Abstract
Fidelity during chromosome segregation is essential to prevent aneuploidy. The proteins and chromatin at the centromere form a unique site for kinetochore attachment and allow the cell to sense and correct errors during chromosome segregation. Centromeric chromatin is characterized by distinct chromatin organization, epigenetics, centromere-associated proteins and histone variants. These include the histone H3 variant centromeric protein A (CENPA), the composition and deposition of which have been widely investigated. Studies have examined the structural and biophysical properties of the centromere and have suggested that the centromere is not simply a ‘landing pad’ for kinetochore formation, but has an essential role in mitosis by assembling and directing the organization of the kinetochore.
The canonical cell cycle is divided into interphase and mitosis (FIG. 1). During interphase, cells undergo growth (G1phase) and DNA replication (Sphase). After Sphase, cells undergo another phase of growth (G2phase) and prepare to enter mitosis (Mphase). During mitosis, the sister chromatids need to be accurately segregated to each daughter cell, thereby ensuring survival from one generation to the next. This is facilitated by a complex array of proteins that regulate the timing and accuracy of chromosome segregation. Chromosome segregation is directed by the centromere, a chromosomal locus that is required for mitosis and acts as the site of kinetochore formation. The proteinaceous kinetochore (BOX 1) ensures proper segregation by linking the chromosome to dynamic microtubules (composed of tubulin dimers), thereby forming the mitotic spindle. Because the centromere mediates chromosome segregation, it is essential that the cell forms only one centromere and associated kinetochore attachment per chromosome to prevent breakage, although organisms with holocentric chromosomes, such as Caenorhabditis elegans, have attachment sites spread throughout the length of the chromosome.
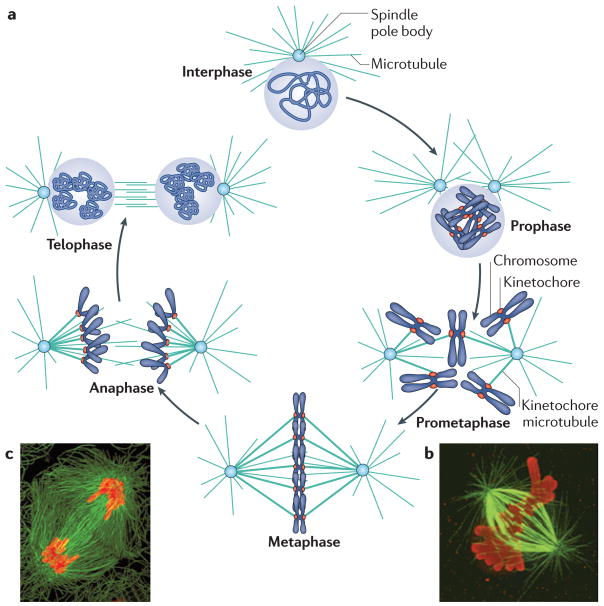
Figure 1. Chromosome segregation in the cell cycle.
a | The various stages of the cell cycle are depicted. During interphase, the cell undergoes growth and replication of the DNA. Upon replication of the spindle pole body and DNA, the cell undergoes a second round of growth and subsequently enters mitosis. Mitosis is divided into prophase (when the chromatin is condensed), prometaphase (when kinetochore microtubules start to interact with kinetochores), metaphase (when chromosomes become bi-oriented), anaphase (when the sister chromatids segregate to opposite spindle poles) and telophase (when chromosomes decondense). In most eukaryotes, the nuclear membrane degrades during mitosis and reforms during telophase, but this does not occur in budding yeast. b,c | Images of metaphase (b) and anaphase (c) cells. Chromosomes are shown in red and microtubules forming the mitotic spindle are shown in green. The proteinacious kinetochore forms at the centromere and mediates attachment to the spindle. Image in part b is reproduced, with permission, from REF. 173 © (2010) The Rockefeller University Press. Image in part c is reproduced, with permission, from REF. 174 © (2010) Macmillan Publishers Ltd. All rights reserved.
Box 1. The kinetochore.
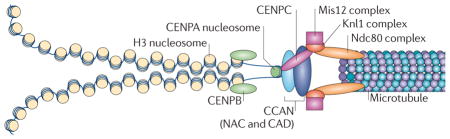
The kinetochore is a large, multiprotein complex that is needed to link the sister chromatids to the mitotic spindle during chromosome segregation. The physical organization of the kinetochore into a trilaminar structure170,171 is visible in electron microscopy images of vertebrate kinetochores, and the molecular composition follows this three-layer organization3,5,14,147,148,172. The kinetochore forms on the microtubule plus end as a ‘basket’ of elongated molecules (namely Ndc80) that recruit the outer- kinetochore components of the KMN network (which comprises the Knl1 complex, the Mis12 complex and the Ndc80 complex) (see the figure). These outer-kinetochore proteins dangle from the expanded basket surface generated by Ndc80 and can interact with the chromatin and proteins of the constitutively centromere-associated network (CCAN). The purpose of the basket is to allow the outer-kinetochore components to move over a greater distance and increase the likelihood of encountering an unattached centromere. The geometry of the centromeric protein A (CENPA)-containing chromatin predisposes CENPA to be at the surface (see main text) and to recruit the CENPA-containing nucleosome-associated complex (NAC) (TABLE1). The two halves of the kinetochore can then interact and form a stable attachment, connecting the chromosome to the microtubule. The kinetochore serves several important roles during chromosome segregation: it links chromosome movement to microtubule dynamics, monitors chromosome bi-orientation and serves as a site of catalysis for synchronizing chromosome segregation with cell cycle events.
CAD, CENPA-containing nucleosome distal; H3, histone H3.
Upon entry into mitosis, chromosomes condense, and the primary constriction forms at the centromere, the region of the chromosome defined by the incorporation of a histone H3 variant, centromeric protein A (CENPA)1,2. The kinetochore is recruited to the centromere, and signalling proteins such as cyclin-dependent kinases (CDKs) signal to microtubules originating at the centrosomes to form a bipolar spindle and attach to the kinetochores. This links the microtubules and chromosomes mechanically and through signalling processes that sense attachment and force to ensure that all chromosomes are bi-oriented and bound before anaphase onset (for more details on mitosis, see REFS 3–6).
Although much is known about the organization and structure of the kinetochore, the physical structures of centromeric chromatin and pericentromeric chromatin, and their contribution to fidelity during chromosome segregation, are not as clear. Recent studies have shed new light on a range of topics, including the composition of the centromeric nucleosome, the histone modifications and variants that are unique to the centromeric chromatin, and the physical organization of this region during mitosis. These studies have expanded our understanding of how the centromere functions to ensure segregation fidelity and prevent errors that lead to aneuploidy, which can ultimately lead to cancer and diseases such as Down’s syndrome, Edwards’ syndrome and Patau’s syndrome.
In this Review, we describe the distinct properties that define centromeric chromatin, including associated proteins, epigenetics and histone variants. We also examine the physical architecture of the chromatin and its spatial geometry, and discuss the mechanical and physical properties of the pericentric chromatin and the importance of maintaining the balance of forces during mitosis. From this work, it is evident that the centromere does not serve passively as a site (or ‘landing pad’) for kinetochore formation, but that the underlying chromatin itself dictates, in part, the geometry and function of the kinetochore and mitotic spindle and therefore has an essential role in mitosis. We do not focus on the kinetochore, which has been reviewed extensively elsewhere3,6,7.
The centromere
The centromere is a unique region on the chromosome that is required for attachment to the mitotic spindle and chromosome segregation. Simple eukaryotic organisms, such as budding yeast, have point centromeres, defined by a specific DNA sequence found on all chromosomes (see below), whereas more complex eukaryotes have larger regional centromeres that are defined by hierarchical arrays of satellite repeats. Many similarities between centromeres in different species have been observed that highlight the unique nature and essential functions of the centromere.
DNA at the centromere
The Saccharomyces cerevisiae centromere DNA was the first eukaryotic centromere to be isolated8 and is now known to have three conserved DNA sequences common to all chromosomes9, a feature not seen in higher eukaryotes (FIG. 2). These are termed centromere DNA element I (CDEI), CDEII and CDEIII, and together they form a 116–120 bp sequence that is sufficient to confer mitotic stability when introduced into plasmids8,10,11. CDEI is 8 bp long and is required for high-fidelity chromosome segregation; CDEII is a 78–86 bp AT-rich region that is required for chromosome segregation; and CDEIII is 26 bp long and contains seven invariant nucleotides conserved across all chromosomes, the mutation of which abolishes centromere function12. Because they are small, not complex and have a single microtubule attachment per chromosome, the centromeres of budding yeast and of the yeast Kluyveromyces lactis have been termed point centromeres.
