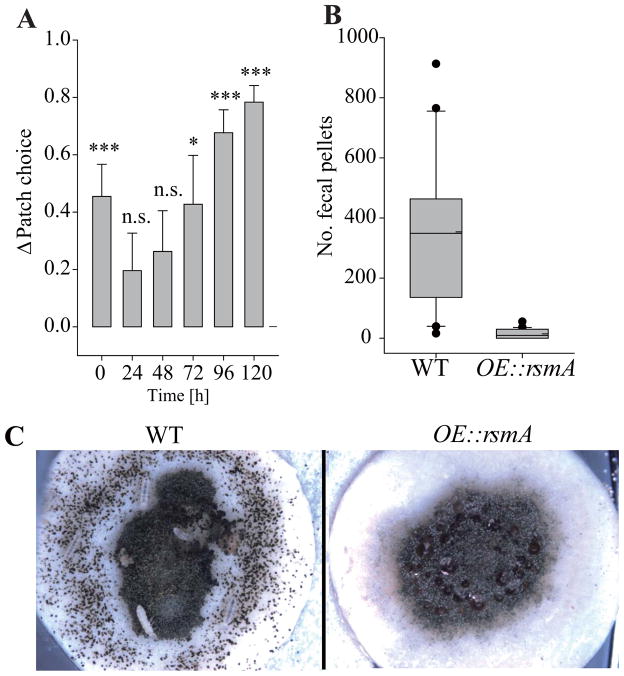
Several studies have shown VeA and LaeA driven secondary metabolites, particularly ST, to be involved in resistance against fungivores (Rohlfs et al. 2007, Staaden et al. 2011, Trienens and Rohlfs, in press). To assess any impact of OE::rsmA on fungivore selectivity, we presented both the WT and the OE::rsmA strain to fungivorous collembolans, Folsomia candida, in a food choice assay. While patch choice was inconsistent during the first two days, collembolans showed an increasing, statistically significant tendency to stay on the WT patches during the following three days (Fig. 2A). Even though animals were moving between the WT and OE::rsmA patches they were almost exclusively feeding on the WT colonies. The latter is indicated by extensive feeding damage and accumulation of fecal pellets around the WT colonies but not at the OE::rsmA colonies (Fig. 2B and C). As this food choice test was conducted from fungus grown on malt extract agar, we assessed SM on this medium as well and again found OE::rsmA greatly increased ST over WT (Fig. S1).
Figure 2. Fungivorous collembolans, Folsomia candida, display strong avoidance of the OE::rsmA strain.
Panel A The tendency of fungivores to stay on WT or OE::rsmA fungal patches measured as Δ patch choice (± SE). Values around zero indicate no preference for either patch, whereas positive values indicate preference for the WT and negative ones preference for the OE::rsmA strain (see Experimental procedures for details). Patch choice behavior varied with time (time series mixed model: F5,94 = 3.06, P = 0.0132); yet with increasing time fungivores showed an increasing preference for the WT strain (n.s. = not significant, * P ≤ 0.05, *** P < 0.001; for deviation from Δ patch choice = 0).
Panel B. Box plots depicting the number of fecal pellets as an indicator of local fungivores feeding activity. Box plots display the sample minimum and maximum, the upper and lower quartile, and the median (solid line) as well as the mean (dashed line). Filled circles may be considered as outlier. Paired t-test on the number of fecal pellets (P < 0.0001) indicates strong feeding activity on WT versus only very little feeding on the OE::rsmA strain.
Panel C. Representative images of colonies showing the differences in feeding activity (note all the fecal pellets surrounding the left colony) and damage caused to the WT (left) and the OE::rsmA strain (right).
In addition to the impact on SM, we reasoned that RsmA, which bears sequence similarity to a Candida bZIP with enhanced resistance to antifungal drugs (Alarco et al. 1999), could be involved in some drug or canonical YAP-like stress response. However, an examination of the OE::rsmA strain in comparison to WT revealed no altered response to antifungals, oxidative stressors or heavy metals (data not shown).
RsmA is a bZIP that binds two sites in the divergent aflR aflJ promoter
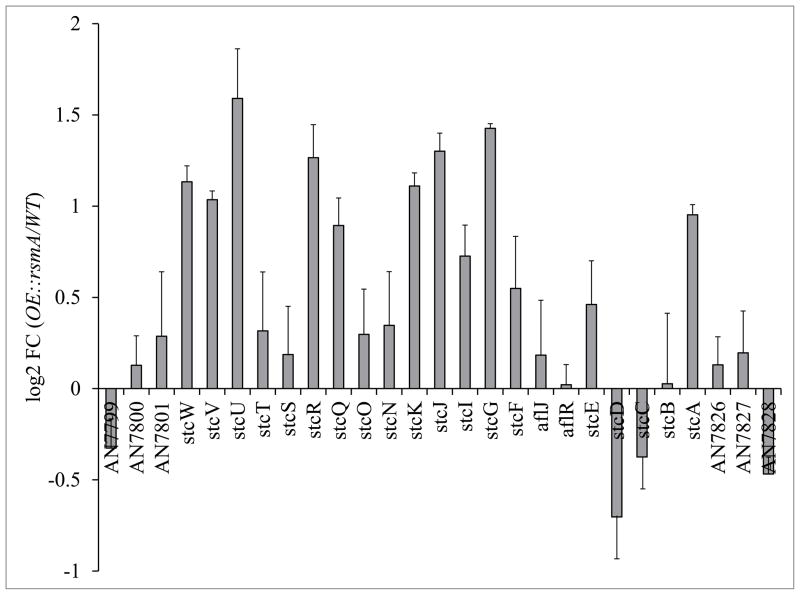
To address the hypothesis that RsmA is a bZIP protein that regulates SM genes through transcriptional activation, microarrays comparing an OE::rsmA with WT were assessed for common motifs in promoters of differentially regulated genes. Quality of the microarrays was confirmed by examining the expression of ST cluster genes. As expected, the majority of the ST cluster genes in OE::rsmA have a higher expression level in comparison with WT (Fig. 3). The two genes downregulated in the OE::rsmA strain, stcC and stcD, have not been found to have a role in ST biosynthesis.
