Abstract
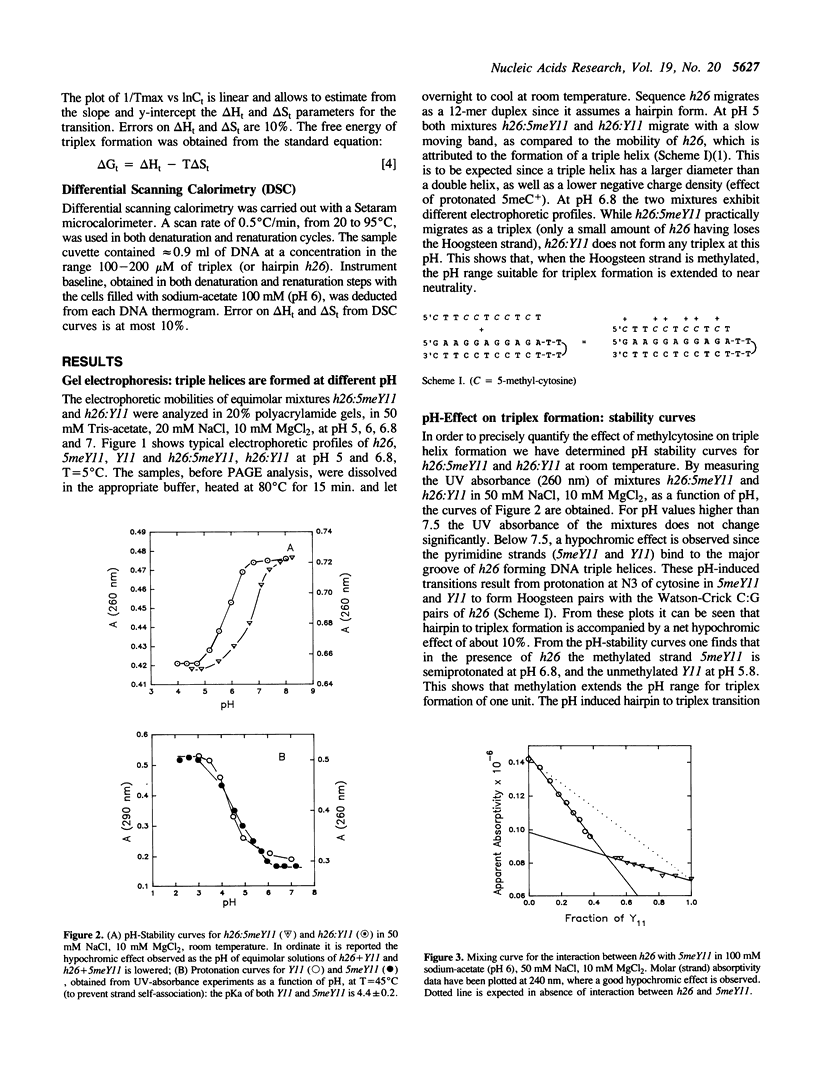
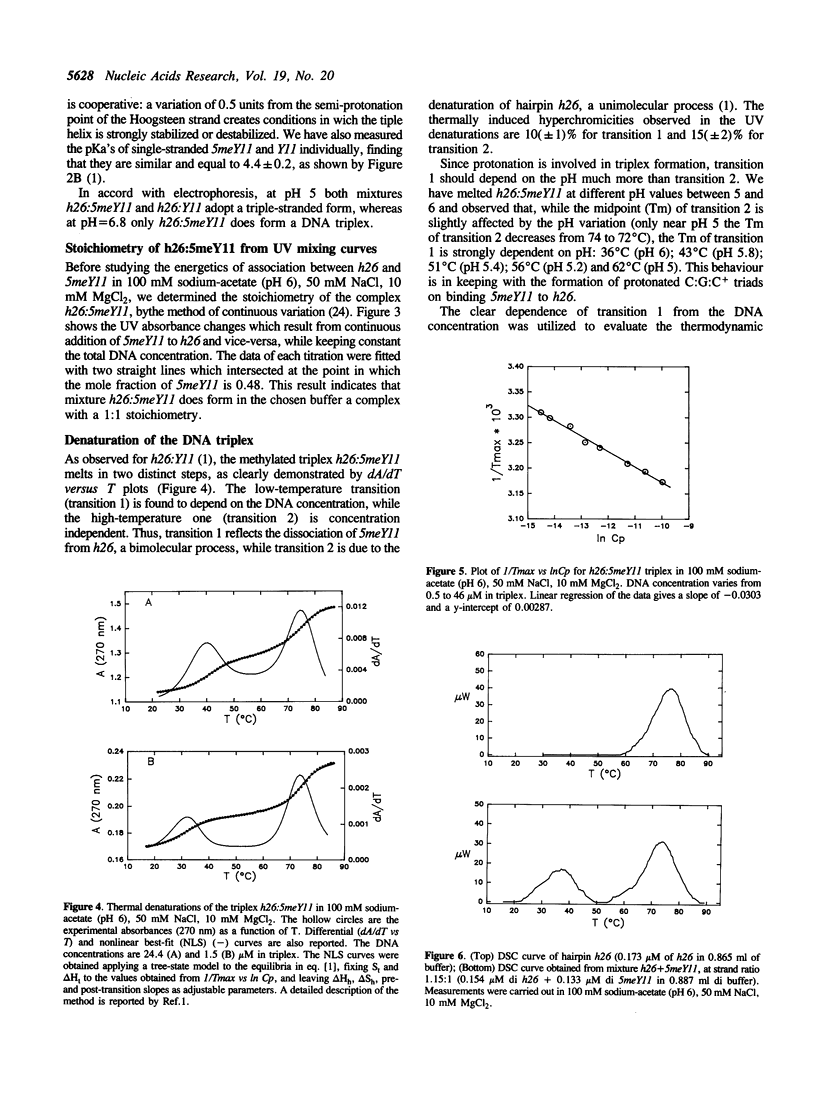
We have previously shown that the pyrimidine oligonucleotide 5'CTTCCTCCTCT (Y11) recognizes the double-helical stem of hairpin 5'GAAGGAGGAGA-T4-TCTCCTCCTTC (h26) by triple-helix formation (1). In this paper, we report the effect on triplex formation of substituting the cytosine residues of Y11 with 5-methylcytosines (5meY11). In addition, we have studied the thermodynamics of the interaction between h26 and 5meY11. The results can be summarised as follows: (i) gel electrophoresis shows that at T = 5 degrees C and pH 5, both Y11 and 5meY11 form DNA triple helices with h26, whereas at pH 6.8 only the methylated strand binds to h26; (ii) pH-stability curves of the DNA triplexes formed from h26 + Y11 and h26 + 5meY11 show that Y11 and 5meY11 are semi-protonated at pH 5.7 and 6.7, respectively. Thus, it is concluded that cytosine methylation expands the pH range compatible with triplex formation by one pH unit; (iii) as the unmethylated triplex (h26:Y11), the methylated one (h26:5meY11) denatures in a biphasic manner, in which the low temperature transition results from the dissociation of 5meY11 from h26. The Tm of the triplex to h26 plus 5meY11 transition is strongly enhanced (about 10 degrees C) by cytosine methylation. A van 't Hoff analysis of denaturation curves is presented; (iv) DSC experiments show that triplex formation between 5meY11 and h26 is characterized by delta H = -237 +/- 25 kJ/mol and delta S = -758 +/- 75 J/Kmol, corresponding to an average delta H of -21 kJ/mol and delta S of -69 J/Kmol per Hoogsteen base pair; (v) the thermodynamic analysis indicates that the extra stability imparted to the triplex by methylcytosine is entropic in origin.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Glaser R. L., Thomas G. H., Siegfried E., Elgin S. C., Lis J. T. Optimal heat-induced expression of the Drosophila hsp26 gene requires a promoter sequence containing (CT)n.(GA)n repeats. J Mol Biol. 1990 Feb 20;211(4):751–761. doi: 10.1016/0022-2836(90)90075-W. