Abstract
We investigate the folding mechanism of the WW domain Fip35 using a realistic atomistic force field by applying the Dominant Reaction Pathways approach. We find evidence for the existence of two folding pathways, which differ by the order of formation of the two hairpins. This result is consistent with the analysis of the experimental data on the folding kinetics of WW domains and with the results obtained from large-scale molecular dynamics simulations of this system. Free-energy calculations performed in two coarse-grained models support the robustness of our results and suggest that the qualitative structure of the dominant paths are mostly shaped by the native interactions. Computing a folding trajectory in atomistic detail only required about one hour on 48 Central Processing Units. The gain in computational efficiency opens the door to a systematic investigation of the folding pathways of a large number of globular proteins.
Keywords: atomistic simulations, protein folding
Unveiling the mechanism by which proteins fold into their native structure remains one of the fundamental open problems at the interface of contemporary molecular biology, biochemistry, and biophysics. A critical point concerns the characterization of the ensemble of reactive trajectories connecting the denatured and native states, in configuration space.
In this context, a fundamental question which has long been debated (1) is whether the folding of typical globular proteins involves a few dominant pathways; i.e., well defined and conserved sequences of secondary and tertiary contact formation, or if it can take place through a multitude of qualitatively different routes. A related important question concerns the role of nonnative interactions in determining the structure of the folding pathways (2, 3).
In principle, atomistic molecular dynamics (MD) simulations provide a consistent framework to address these problems from a theoretical perspective. However, due to their high computational cost, MD simulations can presently only be used to investigate the conformational dynamics of relatively small polypeptide chains, and are only able to cover time intervals much smaller than the folding times of typical globular proteins.
In view of these limitations, a considerable amount of theoretical and experimental activity has been devoted to investigate the folding of protein subdomains, which consist of only a few tens of amino acids, and fold on submillisecond time scales (4). In particular, a number of mutants of the 35 amino acid WW domain of human protein pin1 have been engineered which fold in few tens of microseconds (5). The mutant's small size and their ultrafast kinetics make them ideal benchmark systems, for which numerical simulations can be compared with a large body of experimental data (5–7).
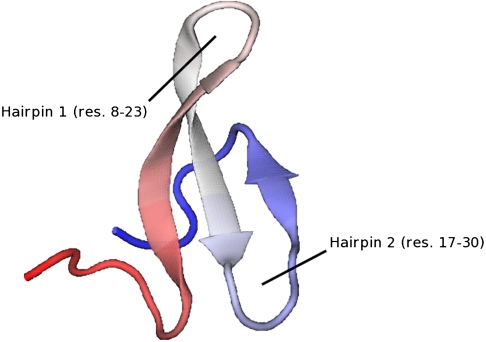
In particular, a MD simulation was performed to investigate the dynamics of a mutant named Fip35 (see Fig. 1), for a time interval longer than 10 μs. Unfortunately, in that simulation no folding transition was observed (8, 9).
Fig. 1.
Native structure of Fip35, a WW domain of the fip mutant of protein human pin1 (pdb code: pin1). The primary sequence of fip35 is: EEKLPPGWEKRMSADGRVYYFNHITNASQWERPSG.
The folding of this WW domain was later investigated by Pande and coworkers, using a world wide distributed computing scheme (10). According to this study the transition proceeds in a very heterogeneous way; i.e., through a multitude of qualitatively different and nearly equiprobable folding pathways.
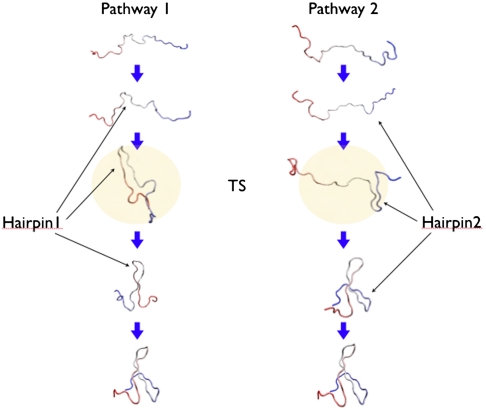
Noé, et al. performed a Markov state model analysis of a large number of short (≲200 ns) nonequilibrium MD trajectories (11) performed on the WW domain of human Pin 1 protein. In their paper the authors reported a complex network of transition pathways, which differ by the specific order in which the different local meta-stable states were visited. On the other hand, in all pathways the formation of hairpins takes place in a definite sequence (see e.g., Fig. 2). In particular, from the statistical model it was inferred that in about 30% of the folding transitions, the second hairpin forms first, as in the right box.
Fig. 2.
Schematic representation of the structure of the two folding pathways obtained in our DRP simulations.
A different conclusion has been reached by Shaw, et al., by analyzing a ms-long MD trajectory with multiple unfolding/refolding events, obtained using a special-purpose supercomputer (12). In that simulation the WW domain of Fip35 was found to fold and unfold predominantly along a pathway in which hairpin 1 is fully structured, before hairpin 2 begins to fold, as shown in the left box of Fig. 2. In a recent paper (13), Krivov reanalyzed the same ms-long MD trajectory in order to identify an optimal set of reaction coordinates. His conclusion was that the folding of this WW domain is thermally activated rather than incipient downhill and that the transition also occurs through a second pathway, in which hairpin 2 forms before hairpin 1. The statistical weights of the two pathways estimated from the number of folding events are 80% ± 20% and 20% ± 10%.