Figure 3. Microarray analysis of sterigmatocystin (ST) gene cluster.
Mean (n = 3) expression ratios (log2, OE::rsmA vs. WT) for genes on Chromosome IV in the region including the ST cluster (AN7899.4 AN7828.4), as measured by microarray.
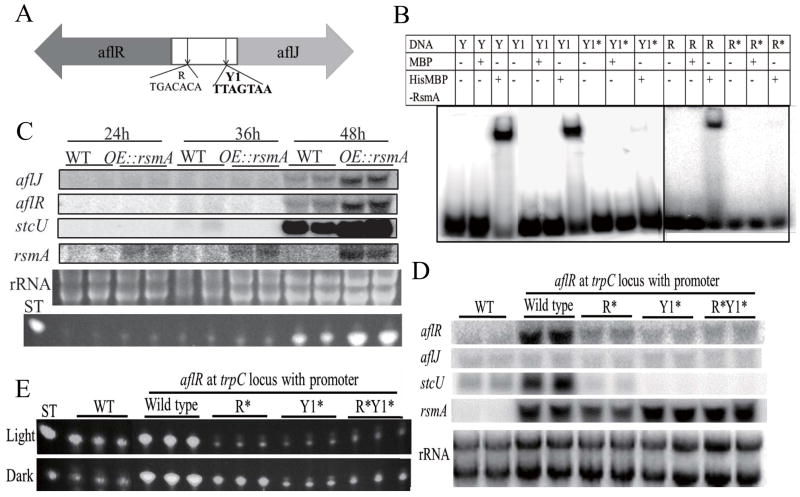
Bioinformatic assessment of the microarray coupled with predictions from yeast YAP binding sites identified two candidate binding sites, TGACTCA and TT(G)ACTAA (Fig. S2). Two of these sites, TGACACA (R) with one base variation (underlined letter) and TTAGTAA(Y), were identified in the bidirectional promoter region of aflR and aflJ (Fig. 4A). AflR is necessary for activation of ST and aflatoxin cluster genes (Fernandes et al. 1998; Chang et al. 1995). AflJ, which shares some similarities to methyltransferases, is required for and enhances AflR activity (Du et al. 2007). RsmA bound to both candidate binding sites, and both aflR and aflJ are overexpressed in the OE::rsmA strain (Fig. 4B and C).
Figure 4. RsmA activates aflR and aflJ expression by specific DNA-binding in the aflR/aflJ divergent promoter.
Panel A RsmA binding sites in the promoter region of aflR/aflJ. Y1: yeast Yap 1 site, R: RsmA site.
Panel B. Specific binding of RsmA to the aflR/aflJ promoter. The purified maltose binding protein (MBP)-tagged RsmA or MBP alone was incubated with the following 32P-labeled sequences: Yap 1 binding site (Y: TTACTAA), Yap 1 binding site in the promoter region of aflR/aflJ (Y1: TTAGTAA), a mutation in this Y1 site (Y1* = TAAGTTA) and RsmA site (R = TGACACA and R*=AAACAGG). *=mutated sequences.
Panel C. rsmA overexpression results in increased aflR, aflJ and stcU expression. RNA was extracted after 24, 36 and 48 h of growth in liquid shake cultures. ST production from the same samples, as analyzed by TLC, is shown below rRNA. Ethidium bromide-stained rRNA is shown as a loading control.
Panel D. RsmA binding sites are required for aflR expression in vivo. aflR, aflJ, rsmA and stcU expression in five strains: WT (RDIT9.32), aflR with WT promoter at trpC locus (TWY18.14), aflR and promoter with R* mutation at trpC locus (TWY19.15), aflR and promoter with Y1* mutation at trpC locus (TWY20.4) and aflR and promoter with R* and Y1* mutations at trpC locus (TWY21.20). RNA was extracted after 48 h of growth in liquid shake cultures. Ethidium bromide-stained rRNA is shown as a loading control.
Panel E. ST production from strains shown in Panel D.
The requirement for these sites was confirmed in vivo by comparing the expression of ectopic copies of aflR with either an intact promoter sequence or with one or both RsmA site mutation sequences in a strain lacking native aflR but containing an OE::rsmA allele at the trpC locus. Mutations at either site significantly reduced aflR and stcU (a biosynthetic gene in the ST pathway) expression (Fig. 4D) as well as ST synthesis (Fig. 4E and Fig. S3). As expected, aflJ expression remained constant as this gene remained at its native locus.
