Abstract
Amsacrine (m-AMSA) is an anticancer agent that displays activity against refractory acute leukemias as well as Hodgkin’s and non-Hodgkin’s lymphomas. The drug is comprised of an intercalative acridine moiety coupled to a 4’-amino-methanesulfon-m-anisidide head group. m-AMSA is historically significant in that it was the first drug demonstrated to function as a topoisomerase II poison. Although m-AMSA was designed as a DNA binding agent, the ability to intercalate does not appear to be the sole determinant of drug activity. Therefore, to more fully analyze structure-function relationships and the role of DNA binding in the action of m-AMSA, we analyzed a series of derivatives for the ability to enhance DNA cleavage mediated by human topoisomerase IIα and topoisomerase IIβ and to intercalate DNA. Results indicate that the 3’-methoxy (m-AMSA) positively affects drug function, potentially by restricting the rotation of the head group in a favorable orientation. Shifting the methoxy to the 2’-position (o-AMSA), which abrogates drug function, appears to increase rotational freedom of the head group and may impair interactions of the 1’-substituent or other portions of the head group within the ternary complex. Finally, the non-intercalative m-AMSA head group enhanced enzyme-mediated DNA cleavage when it was detached from the acridine moiety, albeit with 100-fold lower affinity. Taken together, our results suggest that much of the activity and specificity of m-AMSA as a topoisomerase II poison is embodied in the head group, while DNA intercalation is used primarily to increase the affinity of m-AMSA for the topoisomerase II-DNA cleavage complex.
Amsacrine (m-AMSA) is an acridine derivative with antineoplastic activity (1, 2). The drug is in multiple clinical trials for the treatment of hematological cancers in the United States (3) and is used to treat refractory acute lymphocytic and non-lymphocytic leukemias as well as Hodgkin’s and non-Hodgkin’s lymphomas in other countries (1, 4–6). m-AMSA kills cells by acting as a topoisomerase II poison and increases levels of covalent enzyme-cleaved DNA complexes primarily by decreasing rates of ligation (1, 7–13). In vitro, m-AMSA displays similar activity toward the two isoforms of human topoisomerase II, α and β (14, 15). However, evidence suggests that the β isoform may be the more important target for the cytotoxic actions of the drug (16–19).
m-AMSA is a historically significant topoisomerase II-targeted anticancer drug. In a pioneering study published by Zwelling et al. in 1981, the authors proposed that m-AMSA targeted a topoisomerase based on the ability of the drug to induce protein-associated DNA strand breaks in treated human cells (20). Three years later, m-AMSA was the first drug demonstrated to poison mammalian topoisomerase II in vitro or in human cells (7, 21).
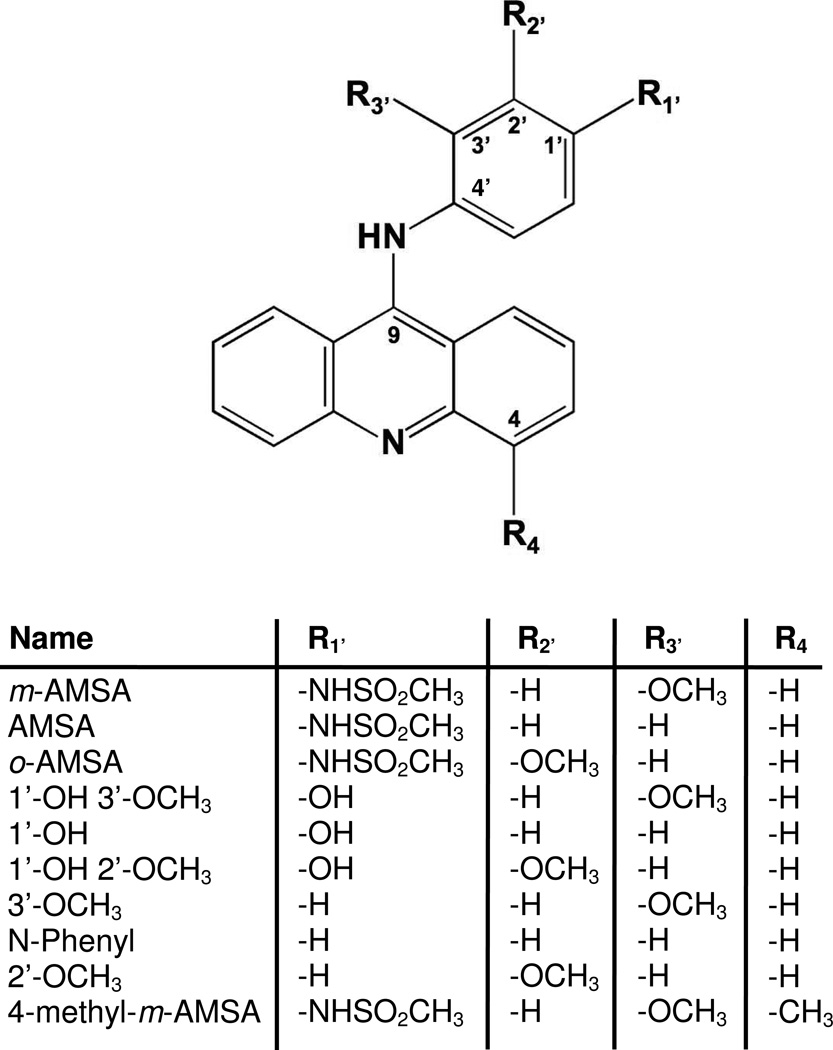
Whereas some topoisomerase II-targeted drugs, such as etoposide, have little if any interaction with DNA in the absence of enzyme (22–24), m-AMSA was designed to be a DNA binding agent (25, 26). To this point, m-AMSA is one of the most widely studied intercalative topoisomerase II poisons (27, 28). The drug is comprised of an acridine moiety coupled to a 4’-amino-methanesulfon-m-anisidide head group. It has long been known that moving the anisidide methoxy group from the meta (3’) to the ortho (2’) position (see Figure 1) attenuates drug activity against mammalian topoisomerase II, despite the fact that the resulting o-AMSA is a stronger intercalator than m-AMSA (7, 20, 29–31).
