Abstract
The circadian clock orchestrates many aspects of human physiology, and disruption of this clock has been implicated in various pathologies, ranging from cancer to metabolic syndrome and diabetes. Although there is evidence that metabolism and the circadian clockwork are intimately linked on a transcriptional level, whether these effects are directly under clock control or are mediated by the rest–activity cycle and the timing of food intake is unclear. To answer this question, we conducted an unbiased screen in human subjects of the metabolome of blood plasma and saliva at different times of day. To minimize indirect effects, subjects were kept in a 40-h constant routine of enforced posture, constant dim light, hourly isocaloric meals, and sleep deprivation. Under these conditions, we found that ∼15% of all identified metabolites in plasma and saliva were under circadian control, most notably fatty acids in plasma and amino acids in saliva. Our data suggest that there is a strong direct effect of the endogenous circadian clock on multiple human metabolic pathways that is independent of sleep or feeding. In addition, they identify multiple potential small-molecule biomarkers of human circadian phase and sleep pressure.
Keywords: metabolomics, LC/GC-MS, metabolite profiling, sleep–wake regulation
The circadian clock has been shown to modulate many aspects of behavior and physiology (1). It is thought to be an important regulator of metabolism, and disruption of the clock and sleep is associated with obesity, metabolic syndrome, and type 2 diabetes, as well as other disorders (2–4). In the last decade, ample data on the circadian transcriptome (5, 6) and the even larger circadian proteome (7) have been compiled. These datasets are directly dependent on the genome of a particular species and cannot be compared easily between model systems. However, changes in physiology and metabolism governed by these genes and proteins ultimately affect the abundance of small metabolites that are quite conserved among species and fewer in number (50-fold fewer than transcripts and 500-fold fewer than proteins).
The relationship between metabolism and the clock is not unidirectional, and the two processes are intertwined (8). For example, metabolic status feeds back to the clock, so that feeding behavior directly entrains molecular clock function (9). Likewise, obesity is correlated with poor sleep (2), and in mice 80% of circadian transcription in the brain is dependent on the rest–activity cycle (10). Given these feedback mechanisms, it is unclear what proportion of circadian metabolic control is directly clock-regulated and what proportion is controlled by circadian rest–activity and food intake.
In plants, the metabolome approach has been used to characterize the effects of clock disruption on general metabolism (11). The circadian metabolome also has been characterized in CBA/N mice, and ∼20% of the recorded molecules were found to vary in abundance with time of day (12). Similarly, the urine and saliva metabolomes of human subjects differ between morning and evening under real-world conditions (13, 14). However, in mammals, indirect cues from behavior, such as feeding time and body temperature, are well-established and potent timing signals to the circadian clock (9, 15). Thus, whether significant numbers of these metabolites are a consequence of circadian patterns of eating and sleeping remains unclear. This knowledge would be critical to effective chronotherapy of metabolic disorders.
To answer this question in human subjects, we used GC/LC coupled with MS in an unbiased screen of the human metabolome of blood plasma and saliva at different times of day. We found that 15% of all identified metabolites in plasma and saliva were under circadian control, independent of sleep or food intake. This indicates that the clock directly controls multiple metabolic pathways, most notably fatty acids in plasma and amino acids in saliva.
Results
Metabolome from Human Plasma Shows That Circadian Metabolism Is Independent of Rhythmic Feeding or Rest–Activity Cycles.
The contribution of rest–activity and feeding patterns to circadian metabolomic variation is most easily investigated using human subjects, whose patterns of eating and sleeping can be precisely and rigorously controlled. Subjects were kept in a 40-h “constant routine” of enforced posture, constant dim light, hourly isocaloric meals, and sleep deprivation to exclude indirect effects (16, 17). Saliva and blood plasma were collected hourly and subsequently analyzed by GC/LC-MS. These matrices have been shown to yield valuable information regarding human metabolic state (13, 18) and are easy to obtain.