AflR is required for RsmA mediated SM production
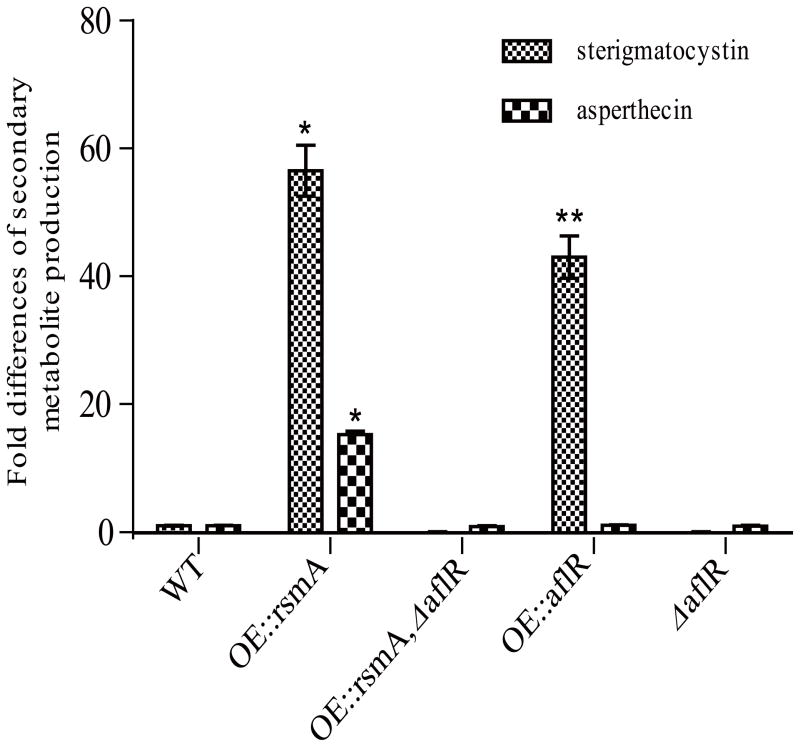
We reasoned AflR could be primarily responsible for the SM phenotype of OE::rsmA. An OE::aflR strain was created with A. nidulans gpdA as promoter at the native locus. Examination of this strain and an OE::rsmA, ΔaflR strain yielded the expected increased and decreased ST phenotype respectively (Fig. 5 and Fig. S4). Unexpectedly, however, we found that asperthecin synthesis was also reduced in the OE::rsmA, ΔaflR double mutant as compared to the OE::rsmA strain despite no increase in the OE::aflR strain alone.
Figure 5. Sterigmatocystin and asperthecin over production require AflR in the OE::rsmA strain.
Both metabolites require the presence of aflR. Statistical differences were analyzed in each secondary metabolite group by using the JMP software package, version 3.2.6 (SAS Institute, Inc., Cary, NC). Statistically significant mean values, indicated with different asterisks in the figures, are significant at P < 0.0001.
Discussion
The study of fungal secondary metabolism (SM) has recently garnered significant interest, and the advent of fungal genomics has lead to new insights into SM regulatory mechanisms. Much of this activity is driven by interests in drug discovery due to the biological properties of these metabolites. Several elegant studies have characterized metabolites from ‘silent’ or ‘repressed’ SM clusters via activation of cluster specific transcription factors (Bergmann et al., 2007; Chiang et al., 2010) and global SM regulators such as LaeA (Bok et al. 2006) or through chromatin remodeling studies (Bok et al. 2009). However, at the heart of these discoveries lies the largely unexplored topic of why these compounds are generated by the producing fungus.
Studies ranging from Flemings’ serendipitous discovery of penicillin synthesis by Penicillium notatum to recent findings of bacterial induction of a silent A. nidulans gene cluster (Schroeckh et al. 2009) serve to illustrate a possible evolutionary development of protective SM by fungi. In support of a direct conduit from environment stress to SM production is our finding of RsmA regulation of SM, particularly the anti-predation metabolite ST. RsmA belongs to a sub-family of bZIP transcription factors commonly referred to as YAP proteins in Saccharomyces cerevisiae based on their homology and similar DNA binding sites as the mammalian AP-1 factor complex (Rodrigues-Pousada et al. 2010; Fernandes et al. 1997). Specific YAP proteins are activated when exposed to environmental challenges ranging from oxidative to heavy metal stress or iron imbalances. RsmA shares the greatest homology with YAP3 of unknown function in yeast. Because the C. albicans homolog of YAP3, FCR3, exhibits resistance to antifungals when overexpressed (Yang et al. 2001), the A. nidulans OE::rsmA strain was originally assessed for a similar activity but was found to show no difference in susceptibility to antifungals as WT (Shabaan et al. 2010 and data not shown). Indeed, this strain exhibited no stress response typical of the YAP activities in yeast. The most striking phenotype was the greatly elevated production of ST and, to a lesser degree, the anthraquinone asperthecin.
No where has a role for ST been more clearly established than in a recent series of fungivore-Aspergillus confrontation studies. Loss of either laeA or aflR or even various enzymatic genes in the ST cluster yields a fungal strain less resistant to fungivore feeding (Rohlfs et al. 2007; Staaden et al. 2010). In these studies, insect preference is consistently towards the SM deletion mutants with distinct repulsive behavior demonstrated towards WT A. nidulans. Here we saw the opposite behavior where fungivores, F. candida, avoided the OE::rsmA strain. Moreover, as indicated by the appearance of insect fecal pellets and damage caused to the colonies, the insects were almost exclusively feeding on the WT strain. This suggests an important role of RsmA in mediating resistance to fungal predators.
Chemical resistance against predation or competition for food substrates is a common phenomenon in nature. Both ST and aflatoxin (produced from ST through two enzyme conversions in A. flavus and A. parasiticus, Chang et al. 1995) have often been cited for their toxicity towards certain insects (Niu et al. 2009; Labrousse and Matile 1996; Llewellyn et al. 1988, Matasyoh et al. 2011, Gunst et al. 1982). These metabolites are not constitutively produced but require exposure to undefined environmental ligands for expression. We propose that RsmA, a bZIP apparently dedicated to SM, represents a defensive response to a specific set of environmental insults requiring up-regulation of ST and other SMs. Specifically, the environmental distresses which could trigger an RsmA mediated SM response include fungivory and confrontations with competitive microbes.