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6292–6296. doi: 10.1073/pnas.85.17.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Klump H. H., Löffler R. Reversible helix/coil transitions of left-handed Z-DNA structures. Comparison of the thermodynamic properties of poly(dG).poly(dC), poly[d(G-C)].poly[d(G-C)], and poly(dG-m5dC).poly(dG-m5dC). Biol Chem Hoppe Seyler. 1985 Apr;366(4):345–353. doi: 10.1515/bchm3.1985.366.1.345. [DOI] [PubMed] [Google Scholar]
- Le Doan T., Perrouault L., Praseuth D., Habhoub N., Decout J. L., Thuong N. T., Lhomme J., Hélène C. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987 Oct 12;15(19):7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Woodsworth M. L., Latimer L. J., Morgan A. R. Poly(pyrimidine) . poly(purine) synthetic DNAs containing 5-methylcytosine form stable triplexes at neutral pH. Nucleic Acids Res. 1984 Aug 24;12(16):6603–6614. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D., Cantor C. R. A stable complex between homopyrimidine oligomers and the homologous regions of duplex DNAs. Nucleic Acids Res. 1988 Mar 25;16(5):2165–2178. doi: 10.1093/nar/16.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Dervan P. B., Wold B. J. Kinetic analysis of oligodeoxyribonucleotide-directed triple-helix formation on DNA. Biochemistry. 1990 Sep 18;29(37):8820–8826. doi: 10.1021/bi00489a045. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Wold B., Dervan P. B. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989 Aug 18;245(4919):725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- Manzini G., Xodo L. E., Gasparotto D., Quadrifoglio F., van der Marel G. A., van Boom J. H. Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J Mol Biol. 1990 Jun 20;213(4):833–843. doi: 10.1016/S0022-2836(05)80267-0. [DOI] [PubMed] [Google Scholar]