Figure 1.
Structure of m-AMSA and derivatives.
The relative activity of m-AMSA vs. o-AMSA against topoisomerase II indicates that DNA binding cannot be the sole determinant of drug function. Moreover, it raises a number of questions regarding the precise role of DNA intercalation in the action of m-AMSA and the contributions of head group substituents to topoisomerase II poisoning. Therefore, to more fully analyze structure-function relationships and the role of DNA binding in the action of m-AMSA, a series of derivatives was analyzed. Results indicate that much of the activity and specificity of m-AMSA as a topoisomerase II poison is embodied in the head group. DNA intercalation also is important for optimal drug function, being used primarily to increase the affinity of m-AMSA for the topoisomerase II-DNA cleavage complex.
EXPERIMENTAL PROCEDURES
Enzymes and Materials
Human topoisomerase IIα and topoisomerase IIβ were expressed in Saccharomyces cerevisiae (32) and purified as described previously (33). Human topoisomerase I was purchased from Topogen. Negatively supercoiled pBR322 DNA was prepared from Escherichia coli using a Plasmid Mega Kit (Qiagen) as described by the manufacturer. [γ-32P]ATP (~6000Ci/mmol) was obtained from Perkin-Elmer. m-AMSA and derivatives were synthesized as described previously (25, 26). N-(4-amino-3-methoxyphenyl) methane-sulfonamide hydrochloride (m-AMSA head group) and etoposide were obtained from Sigma. The m-AMSA head group was stored at −20 °C as a 0.5 M stock solution in 100% DMSO. All other drugs were stored at 4 °C as 20 mM stock solutions in 100% DMSO. All other chemicals were analytical reagent grade.
Plasmid DNA Cleavage
DNA cleavage reactions were carried out using the procedure of Fortune and Osheroff (34). Topoisomerase II DNA cleavage assays contained 220 nM human topoisomerase IIα or human topoisomerase IIβ and 10 nM negatively supercoiled pBR322 in a total of 20 µL of DNA cleavage buffer [10 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, and 2.5% (v/v) glycerol]. Unless stated otherwise, reaction mixtures were incubated at 37 °C for 6 min, and enzyme-DNA cleavage complexes were trapped by the addition of 2 µL of 5% SDS followed by 2 µL of 250 mM EDTA (pH 8.0). Proteinase K (2 µL of a 0.8 mg/mL solution) was added, and samples were incubated at 45 °C for 30 min to digest the enzyme. Samples were mixed with 2 µL of agarose gel loading buffer [60% sucrose in 10 mM Tris–HCl (pH 7.9), 0.5% bromophenol blue, and 0.5% xylene cyanol FF], heated at 45 °C for 5 min, and subjected to electrophoresis in 1% agarose gels in 40 mM Tris-acetate (pH 8.3) and 2 mM EDTA containing 0.5 µg/mL ethidium bromide. DNA bands were visualized with long-range ultraviolet light and quantified using an Alpha Innotech digital imaging system. DNA cleavage was monitored by the conversion of supercoiled plasmid DNA to linear molecules.
Assays were carried out in the absence of compound, in the presence of 0–50 µM m-AMSA or derivatives, or in the presence of 0–3.5 mM m-AMSA head group. In some cases, assays were carried out in the presence of 3 mM dithiothreitol (DTT).
Molecular Modeling
The Calculate Energy Protocol within the Minimization Module of Discovery Studio 2.1 (Accelrys, Inc), was used for conformational space searching for m-AMSA and o-AMSA. Initially, m-AMSA and o-AMSA were input into Discovery Studio 2.1 using the Builder module. Atoms were assigned using the CHARMm forcefield. Geometries for each of the compounds were optimized using the minimization protocol within the simulation tool. Lowest energy structures for each of the compounds were derived using the conjugate gradient algorithm, 2000 steps, and a RMS gradient of 0.001. A dielectric of 1.0, nonbond list radius of 14.0, and spherical cutoff electrostatics were applied.