Finally, we found that RsmA works primarily through induction of AflR, a C6 transcription factor required for ST and aflatoxin biosynthesis (Brown et al., 1996; Chang 2003). We show that RsmA operates through binding to the shared aflR aflJ promoter region in the ST gene cluster resulting in both aflR and aflJ expression. Loss of RsmA binding sites negates this regulation as does aflR loss itself. Interestingly, while both overexpression of rsmA and aflR greatly increased ST synthesis over WT (57 and 43 fold respectively), only overexpression of the former increased asperthecin synthesis (15 fold). Nevertheless, loss of aflR in an OE::rsmA background not only resulted in cessation of ST synthesis as expected but also reduced asperthecin levels comparable to that of WT. Such interactions could possibly be reflective of SM cluster cross talk recently described in Aspergillus (Bergmann et al. 2010). We note that the asperthecin gene cluster does not contain an in-cluster transcription factor but does contain at least one AflR and one RsmA binding motif, either in an asperthecin gene promoter or a nearby gene (Table S2), possibly reflective of a need for both factors in up regulation of this metabolite.
Experimental procedures
Strains, media and growth conditions
The fungal strains used in this study are listed in Table 1. All strains were grown at 37 °C on glucose minimum medium (GMM) (Shimizu et al. 2001) and when appropriate were supplemented with 0.56 g uracil l−1, 1.26 g uridine l−1, 1.0 g tryptophan l−1, 0.5 μM pyridoxine HCl, 0.01 μM biotin, 100 μM arginine and maintained as glycerol stocks at −80 °C. For preparing RNA used for microarray, WT and OE::rsmA strains were grown in a liquid minimal media at 37 °C for 48 h in dark, with shaking at 250 rpm, three replicates each. Escherichia coli strains DH5α and BL21 (DE3) (Novagen, Madison, WI) were propagated in LB medium with appropriate antibiotics for plasmid DNA and protein expression, respectively.
Table 1.
Plasmids and fungal strains used in this study
| Strain/plasmid | Descrption | Reference |
|---|---|---|
| RDIT9.32 | veA | Tsitsigiannis et al. (2004a) |
| RDIT2.3 | veA1 | Tsitsigiannis et al. (2004a) |
| RDIT55.37 | pyroA4, veA | Tsitsigiannis et al. (2004b) |
| RMS4.25 | gpdAp::rsmA::pyroA4::pyroA, veA | Shaaban et al. (2010) |
| RJH256 | biA1; argB2; ΔaflR::argB; trpC801; veA1 | Bok et al. (2006) |
| RSCS3 | ΔaflR::argB, veA | Wilkinson et. al. (2004) |
| RAMB38 | biA1; methG1; ΔaflR::argB; trpC801; veA1 | Bok et al. (2004) |
| RJMP1.49 | pyrG89, pyroA4, ΔnkuA::argB, veA | Shaaban et al. (2010) |
| TWY5.2 | pyrG89, A. fumigatus pyrG::gpdA(p)::rsmA, pyroA4, ΔnkuA::argB, veA | This study |
| TWY18.14 | A. fumigatus pyrG::gpdA(p)::rsmA, ΔaflR::argB, trpC::aflR, veA | This study |
| TWY19.15 | A. fumigatus pyrG::gpdA(p)::rsmA, ΔaflR::argB, trpC::aflRR, veA | This study |
| TWY20.4 | A. fumigatus pyrG::gpdA(p)::rsmA, ΔaflR::argB, trpC::aflRY1, veA | This study |
| TWY21.20 | A. fumigatus pyrG::gpdA(p)::rsmA, ΔaflR::argB, trpC::aflRR,Y1, veA | This study |
| RWY2.12 | A. fumigatus pyrG::gpdA(p)::rsmA, veA | This study |
| RSA15.2 | A. fumigatus pyrG::gpdA(p)::rsmA, ΔaflR::argB, veA | This study |
| RWY16.76 | A. fumigatus pyrG::gpdA(p)::rsmA, ΔaflR::argB, trpC801, veA | This study |
| TWY16.12 | pyrG89, A. fumigatus pyroA::gpdA(p)::aflR, pyroA4, ΔnkuA::argB, veA | This study |
| RWY20.3 | A. fumigatus pyroA::gpdA(p)::aflR, veA | This study |
| pSH96 | ½ trpC in pBSSK- | Wieser and Adams (1995) |
| pTMH44.2 | gpdA(p)::gfp frag::trpC(t) | McDonald et al. (2005) |
| pKLD116 | 6XHis, MBP in pET21a(+) | Rocco et al. (2008) |
| pWY20 | rsmA in pKLD116 | This study |
| pWY43.23 | aflR and promoter in pSH96 | This study |
| pWY44.6 | aflR and aflR(p)R in pSH96 | This study |
| pWY45.2 | aflR and aflR(p)Y1 in pSH96 | This study |
| pWY46.9 | aflR and aflR(p)R,Y1 in pSH96 | This study |
| pWY25.16 | A.fumigatus pyroA::gpdA in pGEMT easy vector | This study |
R: TGACACA site mutated to AAACAGG; Y1: TTACTAA site mutated to TAAGTTA
RXX = ascospore recombinant, TXX = original transformant
Fungivore food choice
Circular glass fiber filters discs (MN85/70 BF), 1.0 cm in diameter, provided the matrix for the experimental patches. Autoclaved discs were soaked in liquid malt extract agar (30 g l−1 standard malt extract, 5 g l−1 soy peptone, 20 g l−1 agar). After hardening, each disc was inoculated with 2 μl conidia suspension (1000 conidia per μl). The discs were transferred to non-vented Petri dishes and incubated at 20 °C and constant darkness for four days prior to food choice experiments. Choice assays were conducted in non-vented 9 cm Petri dishes. Four days after inoculation WT and OE::rsmA patches were placed in each Petri dish (n = 20). Fungal patches were connected by a wet piece of filter paper (length: 5 cm, width: 1.5 cm). Both the position of the patches relative to one another (left/right) in each arena and the position of the arenas in a 2 × 10 grid were randomized. Then 25 F. candida per arena were released in the middle of the filter paper. Subsequently, fungivores that were found to stay on the WT or the OE::rsmA patch were counted every 24 hours after the release of the collembolans into the arenas. The experiment was stopped after the five days and the number of fecal pellets per patch was counted to provide a measure of feeding intensity.