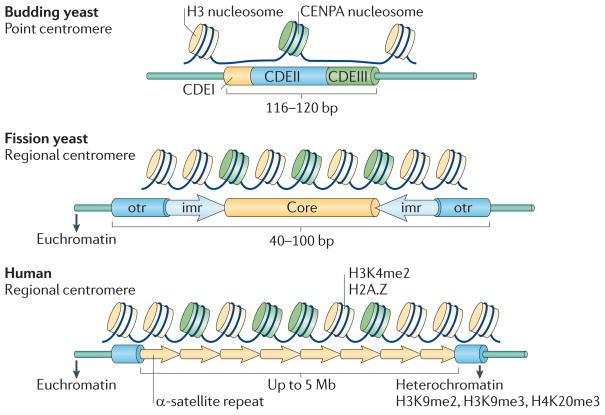
Figure 2. Characteristics of point and regional centromeres.
The point centromeres of budding yeast form a single microtubule attachment per chromosome, whereas larger regional centromeres form multiple attachments. The budding yeast centromere DNA is composed of the conserved centromere DNA element I (CDEI), CDEII and CDEIII. Larger regional centromeres do not contain DNA sequences, but the presence of a centromeric protein A (CENPA)-containing nucleosome is conserved. H3K4me2, histone H3 dimethylated at Lys4; H3K9me, histone H3 methylated at Lys9; H3K20me3, histone H3 trimethylated at Lys20; imr, inverted repeat sequence; otr, outer repeat.
By contrast, the regional centromeres of the fission yeast Schizosaccharomyces pombe are longer (40–100 kb)13, have multiple microtubule attachment sites per centromere14 and do not contain a conserved DNA sequence common to all chromosomes15–17. Instead, these centromeres are composed of central core region1 (cnt1), cnt2 and cnt3 bordered on either side by inverted repeat sequence L (imrL) and imrR, which are flanked by outer repeat L (otrL) and otrR18. These regions do not show DNA sequence homology to the centromere of budding yeast.
Centromeres have been identified in Candida albicans19, Neurospora crassa20, Arabidopsis thaliana21, Drosophila melanogaster22 and Homo sapiens23. The regional centromeres of higher eukaryotes are more difficult to study because they are large and contain arrays of tandem repeats. Indeed, human centromeres can be up to 5 Mb long and contain 1–4 Mb of 171-bp α-satellite repeats18,24,25.
Centromere-associated proteins
Despite large divergence in centromere DNA sequences across organisms, CENPs are highly conserved (see REF. 26 for a discussion of the evolutionary characteristics of centromeres). CENPs are not the only centromere-associated proteins; indeed, many others, for example the budding yeast proteins chromosome transmission fidelity3 (Ctf3) and Mif2, were discovered and named before identifying their centromeric associations. However, we focus on CENPA and CENPC, as they have been extensively studied and shown to be required to form centromeres and associated kinetochores; other constitutively centromere-associated network (CCAN) proteins are briefly described (TABLE1). The CCAN proteins are generally classified as part of either the CENPA-containing nucleosome-associated complex (NAC) or the CENPA-containing nucleosome distal (CAD) proteins27,28.
Table 1.
Centromere-associated proteins
| Subunit | Complex | Subcomplex | Homologues | |||
|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | Schizosaccharomyces pombe | Caenorhabditis elegans | Drosophila melanogaster | |||
| CENPA | NA | NA | Cse4 | Cnp1 | HCP-3 | CID |
| CENPB* | NA | NA | Unknown | Abp1, Cbh1, Cbh2 | Unknown | Unknown |
| CENPC‡ | CCAN|| | NA | Mif2 | Cnp3 | HCP-4 | CENPC |
| CENPM*‡ | CCAN | NA | Iml3 | Mis17 | Unknown | Unknown |
| CENPN‡ | CCAN | NA | Chl4 | Mis15 | Unknown | Unknown |
| CENPU*‡ (CENP50) | CCAN | NA | Unknown | Fta4 | Unknown | Unknown |
| CENPT*‡ | CCAN | CENPT–CENPW | Unknown | SPBC800 | Unknown | Unknown |
| CENPW | CCAN | CENPT–CENPW | Unknown | Unknown | Unknown | Unknown |
| CENPH*‡ | CCAN | CENPH | Mcm16 | Fta3 | Unknown | Unknown |
| CENPI§ | CCAN | CENPH | Ctf3 | Mis6 | Unknown | Unknown |
| CENPK§ | CCAN | CENPH | Unknown | Sim4 | Unknown | Unknown |
| CENPL§ | CCAN | NA | Unknown | Fta1 | Unknown | Unknown |
| CENPO§ | CCAN | CENPO | Mcm21 | Mal2 | Unknown | Unknown |
| CENPP§ | CCAN | CENPO | Ctf19 | Fta2 | Unknown | Unknown |
| CENPQ§ | CCAN | CENPO | Unknown | Fta7 | Unknown | Unknown |
| CENPR§ | CCAN | CENPO | Unknown | Unknown | Unknown | Unknown |
| CENPS§ | CCAN | CENPS | YOL86-A | Unknown | Unknown | Unknown |
| CENPX§ | CCAN | CENPS | Unknown | Unknown | Unknown | Unknown |
| CENPD | CCAN | NA | Unknown | Unknown | Unknown | Unknown |
| CENPG | CCAN | NA | Unknown | Unknown | Unknown | Unknown |
Proteins and protein complexes associated with the centromere in various species6,7,27,28,47,177. Abp1, actin-binding protein 1; Cbh, CENPB homologue; CCAN, constitutively centromere-associated network; CENP, centromeric protein; Chl4, chromosome loss 4; CID, Centromere identifier; Cnp, centromere protein 1; Cse4, chromosome segregation 4; Ctf, chromosome transmission fidelity 3; HCP, holocentric chromosome-binding protein; Iml3, increased minichromosome loss 3; Mcm, minichromosome maintenance; NA, not applicable; Sim4, silencing in the middle of the centromere 4.
Proteins that have been shown to be recruited directly by CENPA-containing nucleosomes27.
Proteins that are proposed to be part of the CENPA-containing nucleosome-associated complex.
Known as Ctf19 complex in S. cerevisiae or Sim4 complex in S. pombe.
CENPA is a centromere-associated protein that acts as a histone variant29 and is required to build a fully functioning kinetochore30–32. CENPA was shown to co-purify with nucleosome core particles33, which indicated that it probably forms a complex with the core histones. Homologues of CENPA have been identified in species from yeast to mammals. Complete loss of CENPA has been found to be lethal in every organism studied to date, although mammalian cells can tolerate a 90% reduction in the levels of CENPA.
CENPC is a DNA-binding protein that associates with the inner-kinetochore plate34. CENPC homologues have been identified in many model organisms, including yeasts, flies, plants and mammals, and it has been shown to be essential for proper progression through mitosis and chromosome segregation1,35–40. Indeed, loss of CENPC at the centromere has been shown in human cells to result in small or absent kinetochores39. CENPC localizes to CENPA-containing chromatin32, and this requires CENPA; reciprocally, CENPA localization requires CENPC40. CENPC binds two different groups of proteins that serve distinct functions: to the Mis12 complex41,42, which is part of the KMN network (which comprises the Knl1 complex, Mis12 complex and Ndc80 complex) of the outer kinetochore that is also needed for recruitment of checkpoint proteins41,42; and to other CCAN components, such as CENPH, CENPI, CENPK, and CENPT43. Furthermore, CENPC, together with CENPN, binds distinct domains in CENPA to direct the assembly of other centromere and kinetochore proteins43.