In the lowest energy structures, torsion angles 1 (rotation angle between C9 and the linking N) for m-AMSA and o-AMSA (−101.78° and −102.82°, respectively) were similar. The lowest energy torsion angles 2 (rotation angle between the linking N and C4’) were −11.69° for m-AMSA and −3.84° for o-AMSA. Using these rotation angles in the starting structures for m-AMSA and o-AMSA, changes to torsion 1 and torsion 2 were evaluated for their contributions to the overall potential energy of the drugs using the energy calculation module within the simulation protocol. Each of the torsion angles was modulated in ± 5° increments from its lowest energy value, and the energy was calculated for each torsion angle change. This method allows the relative stability of the drug to be determined with respect to the lowest energy structure associated with each change in torsion angle.
DNA Cleavage Site Utilization
DNA cleavage sites were mapped using a modification (35) of the procedure of O’Reilly and Kreuzer (36). The pBR322 DNA substrate was linearized by treatment with HindIII. Terminal 5’-phosphates were removed by treatment with calf intestinal alkaline phosphatase and replaced with [32P]phosphate using T4 polynucleotide kinase and [γ-32P]ATP. The DNA was treated with EcoRI, and the 4332 bp singly-end-labeled fragment was purified from the small EcoRI-HindIII fragment by passage through a CHROMA SPIN+TE-100 column (Clontech). Reaction mixtures contained 1 nM labeled pBR322 DNA substrate and 90 nM human topoisomerase IIα in 50 µL of DNA cleavage buffer supplemented with 1 mM ATP in the absence or presence of m-AMSA or derivatives. Reaction mixtures were incubated at 37 °C for 30 s, and enzyme-DNA cleavage complexes were trapped by the addition of 5 µL of 5% SDS followed by 3.75 µL of 250 mM EDTA (pH 8.0). Proteinase K (5 µL of a 0.8 mg/mL solution) was added, and samples were incubated at 45 °C for 30 min to digest the enzyme. DNA products were ethanol precipitated and resuspended in 5 µL of polyacrylamide gel loading buffer [40% formamide, 10 mM NaOH, 0.02% xylene cyanol FF, and 0.02% bromophenol blue]. Samples were subjected to electrophoresis in denaturing 6% polyacrylamide sequencing gels. Gels were dried in vacuo, and DNA cleavage products were visualized with a Bio-Rad Molecular Imager FX.
DNA Intercalation
DNA intercalation was monitored as described previously (34, 37). When used as a substrate, relaxed plasmid DNA was generated by incubation with topoisomerase I (34, 37). Intercalation reaction mixtures contained 20 nM topoisomerase I, 5 nM relaxed or negatively supercoiled pBR322 DNA, and 0–150 µM m-AMSA or derivatives. Ethidium bromide (10 µM) and etoposide (100 µM) were included as positive and negative controls, respectively. Assays were carried out in a total of 20 µL of 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA, 50 mM KCl, 10 mM MgCl2, and 0.5 mM DTT. Mixtures were incubated at 37 °C for 10 min, extracted with a phenol/chloroform/isoamyl alcohol mixture (25:24:1), and added to 3 µL of 0.77% SDS and 77 mM EDTA (pH 8.0). Samples were mixed with 2 µL of agarose gel loading buffer, heated at 45 °C for 5 min, and subjected to electrophoresis in a 1% agarose gel in 100 mM Tris-borate (pH 8.3) and 2 mM EDTA. Gels were stained with 1 µg/mL ethidium bromide, and DNA bands were visualized as described for plasmid DNA cleavage.
Competition with DNA Intercalators
A 50-bp oligonucleotide duplex was designed using a previously identified topoisomerase II cleavage site from pBR322 (38). Oligonucleotide sequences were generated using an Applied Biosystems DNA synthesizer. The 50-mer top and bottom sequences were 5’-TTGGTATCTGCGCTCTGCTGAAGCC↓AGTTACCTTCGGAAAAAGAGTTGGT-3’ and 5’-ACCAACTCTTTTTCCGAAGGT↓AACTGGCTTCAGCAGAGCGCAGATACCAA-3’, respectively (arrows denote cleavage sites). The bottom strand was labeled on the 5’-terminus with [γ-32P]ATP using T4 polynucleotide kinase. Following labeling and gel purification, complementary oligonucleotides were annealed by incubation at 70 °C for 10 min and cooling to 25 °C.
DNA cleavage by human topoisomerase IIα was determined by a modification of the procedure of Fortune et al. (38). Reaction mixtures contained 220 nM human topoisomerase IIα and 100 nM double-stranded oligonucleotide in 10 µL of DNA cleavage buffer. Assays were carried out in the absence or presence of 25 µM m-AMSA and 0–50 µM acridine, 9-aminoacridine, or ethidium bromide. Reactions were incubated for 10 min at 37 °C. DNA cleavage products were trapped by the addition of 2 µL of 10% SDS, followed by 1 µL of 375 mM EDTA (pH 8.0). Samples were digested with proteinase K, and DNA products were ethanol precipitated and resuspended in 5 µL of polyacrylamide gel loading buffer. Samples were subjected to electrophoresis in denaturing 14% polyacrylamide sequencing gels. Gels were dried in vacuo, and DNA cleavage products were visualized as described above.