By subtracting the proportion of fungivores found on OE::rsmA patches from the proportion of animals on WT patches for each arena we obtained “Δ patch choice” values. Values around zero indicate no preferences, whereas positive (max. +1) or negative (min. −1) values indicate preference for the WT or the OE::rsmA respectively. We ran a time series mixed model using the MIXED procedure provided by SAS 9.2 (see Trienens et al. 2010). As a post hoc procedure we tested for significant deviation from “Δ patch choice” values = 0, i.e. no preference for either patch. Variation in the number of fecal pellets was analyzed by means of a paired t-test as data were found to be normally distributed.
Microarray analysis
Sample labeling was performed as previously described (Gasch, 2002) using cyaninedyes (Amersham), Superscript III (Invitrogen, Carlsbad, CA), and amino-allyl-dUTP (Ambion, Austin, TX). Custom A. nidulans FGSC A4 4X expression arrays were designed by NimbleGen. Arrays were hybridized in a NimbleGen hybridization system 12 (BioMicro, Salt Lake City, UT), washed, and scanned using a scanning laser (GenePix 4000B, Molecular Devices). Hybridization, washing and scanning were performed according to NimbleGen protocols (http://www.nimblegen.com/). Data normalization and statistical analyses on biological triplicates were performed using Bioconductor (Gentleman et al. 2004) and custom perl scripts. The affy package (Gautier et al. 2004) was used to apply probe-level quantile normalization to the log2 signal of RNA versus genomic DNA control. Gene-level expression changes were summarized with the median value of each probe set contained completely within each predicted ORF (open reading frame). Genes with significant expression differences in OE::rsmA as compared to WT were identified by performing paired t-tests using the Bioconductor package Limma v2.9.8 (Smyth, 2004) with a false discovery rate (FDR) correction of 0.05 (Storey and Tibshirani, 2003). The group of genes significantly induced in OE:rsmA was analyzed for enrichment of upstream sequence motifs using multiple em for motif elicitation (MEME; Bailey and Elkan 1994). MEME parameters were motif distribution, zoops; minimum width, 6; maximum width, 12; maximum number of motifs, 5.
Gene cloning, plasmid construction and genetic manipulation
The plasmids utilized in this work are listed in Table 1. The oligonucleotide sequences for PCR primers and gel shift assay are given in Table S1 and Table 2, respectively. PCR amplification was carried out on a C1000™ Thermal Cycler from Bio-Rad (Hercules, CA). For creation of rsmA overexpression (OE) strain at native locus, the OE cassette was constructed by using single and double joint PCR procedures (Yu et al. 2004). Single joint PCR reaction was set up for fusion of the maker gene A. fumigatus pyrG (1.97 kb) which was amplified from genomic DNA of A. fumigatus and A. nidulans glyceraldehyde-3-phosphate dehydrogenase gene (gpdA) promoter (1.5 kb) which was amplified from pTMH44.2 (See Table S1 for primer sequences). Then, the 1.03 kb fragment upstream of rsmA and 1.1 kb fragment containing rsmA ORF and downstream were amplified from genomic DNA of A. nidulans using designated primers (Table S1), respectively. These three amplified PCR products were purified with a QIAquick gel extraction kit (Qiagen, Valencia, CA), quantified, and fused using double joint PCR procedures. The final PCR product was amplified with the primer pairs OErsmA_NEST_for and _rev, confirmed with endonuclease digestion and purified for fungal transformation. All PCR steps were performed using an Expand long template PCR system (Roche, Indianapolis, IN) according to the manufacturer’s instructions. Using the same strategy as creation of OE::rsmA cassette, OE::aflR cassette was constructed with A. fumigatus pyroA as select maker gene and A. nidulans gpdA as promoter. To construct the bacterial overexpression plasmid, the entire coding region of rsmA was amplified from cDNA library of A. nidulans by using primers HisMBPRsmA_for and rev. The rsmA fragment was then integrated into plasmid pKLD116 (Rocco et al. 2008) (Table 2) to create pWY20 by using QuickChange strategy (Stratagene, Wilmington, DE). This construct was confirmed by PCR with primers pKLD116_for and _rev and then sequenced. RsmA has in-frame codons for six histidines maltose binding protein (MBP) at the N-terminus as well as a TEV (tobacco etch virus) site for cleaving fusion protein. To detect RsmA binding DNA activity in vivo, we created auxotrophic strain RWY16.76 (A. fumigatus pyrG::gpdA(p)::rsmA, ΔaflR::argB, trpC801, veA) by crossing TWY5.2 (OE::rsmA) with RJH256 (ΔaflR) according to standard methods (Pontecorvo et al. 1953). Similarly, prototrophic strain was constructed by crossing RAMB38 to TWY5.2 to construct RSA15.2 (OE::rsmA, ΔaflR). Crossing TWY16.12 with RDIT55.37 created RWY20.3 (OE::aflR) prototroph. The progeny’s genotypes were determined by growth on select media and PCR confirmation with designated primers (Table S1). The plasmids which were used for ΔaflR complementation in the OE::rsmA background were constructed in two steps. Firstly, a 2.4-kb fragment including the original promoter of aflR, ORF and 0.5 kb downstream of the stop codon was amplified by using designated primers (Table S1) and digested by using restriction enzymes BamH I and EcoR I. Then, the BamH I-EcoR I fragment of aflR was cloned into BamH I and EcoR I sites of the half trpC-containing plasmid pSH96 to create pWY43.23. Secondly, plasmids pWY44.6, pWY45.2 and pWY46.9 were constructed by multiple-site mutation in the promoter of aflR using designated primers (Table S1) according to QuickChange Multi Site-directed Mutagenesis approach (Stratagene, Wilmington, DE). All plasmids were confirmed by sequencing. Fungal protoplast preparation and transformation were carried out as described by Bok and Keller (2004). Each five micrograms of the double-joint cassette was used to overexpress rsmA and aflR by using A. nidulans strain RJMP1.49 as the recipient host. For RsmA binding assay in vivo, each ten micrograms of plasmids pWY43.23, pWY44.6, pWY45.2 and pWY46.9 was used for complementation of ΔaflR using A. nidulans strain RWY16.76 as the recipient host, respectively. Overexpression and complementation strains were verified by PCR and Southernblot analysis.