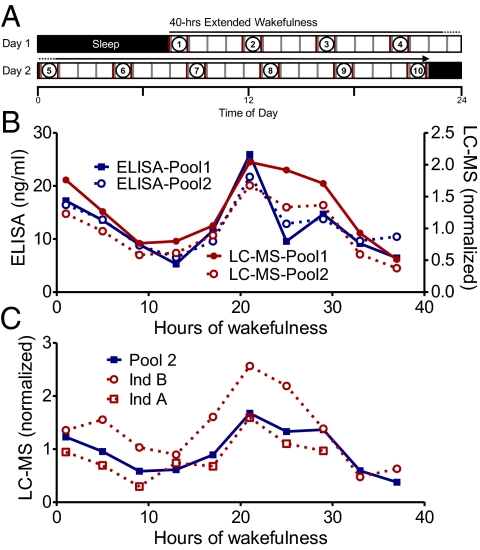
A total of 10 age-matched male subjects underwent the protocol (Fig. 1A) individually in isolation. Samples were pooled in 4-h intervals for two groups of five subjects, and relative levels of metabolites were determined (Fig. 1 and Methods). As a first step, our GC/LC-MS measurement procedure was independently validated with separate ELISA analyses to determine the subjects’ cortisol levels. The pattern of cortisol levels was highly correlated between ELISA and GC/LC-MS (Pearson's r = 0.8; P < 0.0001) (Fig. 1B), confirming the validity of GC/LC-MS quantification. Second, to assess the degree of interindividual variation, we used GC/LC-MS to measure the plasma metabolome of two subjects from one pool and compare the findings with each other and with the average of the pool. Cross-correlation analysis showed highly similar circadian cortisol profiles from the two pools (pool 1 vs. pool 2, r = 0.9784, P < 0.0001). Similarly, each subject was globally similar to the pool (pool 2 vs. individual A, r = 0.9, P < 0.001; pool 2 vs. individual B, r = 0.9, P < 0.002) (Fig. 1C), and subjects were similar to one another (individual A vs. individual B, r = 0.8, P < 0.02). Overall, of the 258 metabolites identified in both individuals, 219 metabolites were found in each of the individuals and the pools, consistent with results of previous studies of interindividual metabolomic variation (18).
Fig. 1.
(A) Experimental design. Scheme of the 40-h extended wakefulness constant routine, sampling, and pooling of samples. For detailed explanations of the sampling and pooling procedures, see Methods. Black bars indicate sleep; open bars, extended wakefulness; red lines, sampling of blood and saliva; gray lines, isocaloric meals. Samples on either side of the circled numbers were pooled together for MS analysis. (B) Comparison of rhythmic cortisol levels measured by LC-MS (red) and ELISA (blue) from plasma and saliva, respectively, of the same subject pools. (C) Comparison of individuals with the pool in which they were used.
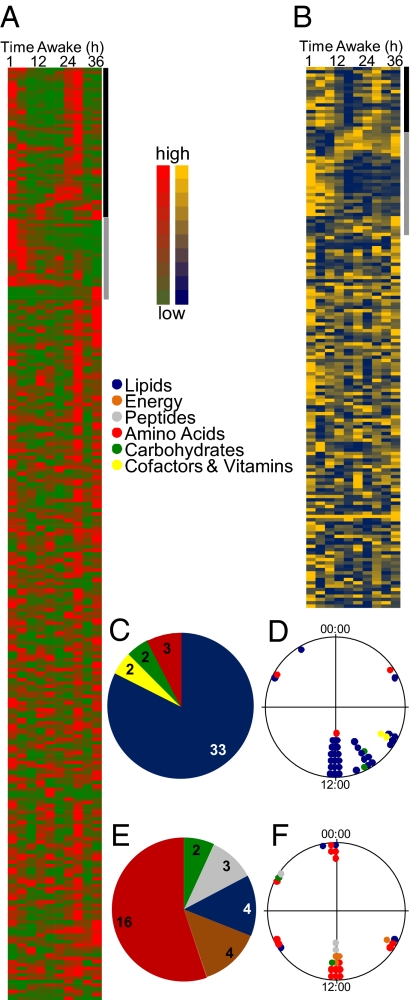
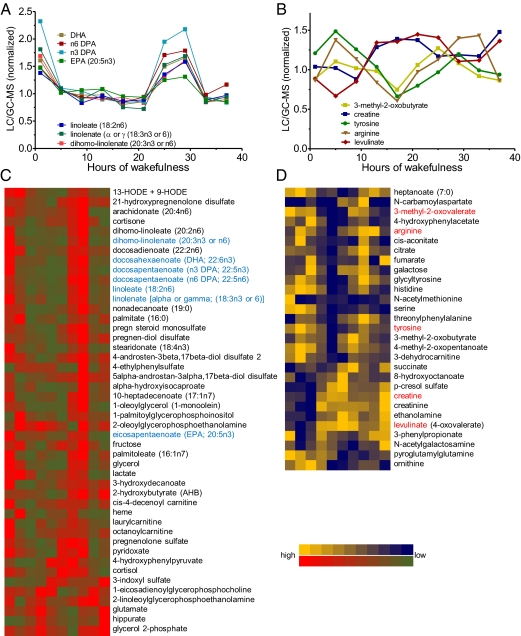
We next measured all identifiable metabolites in all samples by GC/LC-MS. In the plasma pools, ∼15% of the metabolites (41 of 281) displayed a circadian profile (Figs. 2A and 3C). Interestingly, a high proportion of these rhythmic metabolites were fatty acids (Figs. 2C and 3A). The levels of nearly all lipid products were highest at midmorning to noon and were significantly lower at other times of the day (Figs. 2D and 3), even though food intake was constant and sleep did not occur.