While all these theoretical studies yield folding times in rather good agreement with available experimental data on folding kinetics, they provide different pictures of the folding mechanism and raise a number of issues.
Firstly, it is important to assess the degree of heterogeneity of the folding mechanism and to clarify whether the most statistically significant folding pathways are those in which the hairpins form in sequence. Important related questions are also whether the folding mechanism is correlated with the structure of the initial denatured conditions from which the reaction is initiated and with the temperature of the heat bath. Finally, it is interesting to address the problem of the relative role played by native and nonnative interactions in determining the structure of folding pathways. Indeed, while native interactions are arguably shaping the dynamics in the vicinity of the native state, nonnative interactions may in principle play an important role in the transition region and at the rate limiting stages of the reaction.
In order to tackle these questions, in this work we use the Dominant Reaction Pathways (DRP) approach (14–18), a framework which allows to very efficiently compute the statistically most significant pathways connecting given denatured configurations to the native state at an atomistic level of detail, with realistic force fields. To further support our results and to study the role of native and nonnative interactions we map the free energy landscape by performing Monte Carlo simulations in two coarse-grained models.
Our study confirms that Fip35 folds mostly through the two folding channels discussed above and shows that the relative weight of the two channels changes with temperature. In addition, we find that the folding pathways are correlated with the initial condition from which the transition is initiated. The studies based on the coarse-grained model suggest that the folding dynamics in the transition region is not significantly influenced by nonnative interactions.
Methods
Atomistic Force Field.
Our atomistic simulations of the dominant folding trajectories of the Fip35 WW domain were performed using the AMBER ff99SB force field (19) in implicit solvent with Generalized Born formalism implemented in GROMACS 4.5.2 (20). The Born radii were calculated according to the Onufriev-Bashford-Case algorithm (21).
In a recent work based on the DRP method, the dominant pathway in the conformational transition of tetra-alanine obtained using the same version of the AMBER force field was found to agree well with the results of an analogous calculation in which the molecular potential energy was determined ab initio; i.e. directly from quantum electronic structure calculations (22).
Coarse-Grained Model.
To study the equilibrium properties of the folding of the Fip35 WW domain we used the coarse-grained model recently developed in refs. 23, 24. In that model, amino acids are represented by spherical beads centered at the Cα positions. The nonbonded part of the potential energy contains both native and nonnative interactions. The former are the same used in the G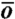 -type model of ref. 25, while the latter consist of a quasi-chemical potential, which accounts for the statistical propensity of different amino acids to be found in contact in native structures, and of a Debye-screened electrostatic term. In this model, the average potential energy due to native interactions in the folded phase is typically one order of magnitude larger than that due to nonnative interactions. Above the folding temperature, this ratio drops to about four.
-type model of ref. 25, while the latter consist of a quasi-chemical potential, which accounts for the statistical propensity of different amino acids to be found in contact in native structures, and of a Debye-screened electrostatic term. In this model, the average potential energy due to native interactions in the folded phase is typically one order of magnitude larger than that due to nonnative interactions. Above the folding temperature, this ratio drops to about four.
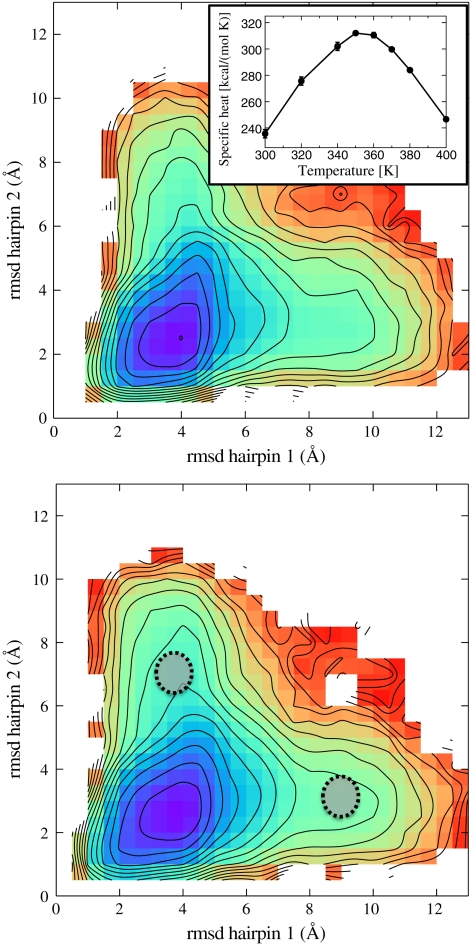
This model was shown to provide an accurate description of protein-protein complexes with low and intermediate binding affinities (23). In the insert of the upper box of Fig. 3 we plot the specific heat, evaluated from Monte Carlo (MC) simulations at different temperatures, which indicates that this model yields the correct folding temperature for this WW domain.
Fig. 3.
The free energy at T = 300 K as a function of the rmsd to native of the two hairpins, obtained from the Monte Carlo simulations in two coarse-grained models, described in Methods . In the upper box the model accounts for both native and nonnative interaction, in the lower box the model contains only native interactions. In the insert of the upper box, we show the corresponding plot of the specific heat. The two shaded regions in the lower box identify the average location of the transition states obtained from DRP simulations.
The Dominant Reaction Pathways Method.