Table 2.
Oligonucleotides utilized for gel shift assay
| Name | Oligonucleotide sequence (5′-3′) | Uses |
|---|---|---|
| AflR EMSA_F | CGACTGACACAAGAAATAACAATTC | RsmA site probe |
| AflR EMSA_R | GAATTGTTATTTCTTGTGTCAGTCG | RsmA site probe |
| AflR EMSA mu_F | CGACAAACAGGAGAAATAACAATTC | Mutated RsmA site probe |
| AflR EMSA mu_R | GAATTGTTATTTCTCCTGTTTGTCG | Mutated RsmA site probe |
| TRX2 site2_for | GATCCTCTTTTCTTACTAAGCGCG | Yeast Yap 1 probe |
| TRX2 site2_rev | GATCCGCGCTTAGTAAGAAAAGAG | Yeast Yap 1 probe |
| aflR_Yap_for | ACAGACTTTTTAGTAATGGCGACTC | A. nidulans Yap 1 probe |
| aflR_Yap_rev | GAGTCGCCATTACTAAAAAGTCTGT | A. nidulans Yap 1 probe |
| aflR_Yapmu2_for | ACAGACTTTTAAGTTATGGCGACTC | A. nidulans mutated Yap 1 probe |
| aflR_Yapmu2_rev | GAGTCGCCATAACTTAAAAGTCTGT | A. nidulans mutated Yap 1 probe |
Nucleic acid analysis
Plasmid preparation, digestion with restriction enzyme, gel electrophoresis, blotting, hybridization, and probe preparation were performed by standard methods (Sambrook et al. 1989). Aspergillus DNA for diagnostic PCR was isolated using the described method previously (Lee and Taylor, 1990). Sequence data were analyzed using the LASERGENE software package from DNASTAR.
Expression and purification of RsmA
E. coli BL21(DE3) cells were transformed with plasmid pWY20. Cells were grown at 37°C in LB medium containing 25 mg ml−1 kanamycin to an OD600 of 0.6. Then, 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added into media for induction at 4 time points (1.5 h, 3 h, 4.5 h and 16 h) under two different temperatures (25 °C and 37 °C). The bacterial cells were harvested and lysed by freeze-thawing and cell debris removed by centrifugation. Cell lysates were examined for recombinant RsmA expression by SDS-PAGE followed by Coomassie blue staining. The best expression condition (3 h induction at 25 °C) was used for RsmA protein preparation (Fig. S5). Recombinant RsmA was purified by using the Ni-NTA resin (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Gel shift assay
The complementary oligonucleotide pairs (Table 2) were annealed in a Thermal Cycler (Bio-Rad, Hercules, CA) following heating at 95 °C for 15 min by cooling 1 °C per minute until the temperature dropped to 25 °C. MBP protein was obtained by cleavage of recombinant RsmA with TEV protease (10127-017) according to the protocol (Invitrogen, Carlsbad, CA) and used as control. The DNA-protein binding reaction was conducted in a 24 μl reaction mixture including 1 μg of poly (dI:dC), 3 μg of purified protein, 3 μg of bovine serum albumin (BSA) and [λ-32P]-ATP oligonucleotide probes with T4 polynucleotide kinase (NEB, Ipswich, MA). The binding buffer contained 8 mM Tris (pH 8.0), 24 mM HEPES, 12 % glycerol, 2 mM EDTA and 1 mM DTT (ditheiothreitol). This mixture was incubated for 20 min at room temperature in the presence of the radiolabeled probe and resolved on a 5% acrylamide gel (Bio-Rad, Hercules, CA) that had been prerun at 110 V for 40 min with 1% Tris-borate-EDTA buffer. The loaded gel was run at 140 V for 60–90 min and then wrapped with plastic and placed on Storage Phosphor screen cassette (GE, Madison, WI) for exposure. After 2 h, the cassette was scanned at Storm 860 phosphoimager (Molecular Dynamics, Sunnyvale, CA) for imaging.
Northern analysis
We examined expression of ST cluster gene transcripts by Northern analysis. Fifty milliliter of liquid GMM were inoculated with 106 spores/ml of OE::rsmA (RWY2.12) and WT (RDIT9.32) strains and incubated with shaking at 250 rpm at 37 C. After 24, 36, and 48 hours, the mycelium was collected and total RNA was extracted using the TRIzol reagent according to the instructions (Invitrogen, Carlsbad, CA). For expression assessment of strains of aflR at trpC locus (TWY18.14, TWY19.15, TWY20.4 and TWY21.20), only the time point of 48 hours was used. Blots were hybridized with fragment from each of of rsmA, aflR, aflJ and stcU which were individually amplified from RDIT9.32 genomic DNA with appropriate primers (Table S1). The rest of the cultures were extracted for ST analysis (See below). All experiments were performed in duplicate. Detection of signals was carried out with a Storm 860 phosphoimager (Molecular Dynamics, Sunnyvale, CA).