Fig. 2.
Heat plots for all identified metabolites in plasma (A) and saliva (B). The black bar indicates circadian metabolites, and the gray bar indicates monotonic increasing/decreasing metabolites. High levels of metabolites are shown in red (plasma) and yellow (saliva), and low levels are shown in green (plasma) and blue (saliva). (C–F) Pathway analyses (C and E) and time-of-day distribution (D and F) of peak phases of rhythmic metabolites in plasma (C and D) and saliva (E and F). Pathways are color-coded as follows: blue, lipids; orange, energy metabolism; gray, peptides; red, amino acids; green, carbohydrates; yellow, cofactors and vitamins.
Fig. 3.
Rhythmic metabolites in plasma and saliva. (A and B) Profiles of substances previously implicated in sleep–wake regulation. (C and D) Heat maps of all such substances. High levels of metabolites are shown in red (plasma) and yellow (saliva) whereas low levels are shown in green (plasma) and blue (saliva). The metabolites in blue are exemplified in A, and those in red are exemplified in B.
Human Salivary Metabolome also Shows Circadian Metabolism Independent of Sleep and Food.
We determined the salivary metabolome from the same pools of subjects as for plasma (Fig. 2B). Saliva is easily obtained through noninvasive means, and thus has great potential for screening large cohorts. Although we identified fewer metabolites in saliva than in plasma (178 vs. 281), as in plasma, ∼15% of these metabolites (29 of 178) had a circadian profile (Fig. 3D). Not surprisingly, only approximately half of all saliva metabolites (88 compounds) were also detected in plasma. Although parotid saliva directly reflects the contents of blood (19), the overall content of metabolites in globally collected saliva also depends on other glands, most prominently the submandibular glands (20). Among the common metabolites detected, 27 were rhythmic in either plasma or saliva. In contrast to the plasma samples, the largest group of compounds in saliva was amino acids (Figs. 2E and 3B). These amino acids exhibited a large phase distribution around the clock (Fig. 3 B and D), likely related to the diverse pathways in which they are involved.
Discussion
Several previous pathway-specific studies of amino acids have suggested that aspects of human metabolism are circadian (21–23). Our global circadian metabolomic analysis has shown that in fact this control extends to 15% of the human circadian metabolome in two different matrices. Moreover, we have demonstrated that this circadian control occurs independent of scheduled sleep and feeding. A similar proportion of rhythmic metabolites has been found to cycle in mice under ad libitum feeding conditions (12). In that earlier study and the present study, this proportion is likely a conservative estimate, given that the procedure used for detection in these protocols can possibly result in degradation of unstable substances (which are more likely than stable substances to show the effects of upstream circadian modulation). Nevertheless, the percentage of rhythmic substances that we found in the metabolome is roughly comparable to that found in previous transcriptome (10–20%) (5, 6) and proteome (20%) (7) analyses.
Although the percentage of circadian metabolites is comparable to the percentages of circadian transcripts and proteins in both mice and humans, in one respect our results are quite surprising. Previous work has shown that on the transcriptional level, >75% of genes expressed rhythmically under ad libitum or restricted feeding conditions do not remain rhythmic when mice are fasted (24). Similarly, in the brain, only 20% of circadian transcripts remain so when mice are sleep-deprived (10). Whereas the latter result can be explained by the fact that few cortical transcripts are likely to affect the plasma or saliva metabolome, the former results offer no ready explanation, given that the authors specifically examined transcription of metabolic regulators in the most relevant tissue (liver). We speculate that the restricted feeding conditions of that study might have provided a systemic metabolic signal very different from that provided by the frequent isocaloric meals consumed by the much larger human subjects in the present study, because 24 h of food deprivation has significant consequences for a small rodent, typically resulting in a loss of 10–15% of body weight (25).