The CENPT–CENPW subcomplex is recruited to H3-containing centromeric chromatin and has been proposed to be an alternative to CENPC for the connection between the centromere and the kinetochore. The CENPT–CENPW subcomplex functions upstream of the CENPH subcomplex44, which comprises CENPH, CENPI and CENPK. These proteins are recruited to the centromere by CENPC43. They are essential for kinetochore function in vertebrates and have been found to play a part in CENPA loading and the recruitment of other, more distal centromere complexes45. The CENPO subcomplex (made up of CENPO, CENPP, CENPQ and CENPR) and the similar CENPU protein (also known as CENP50) are needed to prevent premature separation upon spindle damage46. The CENPS subcomplex (comprising CENPS and CENPX) is required for proper and stable formation of the outer kinetochore, and localization of these proteins to the centromere requires CENPT orCENPK47.
Histones at the centromere
Histone modifications and histone variants serve to demarcate unique regions of the chromosome, including the centromere. The centromeric DNA sequence is rapidly evolving and, as such, centromere function does not depend solely on primary DNA sequence but also on the presence of essential proteins, such as histone variants (including CENPA and H2A.Z), and on the local chromatin context as defined by distinct histone modifications.
Patterns of histone modifications at the centromere
The epigenetic specification of centromeres was first postulated by Earnshaw and Migeon48, with the observation that the two centromeres placed on a dicentric chromosome differed in their ability to direct chromatid segregation by the presence or absence of centromeric proteins. This indicated that additional proteins at the centromere are required to facilitate segregation. Position effect variegation studies have shown that genes placed adjacent to centromeres lead to stochastic inheritance of gene expression49. Subsequent studies have identified epigenetic factors that are required for centromere function in S. cerevisiae50, C. albicans51, S. pombe52 and H. sapiens53.
Chromatin can be flagged up for transcriptional activation (euchromatin) or repression (heterochromatin) through the modification of the amino-terminal tails of canonical histones by methylation, acetylation and phosphorylation13,54. Heterochromatin surrounding the centromere is known to contribute to sister chromatid cohesion and condensation55–60. At regional centromeres in organisms such as humans, mice, flies and fission yeast, the nucleosomes containing the canonical H3 histone (as opposed to the variant CENPA discussed below) are dimethylated at Lys4 (H3K4me2) (FIG. 2), a modification associated with euchromatin. This modification is thought to be important for the physical organization of the centromere61; indeed, depletion of H3K4me2 has been shown to result in a lack of recruitment of Holliday junction recognition protein (HJURP; see below), leading to failed incorporation of CENPA62.
The pericentric chromatin can also be defined by methylation63,64. For example, H3 and H4 in heterochromatic regions surrounding the centromere can be methylated at Lys9 (H3K9me2 and H3K9me3)2,63 and Lys20 (H4K20me3)65,66, and these modifications serve to recruit proteins such as cohesin67 and to maintain the structure of pericentric chromatin. The formation and maintenance of heterochromatin in pericentric chromatin has recently been shown to require the nucleolar remodelling complex in mice68. Interestingly, loss of the heterochromatic modification H4K20me3 is thought to result in aberrant centromere function, as in humans it has been associated with the presence of cancer cells69, which are characterized by a high degree of aneuploidy.
In addition to methylation (correlating with silenced chromatin), centromeric histones lack acetylation, which denotes actively transcribed chromatin. The hypoacetylation of the histones and the methylation of the DNA at the centromere70 alter the chemical interactions of the histones and DNA and define a region distinct from traditional euchromatin and heterochromatin2,61. These modifications maintain the heterochromatic nature of centromeric chromatin, which is thought to contribute to the physical structure of the centromere, as well as to sister chromatid cohesion and condensation55–60. Histone modifications are also essential for defining and maintaining the centromeric region. Recent evidence has shown that it is possible to engineer a human artificial chromosome (HAC) to assess the contribution of chromatin state adjacent to the centromere without perturbing all the centromeres of the cell71,72. These experiments reveal that altering the chromatin to a more closed state results in loss of essential centromere proteins as well as depletion of the H3K4me2 modification and accumulation of H3K9me3 (found in heterochromatin adjacent to the centromere)71. The loss of centromere function is not solely due to loss of CENPA; instead, the authors observed a hierarchical loss of various components of the inner centromere (including CENPC and CENPH)72.
CENPA, an H3 variant unique to the centromere
CENPA is a variant of histone H3, and the homology is found mainly at the α-helical carboxy-terminal histone-fold domain73. The N-terminal tail of CENPA is highly variable between species74 and is required to recruit kinetochore proteins to the centromere75. The CENPA centromere-targeting domain (CATD) within the histone-fold domain is known to be required for centromere targeting and function76,77. Work in S. cerevisiae has shown that specific residues within the CATD are required for the interaction of CENPA with suppressor of chromosome missegregation3 (Scm3)78. At point centromeres, such as those of budding yeast, a single CENPA-containing nucleosome forms the basis for kinetochore formation and microtubule attachment79. Larger regional centromeres have multiple CENPA-containing nucleosomes interspersed between canonical H3-containing nucleosomes80. Tetrasomes of CENPA and H4 have been found to be more rigid than the H3–H4 tetramer, a feature inherent in the structure of the histones76. Furthermore, CENPA-containing nucleosomes are more prone to unwrapping81 and releasing the H2A–H2B dimer81,82, which suggests that CENPA nucleosomes may be pliable, possibly to allow for easier removal from non-centromeric chromatin. These findings indicate that CENPA-containing nucleosomes are structurally distinct from canonical H3–H4-containing nucleosomes, and that they may facilitate the exposure of CENPA-containing nucleosomes on the outer surface of the centromere, as opposed to becoming buried within the bulk chromatin.
Since the structure of the canonical nucleosome octamer was published73, much work has been done to determine the structure of the centromeric nucleosome. A wide range of possibilities for the structure of the centromeric protein in vivo have been proposed, including an octamer in which histone H3 is replaced by CENPA27,80,81,83–85, a nucleosome with only four histones (tetrasome86 or hemisome87–89) or an alternative structure containing the non-histone protein Scm3 (REFS 88,90) (FIG. 3a). These different theories have been proposed in various model organisms, and more work will be needed to further understand differences in centromeric structure and to develop a cohesive model to reconcile these observations. For example, human centromeric nucleosomes are thought to be composed of eight histones, and most are homotypic octamers containing two copies of CENPA83. More recent work, however, has shown that about 10% of human centromeric nucleosomes form a heterotypic octamer containing one CENPA-containing histone and one canonical H3-containing histone27. Furthermore, work in D. melanogaster has found that different compositions of the centromeric nucleosome are present: a nucleosome containing at least two copies of Centromere identifier (CID; the D. melanogaster homologue of CENPA)40 and a hemisome structure composed of one copy each of H2A, H2B, H4 and CID87,89. These experiments found that purified crosslinked centromeric nucleosomes had a molecular mass equal to half of an octamer, and were calculated by atomic force microscopy to be half as high as would be expected for an octamer87,89. It is possible that these represent different stages of assembly or variations found at different stages of the cell cycle.
Figure 3. The CENPA-containing nucleosome.
a | Work in different organisms on the composition and physical properties (direction of DNA wrapping and supercoiling induction) of the centromeric nucleosome has given rise to a range of different possibilities: homotypic octamer (Saccharomyces cerevisiae84,85, Drosophila melanogaster80 and human cells27,80,81,83), heterotypic octamer (human cells27), reverse octamer (molecular dynamics102,175), homotypic tetramer or tetrasome (Schizosaccharomyces pombe86) and heterotypic tetramer or hemisome (S. cerevisiae88 and D. melanogaster87,88). Furthermore, two alternative structures that contain the non-histone protein suppressor of chromosome missegregation 3 (Scm3) have been proposed: hexasomes (S. cerevisiae90) and trisomes (S. cerevisiae and D. melanogaster88). b | Tension on the right-handed reverse-wrapped centromeric protein A (CENPA)-containing nucleosome may cause it to split into two hemisomes (histone H2A, H2B, CENPA and H4), and could serve as a site for the spindle assembly checkpoint to monitor attachment. HJURP, Holliday junction recognition protein. Images in part a are modified, with permission, from REF. 176 © (2011) Elsevier.