Sterigmatocystin (ST) analysis
Five microliter of 105 spores of A. nidulans strains were point inoculated onto solid glucose minimal medium (GMM) and incubated for 4 days at 37°C (Shimizu et al. 2001). An equal size agar plug, 7 mm in diameter, was removed from the center of each plate culture, homogenized in 3 ml dd. water and extracted with an equal amount of chloroform by agitation for 30 minutes at room temperature. The chloroform extracts were then dried completely at room temperature and resuspended in 100 μl of chloroform. Metabolites were separated in the developing solvent toluene:ethyl acetate:acetic acid (TEA, 8:1:1) on silica-coated thin-layer chromatography (TLC) plates (Shwab et al. 2007) and photographs were taken following exposure to UV radiation at 254 and 366 nm wavelengths.
Fermentation and LC/MS analysis
A. nidulans strains were cultivated at 37°C on solid YAG (5 g/l yeast extract, 15 g/l agar and 20 g/l d-glucose supplemented with 1 ml/l of a trace element solution) at 1.0 × 105 spores per 10 cm plate in the dark or under lights. After 7 days, three 7 mm diameter agar plugs were taken from each strain and transferred to a 10 ml vial. The plugs were extracted with 2 ml of MeOH followed by 2 ml of 1:1 CH2Cl2/MeOH each with 1 h sonication. The organic extract was transferred to a 7 ml new vial, in which the organic solvents were evaporated by TurboVap LV (Caliper Life Sciences) to dryness. The crude extract was then re-dissolved in 0.2 ml of DMSO:MeOH (1:4). After filtration, 10 μl of DMSO/MeOH extract was injected for high performance liquid chromatography-photodiode array detection- mass spectrometry (HPLC-DAD-MS) analysis as described previously (Bok et al., 2009).
For determining fold differences, negative ion electrospray ionization (ESI) was used for the detection of asperthecin and positive mode was used for the detection of ST by using extracted ion chromatogram (EIC) at m/z 317 and 325, respectively. The fold differences were calculated according to the following formula: [Area(Sample) − Area(Blank)]/[Area(WT)−Area(Blank)].
Supplementary Material
Acknowledgments
This work was supported by NIH grant PO1GM084077 from the National Institute of General Medical Sciences to N.P.K. and C.C.W. A. P. G. was supported by NIH grant NIGMS R01GM083989-01. M.R. was supported by a DFG research grant RO3523/3-1. We thank Scott Klasek for helping on physiological experiments.
References
- Alarco AM, Raymond M. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J Bacteriol. 1999;181:700–708. doi: 10.1128/jb.181.3.700-708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, Hagiwara D, Yamashino T, Mizuno T. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci Biotechnol Biochem. 2007;71:1800–1803. doi: 10.1271/bbb.70133. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymer. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Balazs A, et al. AtfA bZIP-type transcription factor regulates oxidative and osmotic stress responses in Aspergillus nidulans. Mol Genet Genomics. 2010;283:289–303. doi: 10.1007/s00438-010-0513-z. [DOI] [PubMed] [Google Scholar]
- Bayram Ö, Braus GH. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev. 2011 doi: 10.1111/j.1574–6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- Bayram Ö, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, Horn U, Hertweck C, Brakhage AA. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol. 2010;76:8143–8149. doi: 10.1128/AEM.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Genomic mining for Aspergillus natural products. Chem Biol. 2006;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Braus GH, Irniger S, Bayram Ö. Fungal development and the COP9 signalosome. Curr Opin Microbiol. 2010;13:672–676. doi: 10.1016/j.mib.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. Twenty-five coregulated transcripts define a ST gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK. Aspergillus parasiticus crzA, which encodes calcineurin response zinc-finger protein, is required for aflatoxin production under calcium stress. Int J Mol Sci. 2008;9:2027–2043. doi: 10.3390/ijms9102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, Ehrlich KC, Yu JJ, Bhatnagar D, Cleveland TE. Increased expression of Aspergillus parasiticus AflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl Environ Microbiol. 1995;61:2372–2377. doi: 10.1128/aem.61.6.2372-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol Genet Genomics. 2003;268:711–719. doi: 10.1007/s00438-003-0809-3. [DOI] [PubMed] [Google Scholar]
- Chiang YM, et al. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol. 2010;76:2067–2074. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Obrian GR, Payne GA. Function and regulation of aflJ in the accumulation of aflatoxin early pathway intermediate in Aspergillus flavus. Food Addit Contam. 2007;24:1043–1050. doi: 10.1080/02652030701513826. [DOI] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M, Keller NP, Adams TH. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol Microbiol. 1998;28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- Gasch AP. Yeast genomic expression studies using DNA microarrays. Methods Enzymol. 2002;350:393–414. doi: 10.1016/s0076-6879(02)50976-9. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy - analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunst K, Chinnici JP, Llewellyn GC. Effects of aflatoxin B1, aflatoxin B2, aflatoxin G and ST on viability, rates of development, and body length in two strains of Drosophila melanogaster (Diptera) J Invertebr Pathol. 1982;39:388–394. doi: 10.1016/0022-2011(82)90064-7. [DOI] [PubMed] [Google Scholar]
- Hagiwara D, Asano Y, Yamashino T, Mizuno T. Characterization of bZip-Type transcription factor AtfA with reference to stress responses of conidia of Aspergillus nidulans. Biosci Biotechnol Biochem. 2008;72:2756–2760. doi: 10.1271/bbb.80001. [DOI] [PubMed] [Google Scholar]