In human blood plasma, lipid metabolites represent >75% of all rhythmic compounds. It has been suggested that fatty acid metabolism is under circadian control (3), but whether this is a direct effect of clocks on fatty acid oxidation or rather an indirect effect mediated via food intake or activity and its associated changes in body temperature remains an open question (23). We found that this variation is not an indirect effect, but rather is directly controlled by the circadian clock. The levels of nearly all lipid products were highest around subjective “lunchtime” and were greatly reduced at other times of the day, even though food intake was constant. Interestingly, metabolic syndrome has been reported to be associated with a flattening of molecular clock rhythms in peripheral tissues (26) and with nighttime eating (27), and thus our results are consistent with the idea that direct clock control can serve to “scavenge” harmful fatty acids from the bloodstream at inappropriate times.
In our saliva samples, more than half of all rhythmic compounds were amino acids and associated metabolites. It has been suggested that the plasma content of branched-chain amino acids is predictive of future diabetes (28). In our samples, the average levels of these amino acids varied by 20–200% over the 24-h day, which is equal to or up to 10-fold greater than the differences considered diagnostic of diabetes. Similarly, in a recent genome-wide analysis of genetic determinants of metabolic variation, an allele of the glucokinase regulator gene was reportedly associated with an increased risk of various chronic diseases and with a 3.3% per copy increase in lactate levels (29). In the present study, lactate levels varied by >20-fold as much over the day, illustrating the magnitude of the effects that can be observed. Interestingly, in the aforementioned study, the proportion of rhythmic metabolites among those associated with the genomic traits was only ∼7% (29), suggesting that rhythmic metabolites are underrepresented, and that the associations could be improved if time of day were taken into account.
Overall, the major circadian pathway signatures of the human metabolome that we have identified are consistent with those previously described in rodents (12), in which amino acids as well as some fatty acid byproducts were identified. Along with the large number of metabolites with circadian expression profiles, we also found 34 metabolites in plasma and 27 in saliva that demonstrated a monotonic increase or decrease across the 40-h constant routine protocol (Fig. S1). Because our constant routine protocol was conducted under sleep deprivation conditions, it is possible that some of these metabolites might be associated with sleep pressure. For example, we found a more than threefold increase in two fragments of the C3 complement (30). Up-regulation of the immune system in general and the C3 complement in particular has been reported in sleep deprivation in humans (31). These polypeptides also have been implicated in the regulation of insulin-like growth factor 1 activity (30), which is also linked to sleep (32). Another metabolite, 3-hydroxybutyrate, found to be up-regulated in saliva by 1.5-fold and in plasma by 2.5-fold has been implicated in a rapid eye movement sleep feature (33). A noninvasive biomarker for sleep pressure would be of great practical use, given the well-characterized detrimental effects of sleep deprivation. Because our study did not explicitly compare subjects under different sleep pressures at the same circadian time, further investigations are clearly needed to confirm whether any of these substances are indeed correlated with time awake.
Taken together, our experiments provide data that could be useful for at least three types of future studies. First, knowledge of the metabolites identified here can help identify novel physiological pathways regulated by the circadian clock, and possibly by time awake, in humans. Second, metabolome profiles from pathophysiological populations can be compared with our data and used to identify pathways in which the circadian clock might play a role. Already metabolome profiling has proven to be a potent tool for detecting human pathologies such as cancer (34, 35), predicting the risk for certain diseases such as diabetes (28), and indicating early signs of insulin resistance (36). Circadian metabolome profiling can provide a convenient way to analyze clock involvement and also can improve the predictive validity of biomarkers. Third, our set of metabolites might be used to validate biomarkers for circadian phase and sleep pressure. Detection of such molecules, if validated, could provide convenient and noninvasive assays for circadian phase (12) and sleep pressure.
Methods
Study Subjects.