RESULTS AND DISCUSSION
Contributions of m-AMSA Head Group Substituents to Drug-Induced DNA Cleavage by Human Type II Topoisomerases
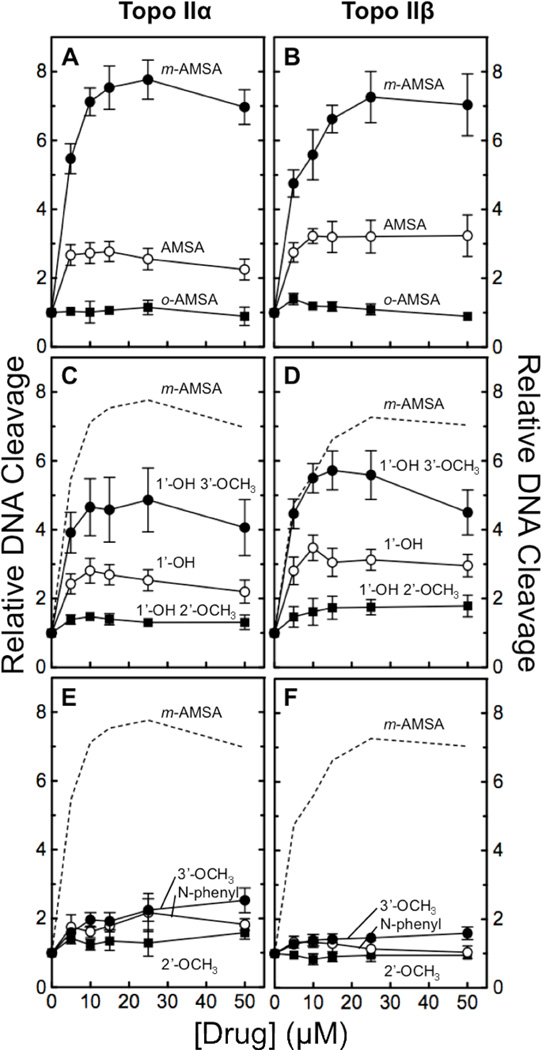
It has long been known that the activity of o-AMSA as a topoisomerase II poison is dramatically lower than that of m-AMSA (see Figure 1 for drug structures) (7, 20, 29–31). As seen in Figure 2, m-AMSA enhanced DNA cleavage mediated by human topoisomerase IIα or topoisomerase IIβ ~7– to 8–fold as compared to no drug reactions, whereas o-AMSA displayed almost no ability to poison either enzyme. This difference is despite the fact that the only change between m- and o-AMSA is the position of the methoxy group (3’ vs. 2’, respectively). The molecular basis underlying this difference in drug activity has not been delineated and several questions have yet to be addressed. For example, does the low activity of o-AMSA reflect the loss of a critical interaction between the 3’-methoxy of m-AMSA and topoisomerase II or DNA, or does the 2’-methoxy of o-AMSA sterically hinder interactions of the drug in the enzyme-DNA complex (or a combination of both)?
Figure 2.
Enhancement of topoisomerase II-mediated DNA cleavage by m-AMSA and derivatives. The effects of m-AMSA (closed circles), AMSA (open circles), and o-AMSA (squares) (panels A and B), 1’-OH 3’-OCH3 (closed circles), 1’-OH (open circles) and 1’-OH 2’-OCH3 (squares) (panels C and D), and 3’-OCH3 (closed circles), N-phenyl (open circles), and 2’-OCH3 (squares) (panels E and F) on the cleavage of negatively supercoiled plasmid DNA by human topoisomerase IIα (panels A, C, and E) and topoisomerase IIβ (panels B, D, and F) were determined. Error bars represent the standard deviation of three independent experiments. Data for m-AMSA are included as dashed lines in panels C–F for comparison.
Therefore, we characterized the ability of AMSA, a derivative of m-AMSA that is lacking the methoxy substituent, to stimulate topoisomerase II-mediated DNA cleavage. If the activity of AMSA were similar to that of m-AMSA, it would suggest that the methoxy group does not enhance drug activity when in the 3’-position, but rather inhibits activity when in the 2’-position. Alternatively, if the activity of AMSA were similar to that of o-AMSA, it would imply that the 3’-methoxy is critical for topoisomerase II poisoning. Results are shown in panels A and B of Figure 2. The activity of AMSA was intermediate to those of m-AMSA and o-AMSA, increasing levels of DNA cleavage ~3–fold. This finding indicates that the presence of the 3’-methoxy positively affects drug function and is necessary for optimal activity, while the presence of the 2’-methoxy impairs drug interactions.
To further explore the role of the methoxy group in drug function, we compared the activities of compounds that contained a 3’-, 2’-, or no methoxy in two additional series: one that replaced the 1’-methanesulfonamide with a 1’-hydroxy moiety (Figure 2, panels C and D) and another that lacked a 1’-substituent (panels E and F). Results were similar to those described above for the 1’-methanesulfonamide series. Compounds with a 3’-methoxy always induced the highest levels of DNA cleavage, compounds with a 2’-methoxy had little effect on enzyme activity, and compounds lacking the methoxy were intermediate.
Although the above relationships regarding the methoxy group were consistent across the three series, the nature of the 1’-substituent had a profound effect on drug activity. Comparing compounds with a 3’-methoxy, the activity of that with a 1’-methanesulfonamide was greater than that with a 1’-hydroxy, which was much greater than that with no substituent at the 1’ position. Thus, it appears that the ability of the 1’-substituent to form hydrogen bonds (or other interactions) is important for drug activity against human type II topoisomerases.