More controversial is the recent work in S. cerevisiae that has revealed a new hexameric nucleosome structure at the centromere that lacks H2A and H2B and instead contains Scm3 (REF. 90). This protein was first identified in a screen for suppressors of chromosome segregation4 (Cse4; the S. cerevisiae homologue of CENPA, which is known as Cnp1 in S. pombe) mutants and it was found to interact with the histone-fold domain of Cse4, but not the essential N-terminal domain84. Various recent studies in both budding and fission yeast have shown that Scm3 is a kinetochore protein that is needed to target Cse4 to the centromeric DNA and to ensure kinetochore function85,86,91–93. As such, it seems likely that the observed hexameric nucleosome represents a transitional state during assembly. Alternatively, it is possible that Scm3 is associated with, but not part of, the centromeric nucleosome, especially in light of recent work in budding yeast suggesting that binding of DNA to a Cse4-containing nucleosome is incompatible with Scm3 binding78. A homologue of Scm3 has not been identified in species other than S. cerevisiae and S. pombe, but functional and sequence analyses have shown that the mammalian HJURP and yeast Scm3 are orthologues that share the same functional domain and are required for proper deposition of CENPA94–98. Interestingly, Scm3 was shown to prevent the ubiquitylation of CENPA by Psh1 (POB3 and SPT16 histone-associated1), an E3 ubiquitin ligase that is thought to control the level of CENPA and remove misincorporated protein99,100.
The physical properties of centromeric nucleosomes are starting to emerge. Intriguing findings were obtained from the crystallization of a protein fragment of CENPA81. These experiments found that CENPA nucleosomes formed canonical octameric nucleosomes that wrapped DNA in a left-handed manner (FIG. 3a), resulting in negative supercoiling. However, the region of CENPA cleaved before crystallization was the flexible, and highly variable, N-terminal region, and it is unclear what effect the removal of this portion of the protein has on the subsequent structure of the nucleosome81. Furthermore, these experiments were carried out using a plasmid lacking a centromeric DNA sequence and without any histone chaperones. Histone chaperones, such as RBAP48 (also known as RBBP4) or Scm3, are needed to assemble CENPA-containing nucleosomes in vitro87,101,102, and this assembly is known to induce positive supercoiling88. Thus, it is likely that the structure of the centromeric nucleosome is not exclusively dictated by the histone particles themselves, but by the underlying DNA and histone chaperones as well.
By contrast, other studies have indicated that CENPA-containing nucleosomes induce positive supercoils87,88, which result from right-handed wrapping of the DNA around the nucleosome. Positive supercoiling would serve to differentiate the centromere from the negatively supercoiled bulk chromatin. The experiments examining the effects of CENPA-containing nucleosomes on chromatin were carried out by in vitro reconstitution of D. melanogaster proteins using circular mini-chromosomes. DNA wrapping around nucleosomes can be detected by changes in plasmid supercoiling following protein removal. The supercoiling is topologically defined by linking number, which is the sum of twist and writhe. In this study, the linking number was changed in the presence of CENPA, which could be explained by altered supercoiling; however, this could also be explained by loss of a nucleosome. If the centromeric nucleosome is positively supercoiled, under tension it may split into two hemisomes88 or become more tightly wrapped in response to the applied force102. The positive supercoiling of this nucleosome would focus the spindle tension on the centromere, and could serve as a place for the checkpoint to monitor attachment (FIG. 3b).
The H2A.Z variant at the centromere
CENPA-containing nucleosomes contain canonical H2A, whereas the variant H2A.Z is associated with nucleosomes containing H3K4me2 (and to a smaller extent H3K9me3) in the centromere of human and mouse cells64 (although the H2A.Z variant is not unique to the centromere). H2A.Z is one of the most studied histone variants, and the structure of the H2A.Z-containing nucleosome has been solved by X-ray crystallography103 and was found to be similar to the canonical H2A-containing nucleo-some73. Interestingly, however, the interaction between H2A.Z and the H3–H4 tetramer is destabilized owing to the differences in amino acid sequence between H2A and H2A.Z. Furthermore, an acidic region on the surface of the H2A.Z-containing nucleosomes allows interactions with non-histone proteins103 and serves as a signpost to direct chromatin-remodelling factors. H2A.Z also functions as a boundary between heterochromatin and euchromatin by antagonizing silencing. The H2A.Z-containing nucleosomes are more resistant to condensation104–107 and form a boundary between heterochromatin and euchromatin. Indeed, loss of the H2A.Z homologue in yeast (Htz1) leads to spreading of the silencing factors silent information regulator2 (Sir2), Sir3 and Sir4, which affects centromere function by perturbing the structural organization of the chromatin and aberrant gene expression throughout the genome.
Assembling the centromere
Various hypotheses have been proposed to explain how the cell identifies and maintains the centromeric region, including the presence of CENPA on the parental chromatid83, chromatin tension and conformation108, and heterochromatin modifications in the pericentromere52,109.
CENPA loading
The timing of CENPA loading varies across different species. In animals such as humans and D. melanogaster, CENPA is loaded between anaphase and G1 (REFS 38,110–112), plants load CENPA in late G2 (REFS 113,114), and fission and budding yeast load CENPA in S–G2 (REFS 115,116). Much work recently has centred on identifying the proteins responsible for loading and maintaining CENPA at the centromere117.
In addition to the CATD, various proteins have known roles in CENPA loading, including the human proteins RBAP46 (also known as RBBP7) and RBAP48 (which is similar to S. pombe Mis16 (REF.118)), CENPH and CENPI45,119. Furthermore, Scm3 and its mammalian orthologue, HJURP (which is part of the CENPA prenucleosomal complex27,120), are also thought to have a role in CENPA loading. Work in S. pombe has shown that Scm3 localization to the centromere requires Mis16 and Mis18 (REFS 86,93), and it is thought that Scm3, Mis16 and Mis18 act as assembly factors to bring and load CENPA at the centromere. These data have led to a multistep model for centromere histone loading, distinguishing between licensing and loading117,121. Licensing is proposed to occur in humans by the recruitment of MIS18 (Mis16–Mis18 in S.pombe), CENPH, CENPI, RBAP46 and RBAP48 (REFS 98,119), followed by the recruitment of loading factors, such as KNL2 (REF.60) and HJURP (Scm3 in yeast)95,98, to load new CENPA into centromeric chromatin, and proper spacing is maintained by the remodelling and spacing factor complex121. Further work into the targeting, loading and proper incorporation of CENPA will continue to develop our understanding of this field.
Histone chaperones serve many functions, including recruiting, loading and removing histone proteins from chromatin, but do not form part of the chromatin itself. The histone chaperone complexes chromatin assembly factor 1 (CAF1) and histone regulator (HIR) are known to be required for H3 and H4 deposition and to affect chromatin structure at silent and centromeric loci in budding yeast122,123. Recently, CAF1 and HIR have been implicated in preventing extra-centromeric incorporation of CENPA by regulating histone eviction124 (the removal of inappropriately incorporated histones). Another histone chaperone, anti-silencing function1 (ASF1), works together with CAF1 and HIR to deposit H3 variants in human cells125, although it is not clear whether this interaction is needed for CENPA loading. Interestingly, Sir1 has been found to be present at budding yeast centromeres and to bind to a component of CAF1, helping retain it there126. This role for Sir1 in budding yeast indicates that, although point and regional centromeres have different mechanisms for forming and maintaining heterochromatin, both form unique chromatin architecture to maintain fidelity in chromosome segregation.
Chromatin remodelling at the centromere
Various chromosome-remodelling complexes function at the centromere to maintain the unique chromatin architecture that underlies the proper functioning of the centromere. Topoisomerase II is thought to function at the centromere in an ATP-dependent manner to remove topological linkages (decatenation), to create these linkages so that sister chromatids stay together (catenation) and to maintain chromatin packaging when the pericentric chromatin is under tension to provide balancing inwards force (discussed below)127. The CCAN (TABLE1) has been shown to affect chromatin structure in the pericentromere independently of CENPA44. The RSC (remodels the structure of chromatin) complex in budding yeast is known to localize to the pericentromere and to maintain the chromatin structure flanking the centromeric nucleosomes128. The loss of this remodelling activity results in loss of fidelity in chromosome segregation and suggests that the architecture found at the centromere is needed to facilitate chromosome segregation. This activity is also observed in humans: the switch–sucrose nonfermentable (SWI–SNFB) remodelling complex is related to the RSC complex of budding yeast and also localizes to kinetochores during mitosis129. Further research into the roles of remodelling and chromatin-modifying complexes at the centromere will advance our understanding of the structure of the centromeric chromatin and how this is maintained.