The study cohort comprised 10 healthy unmedicated male volunteers. A general description of recruitment and demographic and sleep-relevant parameters of the study population are provided in SI Methods and Table S1. After receiving a detailed explanation of the study protocol, each participant provided written informed consent. The study conformed to the Declaration of Helsinki and was approved by the local Ethics Committee (Ethische Kommission beider Basel).
Study Design and Sample Preparation.
For the constant routine study, participants came to the laboratory for a baseline night of sleep, which was followed by 40 h of prolonged wakefulness and then a recovery night, as described previously (37). At all times, participants were in a semireclining position in bed in a room with constant illumination level (<5 l×) and temperature (∼21 °C). Throughout the awake part of the study, participants received small isocaloric meals as well as water once every hour. To prevent thrombosis, all participants received heparin (0.2 mL Fragmin) every 20 h. A technician prevented the participants from falling asleep during the 40 h of wakefulness. No information about clock time was available to the participants.
Hourly samples of blood and saliva were obtained during the period of extended wakefulness. Blood was collected via cannula from the forearm in sterile vacutainers (6-mL BD Vacutainer with 10.8 mg of K2EDTA). Plasma was immediately separated by centrifugation (1650 × g at 4 °C for 10 min) and stored at −20 °C until analysis. Saliva samples were collected with Sarstedt Salivettes and centrifuged before freezing at −20 °C until pooling. This protocol is diagrammed in Fig. 1A.
Metabolites of plasma and saliva were analyzed independently in two pools of five individuals each and also in two individuals from one pool. Each sample was pooled from equal volumes of two consecutive hourly samples, for a total of 10 samples from each case. Sampling was done at 4-h intervals, with sample 1 collected at the beginning of the constant routine and sample 10 collected 40 h later. MS values were verified by direct salivary ELISA (ALPCO) in all samples from all participants.
Small-Molecule Determination.
Metabolites in plasma and saliva were analyzed by Metabolon, as described previously (18). In this method, the total process variability of metabolites was calculated as the relative standard variation of six runs of a pool of equal aliquots from each of the experimental samples. The variation among these six replicates was 8% in plasma and 10% in saliva. In addition, internal standards were injected into each of the samples; here the variability was 5% in plasma and 6% in saliva. Raw peak values for all metabolites were normalized to have a median of 1. Because the absolute levels and thus the lower limit of detection were unknown, missing values were replaced by the minimum values observed for any given metabolite. This procedure prevented overestimation of circadian amplitude in less abundant substances, whose true minimum values are unknown. Overall, <3% of all data points for all metabolites were replaced in this fashion for pooled plasma and saliva samples. Only one metabolite had to be excluded from rhythmic analysis because of missing values. Complete datasets for plasma and saliva pools are provided in Tables S1, S2, S3, S4, S5, and S6.
Statistics.
Rhythmicity of metabolites was assessed using an algorithm previously described for rhythmic transcripts. The JTK-cycle algorithm (38, 39) was used as implemented in R by Kronauer. In brief, this algorithm characterizes samples as rhythmic or nonrhythmic using a nonparametric method based on a combination of the Jonckheere–Terpstra test for monotonic ordering and Kendall's τ test for association of measured quantities. Results were cross-checked by visual inspection. Subsequently, the statistical significance of the circadian rhythmicity of each compound was evaluated with an independent permutation test to identify the false discovery rate for each metabolite (40). A detailed description of this procedure is provided in SI Methods. Pearson correlation coefficients and significance levels were computed using Prism 5 (GraphPad).
Supplementary Material
Acknowledgments
We thank K. Kornacker for supplying the R implementation of the JTK-cycle algorithm. This work was supported by grants from the Hartmann–Müller Foundation (to S.A.B. and R.D.), the Velux Foundation (to C.C. and S.A.B.), and the Swiss National Science Foundation (to S.A.B. and C.C.). R.D. is a Feodor Lynen fellow of the Alexander von Humboldt Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114410109/-/DCSupplemental.