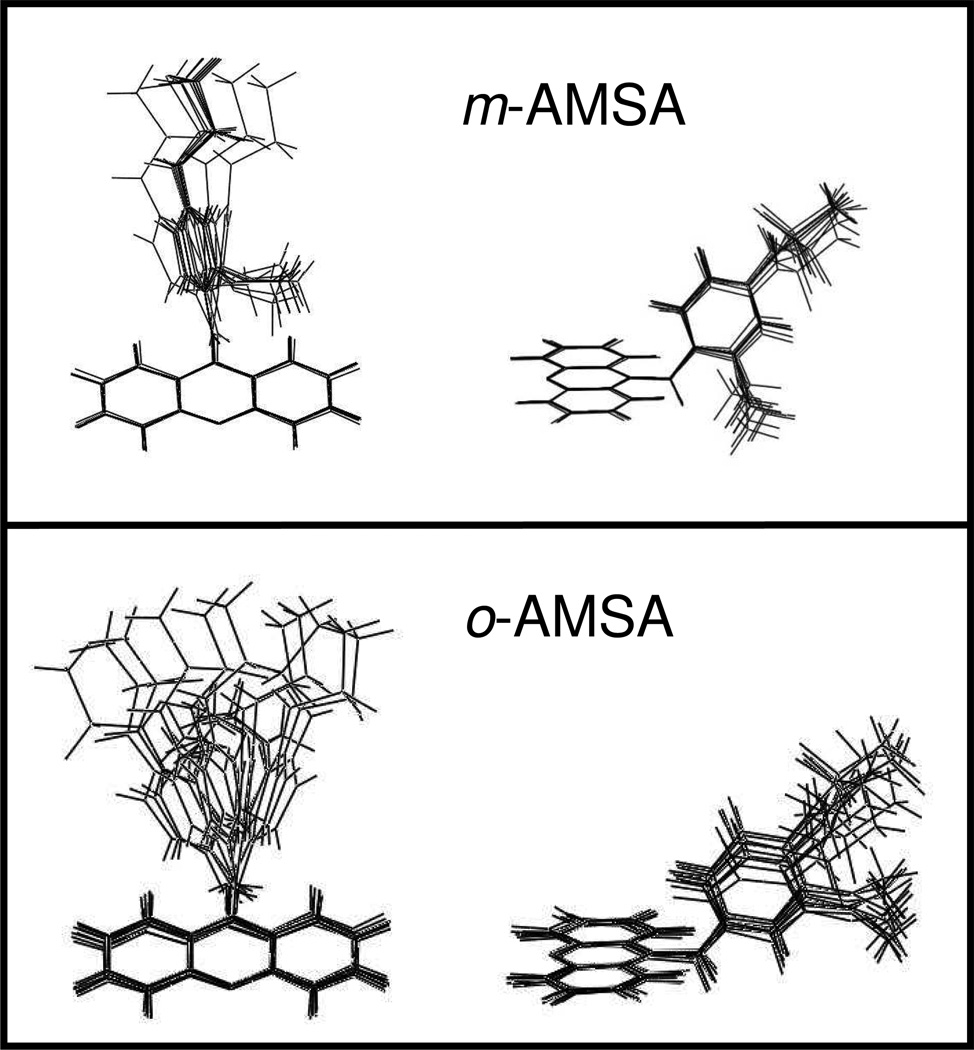
The above results indicate that the 3’-methoxy and 1’-methanesulfonamide positively impact the ability of m-AMSA to poison topoisomerase II, while the 2’-methoxy of o-AMSA impairs this process. However, they do not provide an understanding of the underlying mechanism by which these substituents affect drug activity. Therefore, modeling studies were carried out with m-AMSA and o-AMSA to address this issue (Figure 3).
Figure 3.
Energy minimization models of m-AMSA and o-AMSA. Low-energy structures (within ± 5 kcal/mol of the optimized structures for m-AMSA (top) and o-AMSA (bottom) are shown. Front and side views are shown at left and right, respectively.
As determined by energy minimization calculations, the orientation of the head group in m-AMSA appears to be much more constrained than it is in o-AMSA (Figure 3). The lowest potential energy for m-AMSA (8.85 kcal/mol) was observed when torsion angle 1 (rotation angle between C9 and the linking N, see Figure 1) was set to −101.78°. Torsion angle 1 could be changed by −15° to +30° (a total of 45°) without significant changes in energy (range of 12.63 at −15° to 12.1 kcal/mol at +30°). Rotations past −15° and +30° resulted in a marked increase in the potential energy of the structure. Similar energy profiles were observed for changes to torsion angle 2 (rotation angle between the linking N and C4’). The lowest energy (8.85 kcal/mol) was observed at an angle of 11.69°. Incremental changes to torsion angle 2 followed by energy calculations revealed a significant increase in the potential energy of the molecule if torsion angle 2 was rotated more than −25° and +60° from the starting −11.69°.
In contrast to m-AMSA, o-AMSA appears to have a much broader low energy conformational space. Starting with the lowest energy conformation (−102.82°), torsion angle 1 could be changed by −40° to +45° (a total of 85°) with relatively small fluctuations in the potential energy of the molecule (range of 14.4 kcal/mol at −40° to 14.8 kcal/mol at +45°). The lowest energy for torsion angle 2 was observed at −3.84° and the angle could be rotated with near full rotational freedom.