Architecture of centromeric chromatin
The physical organization of the chromatin plays an important part in preventing aneuploidy during mitosis. The cohesin and condensin complexes, which are enriched in the pericentric region130, have been identified as major contributors to the physical organization and packaging of the mitotic chromosome.
Tension and bi-orientation of centromeric chromatin
Packaging proteins, including cohesin and condensin, serve to organize the large amounts of DNA inside the nucleus into manageable and discrete units that can be successfully segregated. In addition, these proteins allow the cell to sense bi-orientation of the chromosomes and generate tension across the centromeric DNA. Cohesin is composed of structural maintenance of chromosomes1 (SMC1), SMC3, SCC1 and SCC3, and is needed to maintain sister chromatid cohesion. Condensin is composed of SMC2, SMC4 and other non-SMC proteins, and is needed to compact the chromosome and prevent tangles of duplex DNA, termed catenations, which can impede accurate chromosome segregation. Cohesin and condensin have been extensively reviewed recently131–133, so we focus on the contributions of these proteins to organizing the centromeric chromatin.
Cohesin and condensin are enriched at the pericentric region130, a fact that confused researchers for years, as sister centromeres are separated during metaphase when visualized in vivo. How could proteins that specifically bind DNA together be enriched in a region where they seem to be both separated and transiently together? Two models have been proposed to reconcile these observations: transient disassociation of cohesin from pericentric chromatin134, and the formation of intramolecular loops in the pericentromere, as shown in S.cerevisiae135.
Bi-orientation of sister chromatids during mitosis and tension generation by the mitotic spindle are both thought to require cohesin. It is essential that the chromatin does not break when under tension from the mitotic spindle, and this tension is accommodated by cohesin, together with the chromatin itself136,137. In addition to balancing tension, cohesin is important to organize the geometry of the centromere, ensuring that sister centromeres (and associated kinetochores) face opposite spindle poles and bi-orient138. Proper bi-orientation is essential for survival to ensure that the sister chromatids are equally segregated to daughter cells. Two probably overlapping methods have been proposed to promote amphitelic attachment: error correction and geometric bias139,140. However, the relative contributions of these mechanisms remain unclear.
Error correction promotes detachment in the absence of tension, and this is accomplished by Aurora B (increase-in-ploidy1 (Ipl1) in S. cerevisiae). It is thought that Aurora B is spatially confined and that phosphorylation by Aurora B facilitates detachment of incorrect attachments, whereas the tension generated across correct attachments physically separates them from Aurora B activity5,141,142.
The underlying geometry of the centromere (see below) is thought to facilitate segregation by exposing the centromere on the outer surface of the chromosome. Given the high level of conservation of proteins across different species, it is likely that this geometry displays similar conservation.
Geometric organization of the centromere
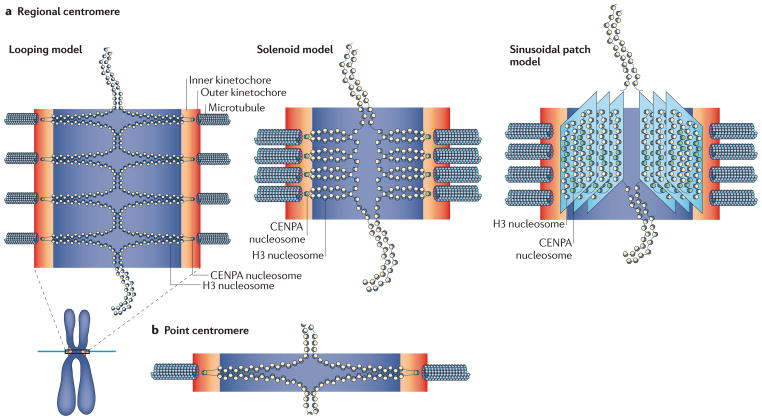
Various models have been proposed for the geometric organization of the eukaryotic centromere, including the looping model87,135,143, the solenoid model2,80,144 and the sinusoidal patch model145 (FIG.4a). The looping model proposes that the pericentric chromatin is looped out from the bulk chromatin towards the spindle pole, whereas the solenoid model proposes that the pericentric chromatin forms a coil with the CENPA-containing nucleosomes facing the spindle pole. The sinusoidal patch model attempts to explain the observed location of various CCAN proteins and the unfolding of the vertebrate kinetochore145.
Figure 4. Chromatin geometry at the centromere.
a | Three models for the organization of the regional centromere have been proposed: the looping model87,135,143, the solenoid model2,80,144 and the sinusoidal patch model145. The looping model proposes that the pericentric chromatin is looped out from bulk chromatin towards the spindle pole. The solenoid model proposes that the pericentric chromatin forms a coil with centromeric protein A (CENPA)-containing nucleosomes facing the spindle pole. The sinusoidal patch model attempts to explain the observed location of various constitutively centromere-associated network (CCAN) proteins and the unfolding of the vertebrate kinetochore. b | The budding yeast pericentromere adopts a cruciform structure, which serves to place the centromere (and therefore the kinetochore) on the poleward-facing side of the chromosomes135. We equate the multiple loops of the looping model in part a to the whole mitotic spindle of budding yeast. H3, histone H3.
These models all propose an organization that would favour CENPA-containing nucleosomes (and therefore kinetochore formation) facing the spindle pole to facilitate microtubule attachment to the kinetochore at the centromere. The sinusoidal patch model also allows for H3-containing nucleosomes to be present on the surface of the centromeric chromatin, where they have been shown to interact with the CENPT–CENPW subcomplex44. The heterochromatin would face inwards towards the sister chromatid, which would provide the physical basis for generating tension across chromosomes and serve as a geometric bias for bi-orientation.
Although geometric orientation increases the likelihood of correct amphitelic attachment, it is not essential and is dispensable for bi-orientation136,140. By contrast, tension is needed to correct erroneous microtubule–kinetochore attachments136. Tension is generated across the centromeres of sister chromatids when they are attached to opposite sides of the mitotic spindle, and this tension allows the cell to sense and correct erroneous attachments. Several lines of evidence show that CENPA is interspersed with canonical H3 and heterochromatin, and that CENPA must be aligned outwards towards the spindle pole bodies to allow kinetochore formation and microtubule attachment, which supports the three models mentioned above.
We propose that the whole budding yeast mitotic spindle serves as a model for a single regional centromere with multiple microtubule attachments per chromosome143, and that the cruciform structure found at budding yeast centromeres is analogous to the looping model for more complex centromeres (FIG. 4b). The cruciform structure of the pericentromere places the centromeres at the apex of the intramolecular loop loaded with cohesin, maximizing the distance between sister centromeres and thus reconciling increased cohesin and maximal spot separation during mitosis135. Further work has revealed that the formation of this structure is promoted by the DNA-binding components of the kinetochore146. This function is likely to be inherent in the structure of the proteins. For example, the S. cerevisiae protein Ndc10 (also known as Cbf3a) is needed to form the looping cruciform structure; it is thought to bind as a dimer, and it is possible that these dimers serve to bring two regions of chromatin together to form a loop. Given the high level of conservation in composition between yeast and higher eukaryotic kinetochores14,147–150, one view is that multiple binding site kinetochores of regional centromeres are repeats of the basic kinetochore of budding yeast, as proposed by the repeated subunit hypothesis143. However, electron microscopy work has suggested that the mammalian kinetochore is disorganized and lacks the recurring subunits proposed by the repeated subunit hypothesis151,152. A view that reconciles these perspectives is that the inner-kinetochore–centromere interface resembles a woven fabric, rather than two separate fixed structures.
In addition to the effects of the kinetochore, histone variants at the centromere also contribute to the geometry of the centromere. Work in human cell lines has shown that the incorporation of H2A.Z and epigenetic modifications contribute to the spatial organization of the centromere, although the authors do not distinguish between a solenoid and a looping model64. Furuyama and Henikoff88 showed that reconstituted CENPA-containing nucleosomes from D. melanogaster wrap DNA in a right-handed manner, opposite to that of canonical H3-containing nucleosomes (FIG. 3a). The reverse-wrap nucleosome and the proposed hemisome structure87,89 combine to suggest that centromeric chromatin is packaged in a manner that is fundamentally different from underlying heterochromatin, promoting the idea that these regions are excluded from the normal packaging of the chromosome144. This exclusion would serve to separate the centromere from the bulk chromatin to allow the formation of the kinetochore and increase the likelihood of microtubule capture. In addition, the relative enrichment of cohesin and condensin at the centromere suggests a unique physical architecture that allows stable attachment to the mitotic spindle and movement to maintain force balance during metaphase. As the microtubules exhibit dynamic instability and are always in a state of lengthening or shortening, the pericentric chromatin must allow this movement while remaining attached. The geometry of the centromere would promote interactions between the centromere and kinetochore, beyond serving simply as a fixed site of attachment.