The high computational cost of MD simulations of macromolecular systems has triggered efforts towards developing alternative theoretical frameworks to investigate their long-time dynamics and reaction kinetics [see e.g. (14, 26, 27, 28, 30, 31, 32) and references therein].
In particular, the DRP approach (14–18, 33) concerns physical systems which can be described by the overdamped Langevin equation. If xk denotes the coordinate of the k-th atom, the Langevin equation in the so-called Ito Calculus reads:
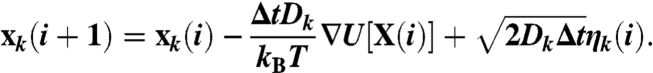 |
[1] |
In this equation, X(i) ≡ (x1(i),…,xN(i)) is the set of atomic coordinates at the i-th time step, Δt is an elementary time interval, Dk is the diffusion coefficient of the k-th atom, kB is the Boltzmann’s constant, T is the temperature of the heat-bath, and U(X) is the potential energy. ηk(i) is a white Gaussian noise with unitary variance, acting on the k-th atom.
The probability for a protein to fold in a given time interval t can be written as
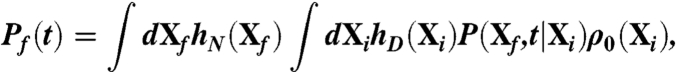 |
[2] |
where hN(D)(X) is the characteristic function of the native (denatured) state (defined in terms of some suitable order parameters), ρ0(Xi) is the initial distribution of micro-states in the denatured state, and P(Xf,t|Xi) is the conditional probability of reaching the (native) configuration Xf starting from the (denatured) configuration Xi, in a time t. If the total time interval t is chosen much smaller than the inverse folding rate, this probability is dominated by single nonequilibrium folding events.
It can be shown that the probability of a given folding trajectory X(t) connecting denatured and native configurations is proportional to the negative exponent of the Onsager-Machlup functional (31, 33), which in discretized form reads
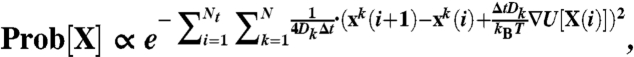 |
[3] |
where Nt is the number of time steps in the trajectory. On the other hand, the paths which do not reach native state before time t do not contribute to the transition probability in Eq. 2. The most probable —or so-called dominant— reaction pathways are those which minimize the exponent in [3]. In principle, these paths may be found by numerically relaxing the effective action functional (14)
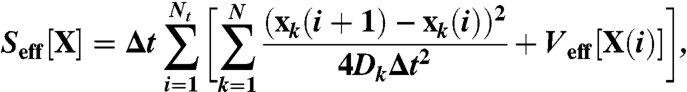 |
[4] |
where Veff(X) is the so-called effective potential, and reads
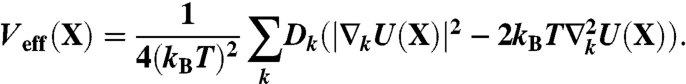 |
[5] |
In practice, for a protein folding transition, directly minimizing the effective action in Eq. 4 is unfeasible, because at least 104–105 time steps are needed to describe a single folding event. On the other hand, for any fixed pair of native and denatured configurations the dominant paths can be equivalently found by minimizing an effective Hamilton-Jacobi (HJ) action in the form (14, 33)
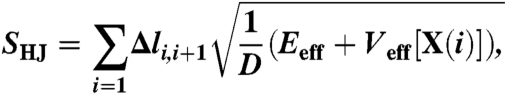 |
[6] |
where 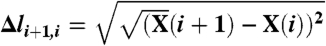 represents the elementary displacement in configuration space, and for sake of clarity, we have assumed that all atoms have the same diffusion coefficient. The parameter Eeff determines the time at which any given frame l of the path is visited (14):
represents the elementary displacement in configuration space, and for sake of clarity, we have assumed that all atoms have the same diffusion coefficient. The parameter Eeff determines the time at which any given frame l of the path is visited (14):
Hence, by adopting the HJ formulation of Eq. 6, it is possible to replace the time discretization with the discretization of the curvilinear abscissa l, which measures the Euclidean distance covered in configuration space during the reaction (33). This way, the problem of the decoupling of time scales is bypassed. As a result, only about 102 frames are usually sufficient to provide a convergent representation of a trajectory. On the other hand, the HJ formalism requires to perform an optimization in the space of reactive pathways of a functional, which can take complex values, which is in general a complicated task.
The DRP approach displays several differences with the SDEL (Stochastic Difference Equation in Length) method (30)—for an application to protein folding see also ref. 36. In particular, while the DRP is based on minimizing the effective HJ action in Eq. 6, in SDEL the folding trajectories are obtained by extremizing the physical HJ action 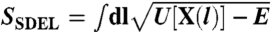 , where U(x) is the potential energy and E is the total mechanical energy, which is assumed to be conserved.
, where U(x) is the potential energy and E is the total mechanical energy, which is assumed to be conserved.
Exploration of the Path Space.
The reliability of the DRP approach in investigating the protein folding transition crucially depends on the efficiency of the algorithm used to find optimum paths. In the analysis of conformational (15, 22) or chemical (18) reactions of relatively small molecules, dominant paths can be found by directly optimizing the HJ action in Eq. 6; e.g. using simulated annealing or gradient-based methods. The DRP calculations for protein folding obtained this way have been extensively tested using reduced models in which the relevant degrees of freedom are individual amino acids and the energy landscape was relatively smooth (34, 35). Unfortunately, when moving from a coarse-grained to an atomistic description, the relaxation algorithms adopted in our previous calculations were found to provide a poor exploration of the space of folding paths in an all-atom calculation.