On the basis of these modeling studies, we propose the following: the 3’-methoxy enhances drug activity by restricting the head group to a narrow range of favorable conformations. Conversely, when the 3’-methoxy is missing in AMSA or moved in o-AMSA, drug activity drops because the head group is no longer constrained to a favored orientation. Finally, the activity of o-AMSA is even lower than that of AMSA because, in addition to the unrestricted head group, the presence of the 2’-methoxy may impose steric constraints that further inhibit interactions of the 1’-substituent or other portions of the head group with the protein or DNA.
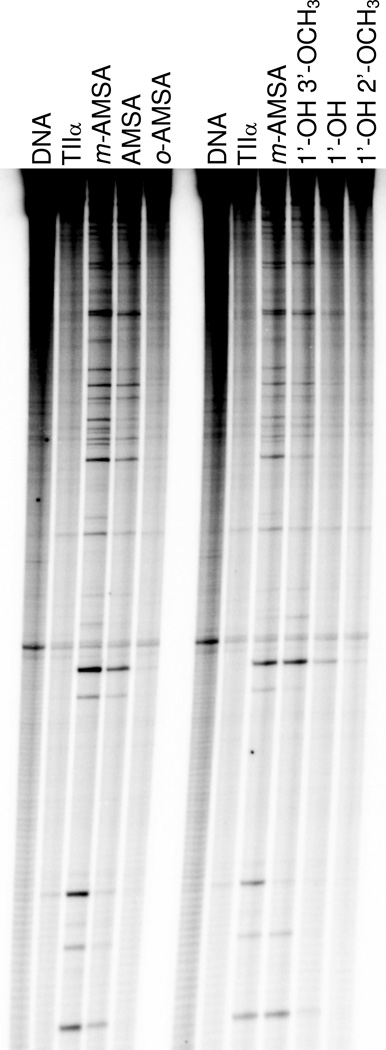
To determine whether changes in the above substituents affect the specificity of the drug class, sites of DNA cleaved by human topoisomerase IIα in the presence of m-AMSA and several derivatives were determined (Figure 4). Similar cleavage maps were observed for m-AMSA, AMSA (which lacks the methoxy group), and 1’-OH 3’-OCH3 (which contains a hydroxy in place of the 1’-methanesulfonamide group of the parent drug). However, minor differences with regard to site specificity and utilization were observed. This result suggests that portions of the m-AMSA head group may have interactions with DNA as well as the protein in the ternary enzyme-drug-DNA complex.
Figure 4.
DNA cleavage site specificity and utilization by human topoisomerase IIα in the presence of m-AMSA and derivatives. A singly end-labeled linear 4332 bp fragment of pBR322 was used as the cleavage substrate. An autoradiogram of a polyacrylamide gel is shown. DNA cleavage reactions were carried out in the absence of drug (TIIα), or in the presence of 10 µM m-AMSA; 25 µM AMSA, 1'-OH 3’-OCH3, or 1’-OH; or 100 µM o-AMSA or 1'-OH 2’-OCH3. DNA standards (DNA) also are shown. Results are representative of three independent experiments.
DNA Intercalation
m-AMSA was originally designed as a DNA-binding drug (25, 26). However, there is no clear correlation between the strength of DNA binding (as determined by intercalation) and drug activity against topoisomerase II. As discussed earlier, o-AMSA, which intercalates more strongly than m-AMSA, displays little ability to poison the type II enzyme (7, 20, 29–31). Therefore, to more fully explore relationships between DNA binding and topoisomerase II poisoning, the ability of the compounds described in Figure 1 to intercalate was determined. Representative intercalation assay gels for m-AMSA, AMSA, o-AMSA and 9-aminoacridine are shown in Figure 5. Representative gels for the other compounds employed are shown in Supplemental Figure S1.
Figure 5.