References
- 1.Weaver DR, Reppert SM. Circadian timekeeping. In: Squire LR, et al., editors. Fundamental Neuroscience. 3rd Ed. New York: Academic; 2008. pp. 931–958. [Google Scholar]
- 2.Wolk R, Somers VK. Sleep and the metabolic syndrome. Exp Physiol. 2007;92:67–78. doi: 10.1113/expphysiol.2006.033787. [DOI] [PubMed] [Google Scholar]
- 3.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 5.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 6.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: Time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 10.Maret S, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci USA. 2007;104:20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukushima A, et al. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA. 2009;106:7251–7256. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertram HC, Eggers N, Eller N. Potential of human saliva for nuclear magnetic resonance-based metabolomics and for health-related biomarker identification. Anal Chem. 2009;81:9188–9193. doi: 10.1021/ac9020598. [DOI] [PubMed] [Google Scholar]
- 14.Slupsky CM, et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 15.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 16.Duffy JF, Dijk DJ. Getting through to circadian oscillators: Why use constant routines? J Biol Rhythms. 2002;17:4–13. doi: 10.1177/074873002129002294. [DOI] [PubMed] [Google Scholar]
- 17.Blatter K, Cajochen C. Circadian rhythms in cognitive performance: Methodological constraints, protocols, theoretical underpinnings. Physiol Behav. 2007;90:196–208. doi: 10.1016/j.physbeh.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Lawton KA, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9:383–397. doi: 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- 19.Takeda I, et al. Understanding the human salivary metabolome. NMR Biomed. 2009;22:577–584. doi: 10.1002/nbm.1369. [DOI] [PubMed] [Google Scholar]
- 20.Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic potential of saliva: Current state and future applications. Clin Chem. 2011;57:675–687. doi: 10.1373/clinchem.2010.153767. [DOI] [PubMed] [Google Scholar]
- 21.Blanco RA, et al. Diurnal variation in glutathione and cysteine redox states in human plasma. Am J Clin Nutr. 2007;86:1016–1023. doi: 10.1093/ajcn/86.4.1016. [DOI] [PubMed] [Google Scholar]
- 22.Breum L, Rasmussen MH, Hilsted J, Fernstrom JD. Twenty-four hour plasma tryptophan concentrations and ratios are below normal in obese subjects and are not normalized by substantial weight reduction. Am J Clin Nutr. 2003;77:1112–1118. doi: 10.1093/ajcn/77.5.1112. [DOI] [PubMed] [Google Scholar]
- 23.Bray MS, Young ME. Regulation of fatty acid metabolism by cell-autonomous circadian clocks: Time to fatten up on information? J Biol Chem. 2011;286:11883–11889. doi: 10.1074/jbc.R110.214643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueno N, Asakawa A, Inui A. Blunted metabolic response to fasting in obese mice. Endocrine. 2007;32:192–196. doi: 10.1007/s12020-007-9016-z. [DOI] [PubMed] [Google Scholar]
- 26.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity. 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suhre K, et al. CARDIoGRAM Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dousset B, et al. Purification from human plasma of a hexapeptide that potentiates the sulfation and mitogenic activities of insulin-like growth factors. Biochem Biophys Res Commun. 1998;247:587–591. doi: 10.1006/bbrc.1998.8834. [DOI] [PubMed] [Google Scholar]
- 31.Hui L, Hua F, Diandong H, Hong Y. Effects of sleep and sleep deprivation on immunoglobulins and complement in humans. Brain Behav Immun. 2007;21:308–310. doi: 10.1016/j.bbi.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Obál F, Jr, et al. Insulin-like growth factor-1 (IGF-1)-induced inhibition of growth hormone secretion is associated with sleep suppression. Brain Res. 1999;818:267–274. doi: 10.1016/s0006-8993(98)01286-4. [DOI] [PubMed] [Google Scholar]
- 33.Tafti M, et al. Deficiency in short-chain fatty acid β-oxidation affects θ oscillations during sleep. Nat Genet. 2003;34:320–325. doi: 10.1038/ng1174. [DOI] [PubMed] [Google Scholar]
- 34.Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers: Blood-based strategies to detect and monitor cancer. Natl Rev. 2011;8:142–150. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- 35.Slupsky CM, et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res. 2010;16:5835–5841. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- 36.Gall WE, et al. RISC Study Group α-Hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS ONE. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czeisler C, Brown E, Ronda J, Kronauer R. A clinical method to assess the endogenous circadian phase (ECP) of the deep circadian oscillator in man. J Sleep Res. 1985;14:295. [Google Scholar]
- 38.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ptitsyn AA, Zvonic S, Gimble JM. Permutation test for periodicity in short time series data. BMC Bioinformatics. 2006;7(Suppl 2):S10. doi: 10.1186/1471-2105-7-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.