In order to overcome this problem, we have used a biased MD algorithm to efficiently produce a large ensemble of paths, starting from a given denatured configuration and reaching the native state (28, 29, 37, 38). In particular, the so-called “ratchet-and-pawl” MD (rMD) algorithm (28, 29) exploits the spontaneous fluctuations of the system along a specific collective coordinate (CC), towards its native configuration. This method is implemented by introducing a time-dependent bias potential VR(X,t), whose purpose is to make it very unlikely for the system to evolve back to previously visited values of the CC. On the other hand, this bias exerts no work on the system when it spontaneously proceeds towards the native state. We emphasize that this approach is quite different from the one used in steered-MD (39), where an external force is continuously applied to the system, in order to drive it towards the desired state.
Following the work of ref. 28 we chose a CC z(t), which defines the distance between the contact map in the instantaneous configuration X(t) from the contact map in the native configuration Xnative. Note that a bias on z does not force nor lock any specific contact, but only imposes a (quasi) monotonic behavior of the total number of native contacts.
In particular, the biasing potential introduced in ref. 28 is defined as
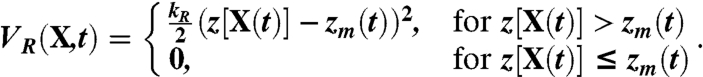 |
[7] |
In these equations, zm(t) is the minimum value assumed by the collective variable z along the rMD trajectory, up to time t.
The value of the collective variable z in the instantaneous configuration X(t) is defined as:
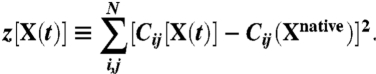 |
[8] |
The entries of the contact map Cij are chosen to interpolate smoothly between 0 and 1, depending on the relative distance of the residues i and j:
| [9] |
where r0 = 7.5 Å is a fixed reference distance. The variable zm(t) is updated only when the system visits a configuration with a smaller value of the CC; i.e., any time z[X(t + δt)] < zm(t). The behavior of the ratchet variable z[X(t)] along two typical folding trajectories is shown in Fig. S1.
The value of the spring constant kR in the ratchet potential —see Eq. 7— controls the amount of bias introduced by the ratchet algorithm. In the SI Text we report on our study on the dependence of our DRP results on the strength of this parameter (see Fig. S2).
The rMD algorithm allows to efficiently generate a large number of trajectories starting from the same configuration and reaching the native state, hence it can be used to explore the folding path space. [3] provides a rigorous way to score such trial trajectories; i.e., to evaluate the probability for each of them to be realized in an unbiased overdamped Langevin dynamics simulation. In particular, the best estimate for the dominant folding pathway is the one with the smallest Onsager-Machlup action. The path may then be used as a starting point for a further refinement based on a local relaxation of the HJ action given by Eq. 6, performed by means of the optimization algorithms described in our previous work (see e.g., refs. 22, 40).
The second refinement step is computationally very expensive, requiring several thousands of CPU hours for each dominant trajectory. However, by performing a number of test simulations, we have found that this step produces only very small rearrangements of the chain, mostly filtering out small thermal fluctuations (see e.g., Fig. S3). Hence, as long as one is concerned mostly with the global qualitative aspects of the folding mechanism, the expensive refinement session of the DRP calculation may be dropped. This choice allows us to reduce the total computational time required to perform the analysis by several orders of magnitude.
Once a dominant path has been found, it is relatively straightforward to identify the configuration which belongs to the transition state ensemble. This identification can be performed by finding the frame XTS in the trajectory such that the probability to reach the native state is equal to that of going back to the denatured configuration (15):
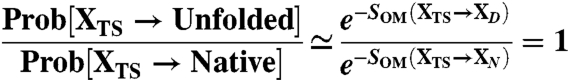 |
[10] |
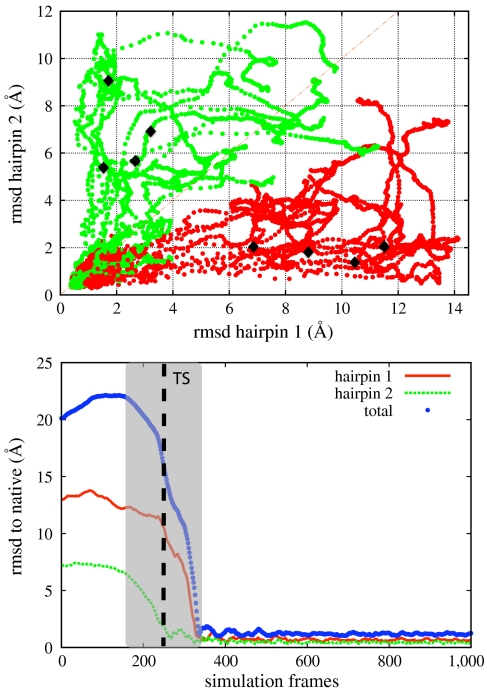
In this equation XN and XD are the first native and denatured configurations visited along the dominant path, starting from XTS. In order to locate these configurations, we need to only take into account the “reactive” part of the path, that is the one which leaves the denatured state and, without recrossing, goes straight to the native. To satisfy this requirement, we considered the total rmsd vs. frame index curve. The typical trend of this curve for most of the dominant trajectories is shown in the lower box of Fig. 4: it consists in an initial plateau, followed by a rather steep fall, and then by another flat region, where the system oscillates in the native state. The reactive part of the path was identified with the region of steep fall in this curve. In particular, the beginning of the transition was set to the frame at which the derivative of the total rmsd curve changes sign, from positive to negative.