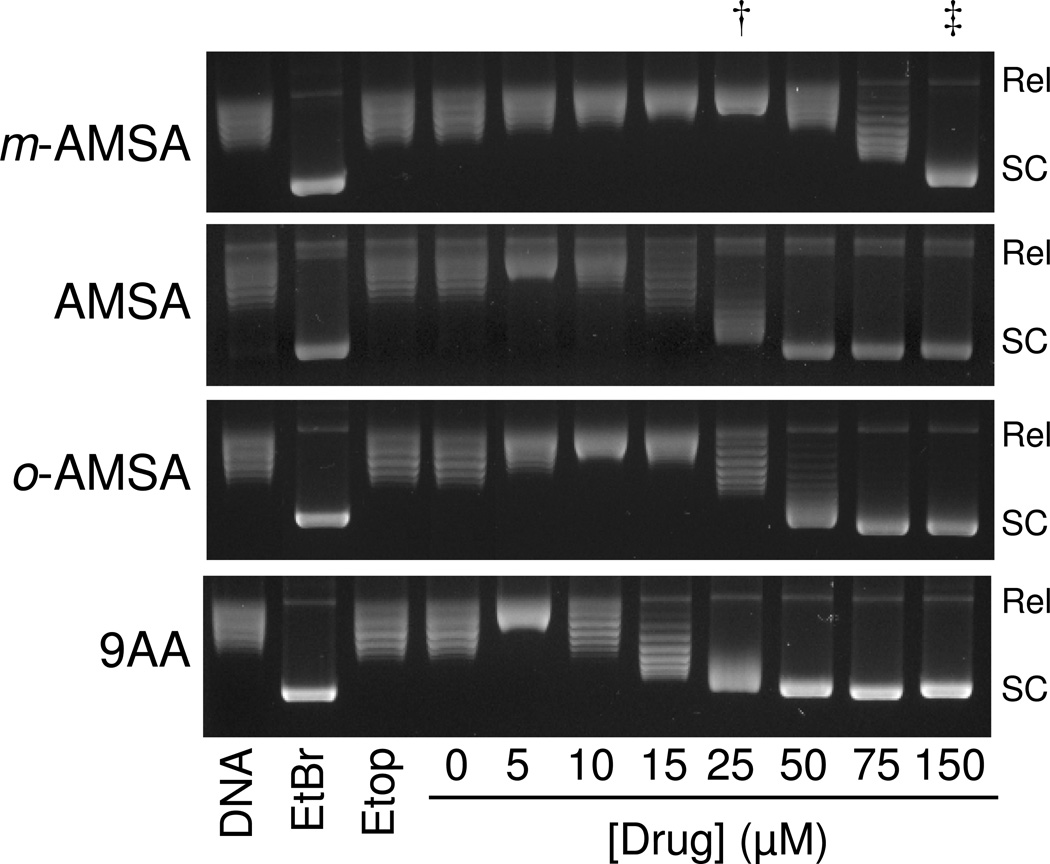
DNA intercalation by m-AMSA and derivatives. The abilities of 0–150 µM m-AMSA, AMSA, o-AMSA, and 9-aminoacridine (9AA) to intercalate into DNA were determined using a topoisomerase I-based supercoiling assay. Representative ethidium bromide (EtBr)-stained agarose gels are shown. The effects of 10 µM EtBr and 100 µM etoposide (Etop) are included as positive and negative controls, respectively. Relaxed DNA standards (DNA) also are shown. Results are representative of three independent experiments. As described in Table 1, concentrations of intercalators required to yield “fully relaxed” (Rel) and “fully supercoiled” (SC) plasmid are used for comparative purposes. Lanes that include these concentrations for m-AMSA are indicated by a dagger (†) and double dagger (‡), respectively.
The DNA intercalation assay is based on the fact that intercalative agents induce constrained negative supercoils and compensatory unconstrained positive superhelical twists in covalently closed circular DNA. Therefore, as the concentration of an intercalative compound increases, a plasmid that is negatively supercoiled or relaxed (i.e., contains an equilibrium distribution of topoisomers whose mean number of superhelical twists is zero) appears to become positively supercoiled. Treatment of an intercalated plasmid with topoisomerase I removes the unconstrained positive DNA supercoils. Subsequent extraction of the compound allows the local drug-induced unwinding to redistribute in a global manner and manifest itself as a net negative supercoiling of the plasmid. Thus, in the presence of an intercalative agent, topoisomerase treatment converts plasmids (through a completely relaxed intermediate population) to a distribution of negatively supercoiled molecules.
In order to compare the relative abilities of compounds to intercalate, two data points were employed: the concentration of drug that converts the population of plasmids to the most relaxed form (i.e., the highest band seen on the gels in Figure 5) following treatment with topoisomerase I and the concentration of drug that converts the initial population to the fully supercoiled form (i.e., the lowest band seen on the gels in Figure 5). These values are given in Table 1 for all of the compounds used in the current study.
Table 1.
DNA Intercalation by m-AMSA and Derivativesa
| Compound | Fully Relaxed | Fully Supercoiled |
|---|---|---|
| Concentration (µM) | Concentration (µM) | |
| m-AMSA | 25 | 150 |
| AMSA | 5–10 | 50 |
| o-AMSA | 10 | 75 |
| 1’-OH 3’-OCH3 | 15 | 150 |
| 1’-OH | 5 | 25 |
| 1’-OH 2’-OCH3 | 5 | 25–50 |
| 3’-OCH3 | 50–75 | >150 |
| N-Phenyl | 15–25 | 75 |
| 2’-OCH3 | 15 | 150 |
| 9-aminoacridine | 5 | 50 |
| 4-methyl-m-AMSA | 15 | 75 |
| Acridine | 75 | ≫150 |
| Ethidium Bromide | 1 | 5–7.5 |
The concentrations of compounds required to convert the plasmid substrate to a “fully relaxed” or “fully supercoiled” population was assessed by the topoisomerase I DNA supercoiling assay described in Figure 5.
The chemical nature of the 1’-substituent had a consistent effect on drug intercalation into DNA, with the strength of intercalation being: 1’-hydroxy series > 1’-methanesulfonamide series > no 1’-substituent series. The presence and position of the methoxy group also affected drug intercalation in a consistent manner, with the strength of intercalation being: no methoxy > 2’-methoxy > 3’-methoxy. Despite these findings, as originally observed for m-AMSA and o-AMSA (7, 20, 29–31), there appears to be little correlation between the strength of DNA binding and enhancement of topoisomerase II-mediated DNA cleavage. For example, while members of the 1’-hydroxy series are stronger intercalators than corresponding members of the 1’-methanesulfonamide series, they are weaker topoisomerase II poisons.