Genome-wide assays for chromatin looping have uncovered a high level of nuclear organization and have shown that centromeric regions tended to not interact over longer ranges and formed a cluster of centromeres lasting throughout, and facilitating progression of, the cell cycle153. Visualization experiments correspond to these results and have shown that centromeres cluster together throughout the cell cycle. These data all suggest that centromeric chromatin forms geometry that promotes clustering of the centromeres at the surface of the chromosome, which reduces the region that the microtubules must ‘search’ to attach to the centromeres.
Polymer physics at the centromere
The molecular architecture of the centromere — the ‘parts list’ — and the in vivo interactions between the many protein complexes that function to load and remodel the centromere, as well as aid in chromosome segregation, continue to be comprehensively studied. In addition to the valuable data these studies provide, polymer physics can be applied to the mitotic spindle to understand chromosome segregation154. Often these concepts seem counterintuitive, and it is essential to remember that the scale and forces at work inside the cell are not immediately obvious to us in our everyday lives.
Chromosome segregation by entropy
Entropy (S) is a measure of the distribution of energy in a system, and can be defined by the equation S= kB ln(W), in which kB is Boltzmann’s constant (4.1 pN × nm at room temperature), ln is the natural logarithm and W is the number of possible ways that the molecule or polymer can occupy the space155. From this equation, we see that, as the number of possible conformations for a polymer increases, the entropy will also increase, which is energetically favourable and pushes the system in that direction.
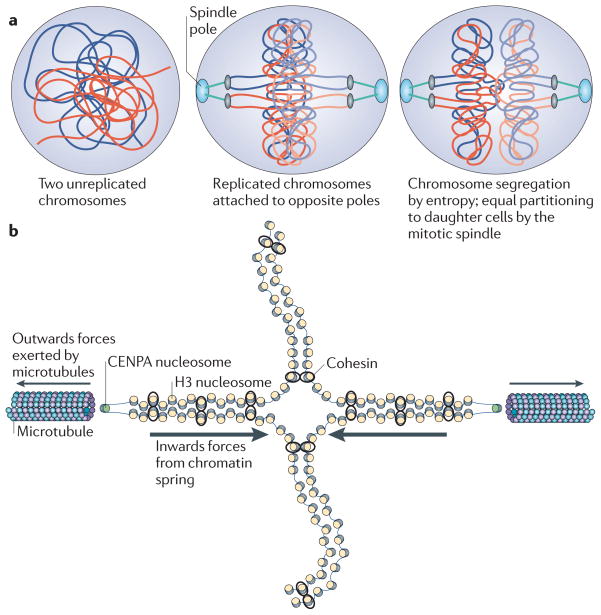
Entropy has been proposed to drive segregation of molecules such as chromatin (for example, in bacteria156,157). This may seem counterintuitive. However, if one considers two long connected chains (such as chromatin) in a confined space (such as the nucleus), the mixing of these chains will reduce the number of possible entropic states, whereas chain segregation will increase the number of possible conformations for each chain, making this an entropically favourable process. It is possible that the mitotic spindle serves to direct chromosome segregation during mitosis, ensuring that the proper complement of chromosomes goes to each daughter cell, but that entropic forces are responsible for segregation of the bulk chromatin (FIG. 5a).
Figure 5. Applying the principles of polymer physics to chromosome segregation.
a | Entropic forces drive the segregation of bulk chromatin. This is because it is energetically favourable for the polymers to segregate, as this allows them to adopt higher entropic states. The mitotic spindle apparatus provides directionality for this segregation and ensures that sister chromatids are equally segregated to daughter cells. b | It is important that the forces present at the mitotic spindle remain balanced to prevent breakage of the chromatin while maintaining tension along the chromatin (to sense bi-orientation). The microtubules exert an outwards force (towards the spindle pole), whereas the chromatin maintains an inwards force and is flexible enough to accommodate microtubule-based tension. CENPA, centromeric protein A; H3, histone H3.
Furthermore, entropy is thought to have a role in removing entanglements between larger eukaryotic chromosomes before anaphase onset158,159. Even though sister chromatids are proposed to form two distinct structures, cohesin continues to hold these together until anaphase, so entropy resolves topological entanglements between chromosomes to ensure that chromosomes can segregate faithfully. Modelling of larger eukaryotic chromosomes has also shown that a force, termed depletion-attraction, results in looping of chromosomes160,161 (for example, during transcription or in a proposed centromeric structure135), which in turn increases the entropic segregation of chromosomes owing to the increased repulsive forces between them162. Depletion-attraction force is generated when two large particles (in this case, separated chromosomal regions) are brought together and interact, which results in an increase in the space available for other smaller particles to occupy.
At the size scale inside the nucleus, forces such as viscosity and thermal motion dominate, whereas weight and inertia are less relevant. The maximal forces generated by microtubules on chromosomes have been measured to be 47 pN of force per microtubule163,164. This number represents the stall force, and the force required to segregate the chromosome is thought to be much lower (~5 pN). Assuming that the mitotic spindle directs chromosome segregation and serves to harness thermal motion to drive segregation, the forces generated by the mitotic spindle on the chromatin must exceed the forces generated by thermal motion. To explain this concept, chromosome movement in thermal motion can be compared to pulling a boat in the ocean. A person (the mitotic spindle) must generate enough force on the boat (the chromosomes) to exceed the forces of the waves (thermal motion) to pull the boat in a certain direction, and the waves will continue to push the boat in that direction.
Maintaining chromatin–spindle force balance
Although much work has been done to examine the roles of packaging complexes, such as cohesin165–167 in maintaining chromosome packaging under tension from the mitotic spindle, the contribution of the chromatin itself in maintaining the balance of forces during mitosis is not well studied (FIG. 5b). Chromatin must be able to resist the forces applied by the microtubules by stretching instead of breaking, and it has been shown that reduction of the chromatin packaging (by depleting histones) is needed to maintain spindle length in response to spindle tension in budding yeast168. The effects of pulling or pushing a material are quantified in its Young’s modulus (E), which represents the relationship of stress to strain and is measured in pascals (1 Pa = 1 N/m2 = 1 kg/m × s2). DNA and microtubules have a similar Young’s modulus, in the order of 1–2 GPa, which is similar to that of hard plastics. Although it has been more challenging to determine the Young’s modulus of the whole chromosome because it is not readily reconstituted outside of the cell, experiments have measured it to be between40 Pa and 400 Pa164,169.
Thus, it seems that the relative flexibility of chromatin could be acting as a buffer or ‘shock absorber’ to temper the forces generated by the spindle on the chromatin to prevent DNA breakage but still allow for tension-sensing mechanisms to correctly establish bi-orientation. Further work into the geometry and packaging of the pericentric chromatin will elucidate the role of the chromatin spring in balancing the outwards forces of the microtubules.
The future
The centromere is an essential site on every eukaryotic chromosome, and errors in this can lead to a wide range of diseases, including cancer and aneuploidy-related disorders such as Down’s syndrome, and to death. Although the level of complexity and underlying chromatin sequence varies across model organisms, a fundamental group of proteins, including the histone variant CENPA, is needed for attachment and function of the centromere. The structure, deposition and regulation of the centromeric nucleosome continue to be studied in great detail. CENPA variants have been identified in a wide range of eukaryotic organisms, but the structure of the nucleosome seems to vary by organism, and it will be important to understand the effects and implications of these differences. The geometry and physical properties of the pericentric chromatin are essential to our understanding of how chromosome segregation occurs, and we must continue to expand our understanding of the forces involved. Future work will incorporate the contributions of various fields to develop a comprehensive model of centromere–kinetochore attachment and the nature of the centromeric and pericentromeric chromatin. Approaching chromosome segregation from an interdisciplinary viewpoint — by combining the molecular and the mechanical properties of the centromere and kinetochore — will allow a better understanding of how fidelity in segregation is maintained.