Fig. 4.
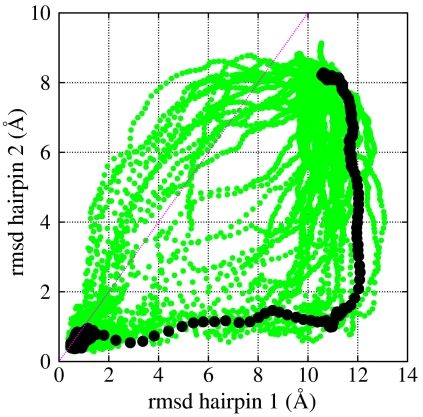
Upper box: the set of dominant folding paths for Fip35, obtained from atomistic DRP simulations, projected on the plane defined by the rmsd of the two hairpins to the corresponding native structures. The dark spots represent a few typical configurations in the two transition states, evaluated by the requesting a probability 1/2 to reach the native state. Lower box: evolution of the rmsd to native of the full protein and of the hairpins along a dominant trajectory. The shaded area is the reactive region and the dashed line identifies the transition state configuration obtained according to Eq. 10.
Further details about the implementation and the computational procedures are given in the SI Text. In particular, the information about the set of parameters used in the simulation and the number of considered trajectories is summarized in Table S1
Results and Discussion
In the upper box of Fig. 4 we show our set of atomistic dominant folding trajectories, projected onto the plane defined by the rmsd to the native structure of the Cα atoms in residues 8–23 (hairpin 1) and 17–30 (hairpin 2). Computing these trajectories required less than 2 d of calculation on 48 CPU’s.
Two distinct folding pathways which differ by the order of formation of the hairpins can be clearly identified: in about half of the computed dominant folding pathways hairpin 1 consistently folds before hairpin 2. In this channel, we find the transition state is located at the “turn” of the paths; i.e., is formed by configurations in which the hairpin 1 is folded while hairpin 2 is largely unstructured (see pathway 1 in Fig. 2). The latter is the mechanism predominantly found in the simulation of ref. 12, performed using the same force field, albeit in explicit solvent.
In about half of the computed dominant paths, we observe that the two hairpins form in the reversed order. In this channel, the transition state is formed by the configurations in which hairpin 2 is folded, while hairpin 1 is unstructured (see pathway 2 in Fig. 2).
Fig. 5 shows that not all the rMD trial trajectories computed starting from a given initial condition follow one of the two folding pathways discussed above. Indeed, many of them involve a simultaneous formation of native contacts in both hairpins. A clear prediction of the DRP formalism is that folding events in which the hairpins form simultaneously are much less frequent than those in which the two secondary structures forms in sequence.
Fig. 5.
The set of trial paths connecting a given denatured configuration to the native state, used in the search for the dominant paths, projected on the plane defined by the rmsd to native of the two hairpins. The darker path is the selected dominant reaction pathways.
Another result emerging from our DRP calculation is the existence of a correlation between the structure of the initial conditions from which the transition is initiated and the pathway taken to fold: if at the beginning of the transition the first hairpin has a rmsd smaller than the second hairpin, then the first pathway is most likely chosen. In the opposite case; i.e., when the second hairpin has a smaller rmsd to native than the first, then the second pathway is generally preferred.
In order to further support these results and gain insight into the folding mechanism, we have performed simulations in an entirely different approach; i.e., by computing equilibrium properties using the coarse-grained models described in Methods. In Fig. 3 we show the free energy landscape at the 300 K, as a function of the rmsd to native of the two hairpins for the two models, which differ by the presence of nonnative interactions. In both cases, we observe the existence of two valleys in the free energy landscape, which correspond to the two folding pathways discussed above. Remarkably, the same structure for this free energy map was obtained by Ferrara, et al. for the 20-residue peptide beta3s—which shares the same native topology of WW domains (42)—by means of equilibrium atomistic simulations based on the CHARMM force field, in implicit solvent. The fact that models with and without nonnative interactions give very similar free energy landscapes suggests that the structure of the two folding pathways of these protein domains is mostly shaped by native interactions.
In general, all experimental data on folding kinetics of WW domains indicate that the formation of the first hairpin is the main rate limiting step (5–7). In particular, the ϕ-values measured by Jäger, et al. display a clear peak in the region associated with hairpin 1, but significant ϕ—values were reported also for residues in the sequence region relative to hairpin 2 (7). This fact indicates that the folding of the latter structure has some rate limiting effect. In addition, it was found that the ϕ-values in the region of the second hairpin grow with temperature, while those in the region of the first hairpin decrease. This observation implies that, at higher and higher temperatures, the second hairpin plays an increasing role in the folding mechanism.