- Michelson A. M., Massoulié J., Guschlbauer W. Synthetic polynucleotides. Prog Nucleic Acid Res Mol Biol. 1967;6:83–141. doi: 10.1016/s0079-6603(08)60525-5. [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Drushlyak K. N., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987 Dec 3;330(6147):495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Ohms J., Ackermann T. Thermodynamics of double- and triple-helical aggregates formed by self-complementary oligoribonucleotides of the type rAxUy. Biochemistry. 1990 Jun 5;29(22):5237–5244. doi: 10.1021/bi00474a004. [DOI] [PubMed] [Google Scholar]
- Pilch D. S., Brousseau R., Shafer R. H. Thermodynamics of triple helix formation: spectrophotometric studies on the d(A)10.2d(T)10 and d(C+3T4C+3).d(G3A4G3).d(C3T4C3) triple helices. Nucleic Acids Res. 1990 Oct 11;18(19):5743–5750. doi: 10.1093/nar/18.19.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum G. E., Park Y. W., Singleton S. F., Dervan P. B., Breslauer K. J. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praseuth D., Perrouault L., Le Doan T., Chassignol M., Thuong N., Hélène C. Sequence-specific binding and photocrosslinking of alpha and beta oligodeoxynucleotides to the major groove of DNA via triple-helix formation. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1349–1353. doi: 10.1073/pnas.85.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature. 1989 Jun 22;339(6226):637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr, Anderson C. F., Lohman T. M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978 May;11(2):103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F. Spectroscopic and calorimetric investigation on the DNA triplex formed by d(CTCTTCTTTCTTTTCTTTCTTCTC) and d(GAGAAGAAAGA) at acidic pH. Nucleic Acids Res. 1990 Jun 25;18(12):3557–3564. doi: 10.1093/nar/18.12.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xodo L. E., Manzini G., Quadrifoglio F., van der Marel G. A., van Boom J. H. The B-Z conformational transition in folded oligodeoxynucleotides: loop size and stability of Z-hairpins. Biochemistry. 1988 Aug 23;27(17):6327–6331. doi: 10.1021/bi00417a019. [DOI] [PubMed] [Google Scholar]
- de los Santos C., Rosen M., Patel D. NMR studies of DNA (R+)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989 Sep 5;28(18):7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]