Acknowledgments
We thank the members of the Bloom laboratory for discussions and editorial suggestions. We also thank the anonymous referees for their useful comments. We apologize to those authors whose work was not included owing to space limitations. This work was supported by the US National Institutes of Health.
Glossary
- Kinetochore
A multiprotein complex that assembles on centromeric DNA and mediates the attachment and movement of chromosomes along the microtubules of the mitotic spindle
- Holocentric chromosomes
Chromosomes lacking a localized centromere and primary constriction site. In holocentric chromosomes, kinetochores are diffuse and kinetochore microtubules attach along the length of the chromosome
- Centrosomes
Specialized organelles that duplicate during interphase and that constitute the centre of the mitotic spindle
- Centromeric chromatin
The chromatin where centromeric protein A is incorporated, underlying the kinetochore
- Pericentromeric chromatin
The chromatin flanking the centromeric chromatin
- Nucleosome
The basic structural subunit of chromatin, which consists of ~147base pairs of DNA wrapped ~1.7times around an octamer of histones (2copies each of H2A, H2B, H3 and H4)
- Satellite repeats
Specific DNA sequences that are repeated many times in long tandem arrays
- Dicentric chromosome
A chromosome that carries two centromeres, which arise from the aberrant fusion of ‘naked’ telomeres or interstitial double-strand breaks. These can also be experimentally generated by inserting a second conditional centromere into a chromosome
- Position effect variegation
Variable expression of DNA sequence based on temporal or quantitative effects from adjacent chromatin; for example, if an active gene is relocated to a heterochromatic region, it can randomly be silenced
- Tetrasomes
(Homotypic tetramers). Proposed nucleosome structures found at the centromere and composed of two copies each of centromeric protein A and H4
- Hemisome
(Heterotypic tetramer). A proposed nucleosome structure found at the centromere and composed of one copy each of H2A, H2B, centromeric protein A and H4
- Homotypic octamers
In the context of nucleosome composition, octamers in which both copies of H3 have been replaced by centromeric protein A
- Heterotypic octamer
In the context of nucleosome composition, an octamer in which only one copy of H3 has been replaced by centromeric protein A
- Amphitelic attachment
Connection of sister kinetochores to microtubules that emanate from opposite spindle pole bodies
- Intramolecular loop
A loop of chromatin formed by bringing distant regions of the same sister chromatid together, as opposed to intermolecular interactions between sister chromatid pairs. It is the proposed structure of the pericentromeric chromatin in budding yeast
- Entropy
A thermodynamic property related to the state of disorder of a system
- Thermal motion
The random motion and collision of particles owing to temperature
- Young’s modulus
(Also known as elastic modulus). A measure of the stiffness of a polymer, measured as stress divided by strain
- Stress
In the context of polymer physics, stress is defined as the force per unit area and measures how a material responds to external force
- Strain
In the context of polymer physics, strain measures the deformation of the material, measured as change in length over length (ΔL/L)
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Kerry Bloom’s homepage:
http://www.bio.unc.edu/bloom/lab
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Moore LL, Roth MB. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nature Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheeseman IM, Desai A. Molecular architecture of the kinetochore–microtubule interface. Nature Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 4.Bouck DC, Joglekar AP, Bloom KS. Design features of a mitotic spindle: balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu Rev Genet. 2008;42:335–359. doi: 10.1146/annurev.genet.42.110807.091620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. A comprehensive review of the structure and functions of the kinetochore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukagawa T, De Wulf P. In: The Kinetochore — From Molecular Discoveries to Cancer Therapy. De Wulf P, Earnshaw WC, editors. Springer Science + Business Media; New York: 2009. pp. 133–191. [Google Scholar]
- 7.Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- 8.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 9.Clarke L, Carbon J. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature. 1983;305:23–28. doi: 10.1038/305023a0. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 11.Hieter P, et al. Functional selection and analysis of yeast centromeric DNA. Cell. 1985;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 12.McGrew J, Diehl B, Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan BA. In: The Kinetochore — From Molecular Discoveries to Cancer Therapy. De Wulf P, Earnshaw WC, editors. Springer Science + Business Media; New York: 2009. pp. 45–76. [Google Scholar]
- 14.Joglekar AP, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chikashige Y, et al. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 16.Clarke L, Amstutz H, Fishel B, Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baum M, Ngan VK, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pidoux AL, Allshire RC. Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res. 2004;12:521–534. doi: 10.1023/B:CHRO.0000036586.81775.8b. [DOI] [PubMed] [Google Scholar]
- 19.Sanyal K, Baum M, Carbon J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci USA. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centola M, Carbon J. Cloning and characterization of centromeric DNA from Neurospora crassa. Mol Cell Biol. 1994;14:1510–1519. doi: 10.1128/mcb.14.2.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Copenhaver GP, et al. Genetic definition and sequence analysis of Arabidopsis centromeres. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 24.Maio JJ. DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J Mol Biol. 1971;56:579–595. doi: 10.1016/0022-2836(71)90403-7. [DOI] [PubMed] [Google Scholar]
- 25.Choo KH. Domain organization at the centromere and neocentromere. Dev Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 26.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 27.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nature Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 28.Panchenko T, Black BE. The epigenetic basis for centromere identity. Prog Mol Subcell Biol. 2009;48:1–32. doi: 10.1007/978-3-642-00182-6_1. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howman EV, et al. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nature Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitoh H, et al. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 35.Dawe RK, Reed LM, Yu HG, Muszynski MG, Hiatt EN. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell. 1999;11:1227–1238. doi: 10.1105/tpc.11.7.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukagawa T, Regnier V, Ikemura T. Creation and characterization of temperature-sensitive CENP-C mutants in vertebrate cells. Nucleic Acids Res. 2001;29:3796–3803. doi: 10.1093/nar/29.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogura Y, Shibata F, Sato H, Murata M. Characterization of a CENP-C homolog in Arabidopsis thaliana. Genes Genet Syst. 2004;79:139–144. doi: 10.1266/ggs.79.139. [DOI] [PubMed] [Google Scholar]
- 38.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 39.Tomkiel J, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erhardt S, et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Okada M, et al. The CENP-H–I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 46.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amano M, et al. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- 49.Ekwall K. Epigenetic control of centromere behavior. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- 50.Mythreye K, Bloom KS. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- 52.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128:647–650. doi: 10.1016/j.cell.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 54.Glynn M, Kaczmarczyk A, Prendergast L, Quinn N, Sullivan KF. Centromeres: assembling and propagating epigenetic function. Subcell Biochem. 2010;50:223–249. doi: 10.1007/978-90-481-3471-7_12. [DOI] [PubMed] [Google Scholar]
- 55.Bernard P, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 56.Giet R, Glover DM. Drosophila Aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendzel MJ, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 59.Jager H, Rauch M, Heidmann S. The Drosophila melanogaster condensin subunit Cap-G interacts with the centromere-specific histone H3 variant CID. Chromosoma. 2005;113:350–361. doi: 10.1007/s00412-004-0322-4. [DOI] [PubMed] [Google Scholar]
- 60.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunleavy E, Pidoux A, Allshire R. Centromeric chromatin makes its mark. Trends Biochem Sci. 2005;30:172–175. doi: 10.1016/j.tibs.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 62.Bergmann JH, et al. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci USA. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 67.Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Naure Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 68.Guetg C, et al. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–2146. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nature Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 70.Zhang W, Lee HR, Koo DH, Jiang J. Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell. 2008;20:25–34. doi: 10.1105/tpc.107.057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakano M, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. Describes the use of a HAC to study the effects of altering centromeric chromatin on kinetochore formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cardinale S, et al. Hierarchical inactivation of a synthetic human kinetochore by a chromatin modifier. Mol Biol Cell. 2009;20:4194–4204. doi: 10.1091/mbc.E09-06-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 74.Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr Opin Genet Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 75.Van Hooser AA, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- 76.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 77.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Z, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011 Mar;16 doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sekulic N, Bassett EA, Rogers DJ, Black BE. reveals physical The structure of (CENP-A–H4)2 features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conde e Silva N, et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 83.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y, et al. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. Reports the right-handed wrapping of CenH3 nucleosomes in vitro using reconstituted D. melanogaster proteins on circular minichromosomes, and proposes that this alternative right-handed wrapping serves to keep the centromere decondensed and accessible for kinetochore formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 91.Camahort R, et al. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 92.Stoler S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pidoux AL, et al. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 96.Aravind L, Iyer LM, Wu C. Domain architectures of the Scm3p protein provide insights into centromere function and evolution. Cell Cycle. 2007;6:2511–2515. doi: 10.4161/cc.6.20.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci USA. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Foltz DR, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hewawasam G, et al. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ranjitkar P, et al. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin invitro. Proc Natl Acad Sci USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lavelle C, et al. Right-handed nucleosome: myth or reality? Cell. 2009;139:1216–1217. doi: 10.1016/j.cell.2009.12.014. author reply 1217–1218. [DOI] [PubMed] [Google Scholar]