An analysis based on ϕ-values alone does not permit to fully characterize the folding mechanism. In particular, such an analysis cannot distinguish between a single-channel folding mechanism in which native contacts in the two hairpins form simultaneously and a multiple-channel folding mechanism in which the reaction rate in each channel is limited by the folding of one of the hairpins. In ref. 41 Weikl has shown that the full body of existing ϕ-value data (taken from refs. 6, 7) can be consistently and quantitatively explained by a simple kinetic model in which the folding of WW domains occurs through alternative channels, which correspond to the two pathways found in our DRP simulations. From a global fit of the experimental data, the author concluded that the relative probability of the first folding pathway for FBP (another WW domain much similar to Fip35) and Pin 1 WW domain are 77% ± 5% and 67 ± 5%, respectively.
Let us now discuss the relative statistical weight of the two folding pathways. To this goal we need to estimate and compare the reaction rates in the two channels. The formalism for evaluating reaction rates in the DRP approach was developed in detail in ref. 17, where it was shown that this method reproduces Kramers theory in the low-temperature regime. Applying that formalism, the ratio of the folding rates in the two channels reads
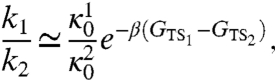 |
[11] |
where the label 1 (2) identifies the channel in which hairpin 1 (2) folds first. In Eq. 11, the exponent contains the difference of the free-energies of the two transition states, defined from the dominant trajectories according to the commitment analysis described in Methods and in ref. 15. In particular, one has
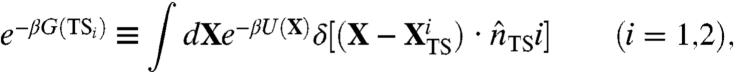 |
[12] |
where 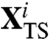 is a point of the transition state which is visited by a typical dominant path in the i—th reaction channel and
is a point of the transition state which is visited by a typical dominant path in the i—th reaction channel and 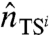 is a versor tangent to the dominant path at
is a versor tangent to the dominant path at 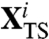 . Using Eq. 10 to identify the transition states, we have found that the two partition functions defined in Eq. 12 are dominated by configurations in which one of the hairpins is fully formed while the other is still completely unstructured (see Fig. 4). The average location of the computed transition states in the plane defined by the rmsd to native of the two hairpins is highlighted in the lower box of Fig. 3.
. Using Eq. 10 to identify the transition states, we have found that the two partition functions defined in Eq. 12 are dominated by configurations in which one of the hairpins is fully formed while the other is still completely unstructured (see Fig. 4). The average location of the computed transition states in the plane defined by the rmsd to native of the two hairpins is highlighted in the lower box of Fig. 3.
The coefficients 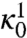 and
and 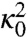 in the prefactor of Eq. 11 are defined in terms of quantities which can be calculated from the dominant paths—see ref. 17 for details. These terms estimate the average flux of reactive trajectories across the isocommittor dividing surface, including the contributions from small thermal fluctuations around the dominant paths. Unfortunately, evaluating
in the prefactor of Eq. 11 are defined in terms of quantities which can be calculated from the dominant paths—see ref. 17 for details. These terms estimate the average flux of reactive trajectories across the isocommittor dividing surface, including the contributions from small thermal fluctuations around the dominant paths. Unfortunately, evaluating 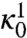 and
and 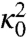 necessarily requires to perform the computationally expensive local optimization of the HJ action. However, if the reaction is thermally activated, the ratio of rates k1/k2 is mostly controlled by the exponential contribution. Hence, we can consider the Arrhenius approximation
necessarily requires to perform the computationally expensive local optimization of the HJ action. However, if the reaction is thermally activated, the ratio of rates k1/k2 is mostly controlled by the exponential contribution. Hence, we can consider the Arrhenius approximation 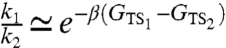 . We emphasize that in this approach the DRP information about the nonequilibrium reactive dynamics is used to define the two transition states. On the other hand, the numerical value of the free energy difference may be obtained from equilibrium techniques; e.g., by sampling the integrals in Eq. 12 by means of computationally very expensive umbrella sampling or meta-dynamics (43) atomistic calculations.
. We emphasize that in this approach the DRP information about the nonequilibrium reactive dynamics is used to define the two transition states. On the other hand, the numerical value of the free energy difference may be obtained from equilibrium techniques; e.g., by sampling the integrals in Eq. 12 by means of computationally very expensive umbrella sampling or meta-dynamics (43) atomistic calculations.
In this first exploratory application of the DRP formalism to a realistic protein folding reaction we choose to perform a much rougher estimate which relies on two main approximations. First, we identify the difference (GTS1 - GTS2) with the difference of the free energy in the two shaded regions of the energy landscape shown in Fig. 3. The centers of these regions represent the average location of the configurations in the two transition states TS1 and TS2 obtained from DRP simulations, projected onto the plane selected by the rmsd to native of the two hairpins. The sizes of the shaded area represents the errors on the average location of the transition states on this plane, estimated from the standard deviation. The second assumption of our model is that such a free energy difference is driven by the balance between energy gain and entropy loss associated to the formation of native contacts in the two hairpins. This native-centric standpoint is supported in part by the fact that free energy landscapes computed in different models with and without nonnative interactions are found to be very similar, as it is clear from comparing the boxes in Fig. 3. Hence, to estimate GTS1 - GTS2 we used the G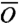 -type model described in Methods. It is important to emphasize that we are not computing the rate directly from a transition state theory formulated in the coarse-grained model, but we are using it only to estimate a free energy difference.
-type model described in Methods. It is important to emphasize that we are not computing the rate directly from a transition state theory formulated in the coarse-grained model, but we are using it only to estimate a free energy difference.