- Keller NP, Hohn TM. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- Labrousse H, Matile L. Toxicological biotest on Diptera larvae to detect ciguatoxins and various other toxic substances. Toxicon. 1996;34:881–891. doi: 10.1016/0041-0101(96)00045-1. [DOI] [PubMed] [Google Scholar]
- Lee SB, Taylor JW. In: In PCR protocols: A guide to methods and applications. Innis MA, editor. Academic Press; San Diego; USA: 1990. pp. 282–287. [Google Scholar]
- Llewellyn GC, Gee CL, Sherertz PC. Toxic responses of developing fifth instar milkweed bugs, Oncopeltus fasciatus (Hemiptera), to aflatoxin B1. Bull Environ Contam Toxicol. 1988;40:332–338. doi: 10.1007/BF01689088. [DOI] [PubMed] [Google Scholar]
- Matasyoh JC, Dittrich B, Schueffler A, Laatsch H. Larvicidal activity of metabolites from the endophytic Podospora sp. against the malaria vector Anopheles gambiae. Parasitol Res. 2011;108:561–566. doi: 10.1007/s00436-010-2098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald T, Brown D, Keller NP, Hammond TM. RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant Microbe Interact. 2005;18:539–545. doi: 10.1094/MPMI-18-0539. [DOI] [PubMed] [Google Scholar]
- Niu G, Siegel J, Schuler MA, Berenbaum MR. Comparative toxicity of mycotoxins to navel orangeworm (Amyelois transitella) and corn earworm (Helicoverpa zea) J Chem Ecol. 2009;35:951–957. doi: 10.1007/s10886-009-9675-8. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Qiao JJ, et al. Afyap1, encoding a bZip transcriptional factor of Aspergillus fumigatus, contributes to oxidative stress response but is not essential to the virulence of this pathogen in mice immunosuppressed by cyclophosphamide and triamcinolone. Med Mycol. 2008;46:773–782. doi: 10.1080/13693780802054215. [DOI] [PubMed] [Google Scholar]
- Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pousada C, Menezes RA, Pimentel C. The Yap family and its role in stress response. Yeast. 2010;27:245–258. doi: 10.1002/yea.1752. [DOI] [PubMed] [Google Scholar]
- Rohlfs M, Albert M, Keller NP, Kempken F. Secondary chemicals protect mould from fungivory. Biol Lett. 2007;3:523–525. doi: 10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze LV, Chanda A, Wee J, Awad D, Linz JE. Stress-related transcription factor AtfB integrates secondary metabolism with the oxidative stress response in Aspergilli. J Biol Chem. 2011;286:35137–35148. doi: 10.1074/jbc.M111.253468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA: 1989. [Google Scholar]
- Schroeckh V, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban MI, Bok JW, Lauer C, Keller NP. Suppressor mutagenesis identifies a Velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot Cell. 2010;9:1816–1824. doi: 10.1128/EC.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab EK, et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Staaden S, Milcu A, Rohlfs M, Scheu S. Fungal toxins affect the fitness and stable isotope fractionation of Collembola. Soil Biol Biochem. 2010;42:1766–1773. [Google Scholar]
- Staaden S, Milcu A, Rohlfs M, Scheu S. Olfactory cues associated with fungal grazing intensity and secondary metabolite pathway modulate Collembola foraging behaviour. Soil Biol Biochem. 2011;43:1411–1416. [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome wide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CC, Oakley BR. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trienens M, Rohlfs M. Insect-fungus interference competition the potential role of global secondary metabolite regulation, pathway-specific mycotoxin expression and formation of oxylipins. Fung Ecol. doi: 10.1016/j.funeco.2011.07.009. (in press) [DOI] [Google Scholar]
- Trienens M, Rohlfs M. Experimental evolution of defense against a competitive mold confers reduced sensitivity to fungal toxins but no increased resistance in Drosophila larvae. BMC Evol Biol. 2011;11:206. doi: 10.1186/1471-2148-11-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trienens M, Keller NP, Rohlfs M. Fruit, flies and filamentous fungi -experimental analysis of animal-microbe competition using Drosophila melanogaster and Aspergillus mould as a model system. Oikos. 2010;119:1765–1775. [Google Scholar]
- Tsitsigiannis DI, Zarnowski R, Keller NP. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J Biol Chem. 2004a;279:11344–11353. doi: 10.1074/jbc.M310840200. [DOI] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Kowieski TM, Zarnowski R, Keller NP. Lipogenic signals act as developmental regulators of spore balance in Aspergillus nidulans. Eukaryot Cell. 2004b;3:1398–1411. doi: 10.1128/EC.3.6.1398-1411.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P, et al. FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol Microbiol. 2010;77:972–994. doi: 10.1111/j.1365-2958.2010.07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser J, Adams TH. flbD encodes a myb-like DNA binding protein that controls initiation of Aspergillus nidulans conidiophore development. Genes Dev. 1995;9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- Wilkinson HH, Ramaswamy A, Sim SC, Keller NP. Increased conidiation associated with progression along the sterigmatocystin biosynthetic pathway. Mycologia. 2004;96:1190–1198. [PubMed] [Google Scholar]
- Yang X, Talibi D, Weber S, Poisson G, Raymond M. Functional isolation of the Candida albicans FCR3 gene encoding a bZip transcription factor homologous to Saccharomyces cerevisiae Yap3p. Yeast. 2001;30:1217–1225. doi: 10.1002/yea.770. [DOI] [PubMed] [Google Scholar]
- Yin W, Keller NP. Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol. 2011;49:329–339. doi: 10.1007/s12275-011-1009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.