- 103.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 104.Bruce K, et al. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 106.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 107.Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nature Struct Biol. 2002;9:172–176. doi: 10.1038/nsb767. [DOI] [PubMed] [Google Scholar]
- 108.Mellone BG, Allshire RC. Stretching it: putting the CEN(P-A) in centromere. Curr Opin Genet Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 109.Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. J Cell Biol. 2001;153:101–110. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lermontova I, et al. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lermontova I, Fuchs J, Schubert V, Schubert I. Loading time of the centromeric histone H3 variant differs between plants and animals. Chromosoma. 2007;116:507–510. doi: 10.1007/s00412-007-0122-8. [DOI] [PubMed] [Google Scholar]
- 115.Takahashi K, Takayama Y, Masuda F, Kobayashi Y, Saitoh S. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Phil Trans R Soc B Biol Sci. 2005;360:595–606. doi: 10.1098/rstb.2004.1614. discussion 606–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pearson CG, et al. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- 117.Mellone BG, Zhang W, Karpen G. Frodos found: behold the CENP-A “Ring” bearers. Cell. 2009;137:409–412. doi: 10.1016/j.cell.2009.04.035. A thorough review of research regarding Scm3, its relationship to HJURP and its role in CENPA loading. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 119.Fujita Y, et al. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 120.Kato T, et al. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- 121.Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lopes da Rosa J, Holik J, Green EM, Rando OJ, Kaufman PD. Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics. 2011;187:9–19. doi: 10.1534/genetics.110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Galvani A, et al. In vivo study of the nucleosome assembly functions of ASF1 histone chaperones in human cells. Mol Cell Biol. 2008;28:3672–3685. doi: 10.1128/MCB.00510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sharp JA, Krawitz DC, Gardner KA, Fox CA, Kaufman PD. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 2003;17:2356–2361. doi: 10.1101/gad.1131103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee MT, Bachant J. SUMO modification of DNA topoisomerase II: trying to get a CENse of it all. DNA Repair (Amst) 2009;8:557–568. doi: 10.1016/j.dnarep.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hsu JM, Huang J, Meluh PB, Laurent BC. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol Cell Biol. 2003;23:3202–3215. doi: 10.1128/MCB.23.9.3202-3215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xue Y, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Glynn EF, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:e259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nature Rev Genet. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 133.Hirano T. At the heart of the chromosome: SMC proteins in action. Nature Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 134.Ocampo-Hafalla MT, Katou Y, Shirahige K, Uhlmann F. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma. 2007;116:531–544. doi: 10.1007/s00412-007-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- 137.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 138.Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. Details the link between cohesion at the centromere and kinetochore geometry in promoting bipolar attachment. [DOI] [PubMed] [Google Scholar]
- 139.Stumpff J, Asbury CL. Chromosome bi-orientation: euclidian euploidy. Curr Biol. 2008;18:R81–R83. doi: 10.1016/j.cub.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 140.Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 141.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J Cell Sci. 2010;123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Birchler JA, Gao Z, Han F. A tale of two centromeres—diversity of structure but conservation of function in plants and animals. Funct Integr Genomics. 2009;9:7–13. doi: 10.1007/s10142-008-0104-9. [DOI] [PubMed] [Google Scholar]
- 145.Ribeiro SA, et al. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci USA. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nature Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. Reports on the protein composition of the budding yeast kinetochore using quantitative fluorescence microscopy and proposes the architecture of a kinetochore–microtubule attachment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Joglekar AP, Bloom K, Salmon ED. Invivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. Determination of the organization of a human kinetochore using two-colour fluorescence light microscopy, proposing a mechanism of tension-sensing within the kinetochore that contributes to spindle assembly checkpoint activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Johnston K, et al. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nature Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McEwen BF, Dong Y. Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell Mol Life Sci. 2010;67:2163–2172. doi: 10.1007/s00018-010-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. Maps whole-genome organization of the budding yeast genome by capturing inter- and intra-chromosome interactions, illustrating the higher-order organization and clustering of the chromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bloom K. Centromere dynamics. Curr Opin Genet Dev. 2007;17:151–156. doi: 10.1016/j.gde.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 155.Grosberg AY, Khokhlov AR. Giant Molecules: Here, There, And Everywhere. Academic Press; San Diego: 1997. [Google Scholar]
- 156.Jun S, Wright A. Entropy as the driver of chromosome segregation. Nature Rev Microbiol. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Finan K, Cook PR, Marenduzzo D. Non-specific (entropic) forces as major determinants of the structure of mammalian chromosomes. Chromosome Res. 2010;19:53–61. doi: 10.1007/s10577-010-9150-y. [DOI] [PubMed] [Google Scholar]
- 158.Marko JF. Linking topology of tethered polymer rings with applications to chromosome segregation and estimation of the knotting length. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79:051905. doi: 10.1103/PhysRevE.79.051905. [DOI] [PubMed] [Google Scholar]
- 159.Koszul R, Kleckner N. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol. 2009;19:716–724. doi: 10.1016/j.tcb.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Marenduzzo D, Micheletti C, Cook PR. Entropy-driven genome organization. Biophys J. 2006;90:3712–3721. doi: 10.1529/biophysj.105.077685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bohn M, Heermann DW. Repulsive forces between looping chromosomes induce entropy-driven segregation. PLoS ONE. 2011;6:e14428. doi: 10.1371/journal.pone.0014428. Recent work that reports on the entropic forces between eukaryotic chromosomes and the effects of chromosome looping on transcriptional regulation, chromosome order and segregation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jannink G, Duplantier B, Sikorav JL. Forces on chromosomal DNA during anaphase. Biophys J. 1996;71:451–465. doi: 10.1016/S0006-3495(96)79247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. Measures forces produced by the mitotic spindle using a flexible glass needle on intact grasshopper spermatocytes to determine the force generated by the mitotic spindle on the chromosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang T, Lim HH, Cheng CS, Surana U. Deficiency of centromere-associated protein Slk19 causes premature nuclear migration and loss of centromeric elasticity. J Cell Sci. 2006;119:519–531. doi: 10.1242/jcs.02757. [DOI] [PubMed] [Google Scholar]
- 166.Chai CC, Teh EM, Yeong FM. Unrestrained spindle elongation during recovery from spindle checkpoint activation in cdc15-2 cells results in mis-segregation of chromosomes. Mol Biol Cell. 2010;21:2384–2398. doi: 10.1091/mbc.E09-07-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tanaka TU, Stark MJ, Tanaka K. Kinetochore capture and bi-orientation on the mitotic spindle. Nature Rev Mol Cell Biol. 2005;6:929–942. doi: 10.1038/nrm1764. [DOI] [PubMed] [Google Scholar]
- 168.Bouck DC, Bloom K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol. 2007;17:741–748. doi: 10.1016/j.cub.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Marshall WF, Marko JF, Agard DA, Sedat JW. Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr Biol. 2001;11:569–578. doi: 10.1016/s0960-9822(01)00180-4. [DOI] [PubMed] [Google Scholar]
- 170.Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell invitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- 171.Jokelainen PT. The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J Ultrastruct Res. 1967;19:19–44. doi: 10.1016/s0022-5320(67)80058-3. [DOI] [PubMed] [Google Scholar]
- 172.Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–655. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 173.Sedwick C. Ted Salmon: kinetochores at the core of it all. J Cell Biol. 2010;191:896–897. doi: 10.1083/jcb.1915pi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Canman JC, et al. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- 175.Bancaud A, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 176.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ohta S, et al. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010;142:810–821. doi: 10.1016/j.cell.2010.07.047. A thorough characterization of centromere-associated proteins using quantitative proteomics and bioinformatic analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]