This way, we obtained an estimate k1/k2 ≃ 2.3 which corresponds to a relative weight of the first folding channel of 70% and 30%. We stress that, such a simple calculation should be considered only a rough estimate. The results indicate that the two channels have more or less comparable weight and that the first channel is the most probable, in qualitative agreement with experimental results and with the simulations of Shaw, et al..
This simple scheme enables us to address the question of the dependence of the relative weights of the two channels on the temperature. Repeating the calculation at a higher temperature of 380 K—assuming that the structure of the transition states is not significantly modified—we find k1/k2 ≃ 1.6, which corresponds to a branching ratio of channel 1 of about 60%. Hence, the rate limiting role of the second channel grows with temperature, in qualitative agreement with experimental kinetic data.
This fact can be understood as follows. The folding of one of the hairpins generates an entropy loss proportional to the number n of native contacts formed. The transition state in the first folding channel involves forming a longer hairpin, hence reaching it produces a larger entropy loss (but also larger gain of native energy). The role of the entropy loss relative to the energy gain in forming the hairpins grows with temperature, hence disfavoring the first folding channel relative to the second.
Conclusions
The folding mechanism of the WW domain which emerges from our atomistic and coarse-grained simulations is not heterogeneous. Instead, the folding proceeds through two dominant channels, defined by a hierarchical order of hairpin formation. Our estimate for the relative rate of the two channels is compatible with both Weikl’s analysis of kinetic data and with Krivov’s analysis of long equilibrium MD simulations. Our results also suggest that the folding pathway is correlated with the structure of the denatured configuration from which the peptide initiates the reaction.
The most important result of this work is to show that, using the DRP approach, it is possible to characterize at least the main qualitative aspects of the folding mechanism at an extremely modest computational cost, in the range of few hundreds of CPU hours. Such a level of computational efficiency opens the door to the investigation of the folding pathways of a large number of single-domain proteins, with sizes significantly larger than that of the small domain studied in the present work.
Supplementary Material
Acknowledgments.
The DRP approach was developed in collaboration with H. Orland, M. Sega, and F. Pederiva. We thank G. Tiana and C. Camilloni for sharing details of their implementation of the ratched-MD algorithm and S. Piana-Agostinetti for an important discussion. All the authors are members of the Interdisciplinary Laboratory for Computational Science (LISC), a joint venture of Trento University and the Bruno Kessler Foundation. S.a.B. and T.Š. have been partially supported by the Provincia Autonoma di Trento, through the AuroraScience project. Simulations were performed on the AURORA supercomputer located at the LISC (Trento) and partially on the TITANE cluster, which was kindly made available by the IPhT of CEA-Saclay.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111796109/-/DCSupplemental.
References
- 1.Snow CD, Sorin EJ, Rhee YM, Pande VS. How well can simulation predict protein folding kinetics and thermodynamics? Annu Rev Bioph Biom. 2005;34:43–69. doi: 10.1146/annurev.biophys.34.040204.144447. [DOI] [PubMed] [Google Scholar]
- 2.Mirny L, Shakhnovich E. Protein folding theory: from lattice to all atom models. Annu Rev Bioph Biom. 2001;30:361–396. doi: 10.1146/annurev.biophys.30.1.361. [DOI] [PubMed] [Google Scholar]
- 3.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struc Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Kubelka J, Hofrichter J, Eaton W. The protein folding ‘speed limit’. Curr Opin Struc Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, et al. An experimental survey of the transition between two-state and downhill protein folding scenarios. Proc Nat'l Acad Sci USA. 2008;105:2369–2374. doi: 10.1073/pnas.0711908105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deechongkit S, et al. Context-dependent contributions of backbone hydrogen bonding to sheet folding energetics. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 7.Jäger M, Nguyen H, Crane JC, Kelly JW, Gruebele M. The folding mechanism of a β-sheet: the WW domain. J Mol Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 8.Freddolino PL, Liu F, Gruebele M, Schulten K. Ten-microsecond molecular dynamics simulation of a fast-folding WW domain. Biophys J. 2008;94:L75–77. doi: 10.1529/biophysj.108.131565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freddolino PL, Park S, Roux B, Schulten K. Force field bias in protein folding simulations. Biophys J. 2009;96:3772–3780. doi: 10.1016/j.bpj.2009.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ensign DL, Pande VS. The Fip35 WW domain folds with structural and mechanistic heterogeneity in molecular dynamics simulations. Biophys J. 2009;96:L53–55. doi: 10.1016/j.bpj.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noé F, Schütte C, Vanden-Eijnden E, Reich L, Weikl TR. Constructing the equilibrium ensemble of folding pathways from short off-equilibrium simulations. Proc Nat'l Acad Sci USA. 2009;106:19011–19016. doi: 10.1073/pnas.0905466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw DE, et al. Atomic level characterization of the structural dynamics of proteins. Science (New York NY) 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 13.Krivov SV. The free energy landscape analysis of protein (FIP35) folding dynamics. J Phys Chem B. 2011;115:12315–12324. doi: 10.1021/jp208585r. [DOI] [PubMed] [Google Scholar]
- 14.Faccioli P, Sega M, Pederiva F, Orland H. Dominant pathways in protein folding. Phys Rev Lett. 2006;97:1–4. doi: 10.1103/PhysRevLett.97.108101. [DOI] [PubMed] [Google Scholar]
- 15.Sega M, Faccioli P, Pederiva F, Garberoglio G, Orland H. Quantitative protein dynamics from dominant folding pathways. Phys Rev Lett. 2007;99:1–4. doi: 10.1103/PhysRevLett.99.118102. [DOI] [PubMed] [Google Scholar]
- 16.Autieri E, Faccioli P, Sega M, Pederiva F, Orland H. Dominant reaction pathways in high-dimensional systems. J Chem Phys. 2009;130:064106. doi: 10.1063/1.3074271. [DOI] [PubMed] [Google Scholar]
- 17.Mazzola G, a Beccara S, Faccioli P, Orland H. Fluctuations in the ensemble of reaction pathways. J Chem Phys. 2011;134:164109. doi: 10.1063/1.3581892. [DOI] [PubMed] [Google Scholar]
- 18.a Beccara S, Garberoglio G, Faccioli P, Pederiva F. Communications: ab initio dynamics of rare thermally activated reactions. J Chem Phys. 2010;132:111102. doi: 10.1063/1.3355866. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 20.Hess B, Kutzner C, van derSpoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 21.Onufriev A, Case D, Bashford D. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 22.a Beccara S, et al. Dominant folding pathways of a peptide chain from ab initio quantum-mechanical simulations. J Chem Phys. 2011;134:024501. doi: 10.1063/1.3514149. [DOI] [PubMed] [Google Scholar]
- 23.Kim YC, Hummer G. Coarse grained models for simulations of multiprotein complexes: application to ubiquitin binding. J Mol Biol. 2008;375:1416–1433. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Best RB, Hummer G. Reaction coordinates and rates from transition paths. Proc Nat'l Acad Sci USA. 2005;102:6732–6737. doi: 10.1073/pnas.0408098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karanicolas J, Brooks CL., III The origins of the asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dellago C, Bolhuis PG, Geissler PL. Transition Path Sampling. Vol 123. Hoboken, NJ: John Wiley & Sons, Inc; 2002. [Google Scholar]
- 27.Chodera JD, Swope WC, Pitera JW, Dill K. Long-time protein folding dynamics from short molecular dynamics simulations. Multiscale Model Sim. 2006;5:1214–1226. [Google Scholar]
- 28.Camilloni C, Broglia RA, Tiana G. Hierarchy of folding and unfolding events of protein G, CI2, and ACBP from explicit-solvent simulations. J Chem Phys. 2011;134:045105. doi: 10.1063/1.3523345. [DOI] [PubMed] [Google Scholar]
- 29.Paci E, Karplus M. Forced unfolding of fibronectin type 3 modules: an analysis by biased molecular dynamics simulations1. J Mol Biol. 1999;288:441–459. doi: 10.1006/jmbi.1999.2670. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh A, Elber R, Scheraga H. An atomically detailed study of the folding pathways of protein A with the stochastic difference equation. Proc Nat'l Acad Sci USA. 2002;99:10394–10398. doi: 10.1073/pnas.142288099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eastman P, Gronbech-Jensen N, Doniach S. Simulation of protein folding by reaction path annealing. J Chem Phys. 2001;114:3823–3841. [Google Scholar]
- 32.Dryga A, Warshel A. Renormalizing SMD: the renormalization approach and its use in long time simulations and accelerated PMF calculations of macromolecules. J Phys Chem B. 2010;114:12720–12728. doi: 10.1021/jp1056122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elber R, Shalloway D. Temperature dependent reaction coordinates. J Chem Phys. 2000;112:5539–5545. [Google Scholar]
- 34.Faccioli P. Characterization of protein folding by dominant reaction pathways. J Phys Chem B. 2008;112:13756–13764. doi: 10.1021/jp805762d. [DOI] [PubMed] [Google Scholar]
- 35.Faccioli P, Lonardi A, Orland H. Dominant reaction pathways in protein folding: a direct validation against molecular dynamics simulations. J Chem Phys. 2010;133:045104. doi: 10.1063/1.3459097. [DOI] [PubMed] [Google Scholar]
- 36.Cardenas E, Elber R. Kinetics of cytochrome C folding: atomically detailed simulations. Proteins. 2003;51:245–257. doi: 10.1002/prot.10349. [DOI] [PubMed] [Google Scholar]
- 37.Marchi M, Ballone P. Adiabatic bias molecular dynamics: a method to navigate the conformational space of complex molecular systems. J Chem Phys. 1999;110:3697–3702. [Google Scholar]
- 38.Zuckerman DM, Woolf TB. Dynamic reaction paths and rates through importance-sampled stochastic dynamics. J Chem Phys. 1999;111:9475–9484. [Google Scholar]
- 39.Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr Opin Struc Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 40.a Beccara S, Garberoglio G, Faccioli P. Quantum diffusive dynamics of macro-molecular transitions. J Chem Phys. 2010:1–12. doi: 10.1063/1.3609244. [DOI] [PubMed] [Google Scholar]
- 41.Weikl TR. Transition states in protein folding kinetics: modeling ϕ -values of small β -sheet proteins. Biophys J. 2008;94:929–937. doi: 10.1529/biophysj.107.109868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrara P, Caflish A. Folding simulations of a three-stranded antiparallel β -sheet peptide. Proc Nat'l Acad Sci USA. 2000;97:10780–10785. doi: 10.1073/pnas.190324897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laio A, Parrinello M. Escaping free energy minima. Proc Nat'l Acad Sci USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.