One caveat regarding the above observations should be noted: all of the m-AMSA derivatives that were examined contained altered substituents on the head group. Since portions of the head group are likely to interact with topoisomerase II or the scissile bond in the cleavage complex, it is possible that the same alterations that strengthen DNA intercalation also interfere with these critical interactions (or vice versa). This could explain the lack of correlation between drug activity and DNA intercalation. Therefore, to address the issue of DNA binding without changing substituents on the head group, the ability of 4-methyl-m-AMSA to intercalate DNA and poison topoisomerase IIα was evaluated. The methyl substituent in this compound is on the acridine ring. A previous crosslinking study using a photoactivated m-AMSA analog (39) demonstrated that the acridine moiety interacts with DNA in the ternary topoisomerase II-drug-DNA complex (40).
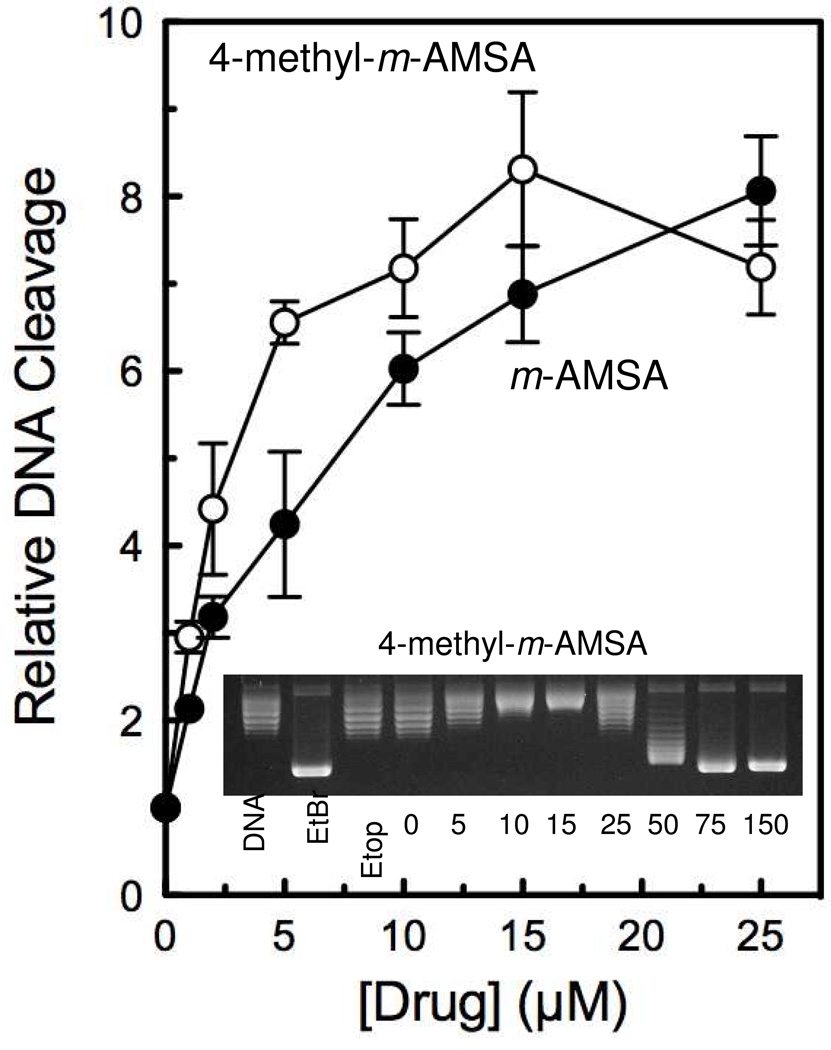
Intercalation studies (Figure 6 inset and Table 1) indicate that 4-methyl-m-AMSA binds DNA with an affinity that is approximately twice that of m-AMSA. Similarly, the potency of 4-methyl-m-AMSA (as determined by a topoisomerase IIα-DNA cleavage assay), was ~2–fold higher than that of m-AMSA. Despite the change in potency, the methylated and unmethylated compounds displayed comparable efficacies (maximal levels of DNA cleavage).
Figure 6.
Enhancement of topoisomerase IIα-mediated DNA cleavage by 4-methyl-m-AMSA. The effects of 4-methyl-m-AMSA (open circles) on the cleavage of negatively supercoiled plasmid DNA by human topoisomerase IIα are compared to those of m-AMSA (closed circles). Error bars represent the standard deviation of three independent experiments. The inset shows a representative topoisomerase I-based intercalation assay for 4-methyl-m-AMSA.
The correlation between DNA binding and cleavage observed for 4-methyl-m-AMSA suggests that intercalation plays an important role in drug action by increasing the affinity of m-AMSA for the ternary complex. If this suggestion is correct, inhibiting the ability of m-AMSA to intercalate should attenuate drug action. To test this prediction, competition experiments were carried out in the presence of acridine (a weak intercalator), 9-aminoacridine (a strong intercalator), and ethidium bromide (a stronger intercalator) (see Table 1 and Figures 4 and S1 for intercalation data).
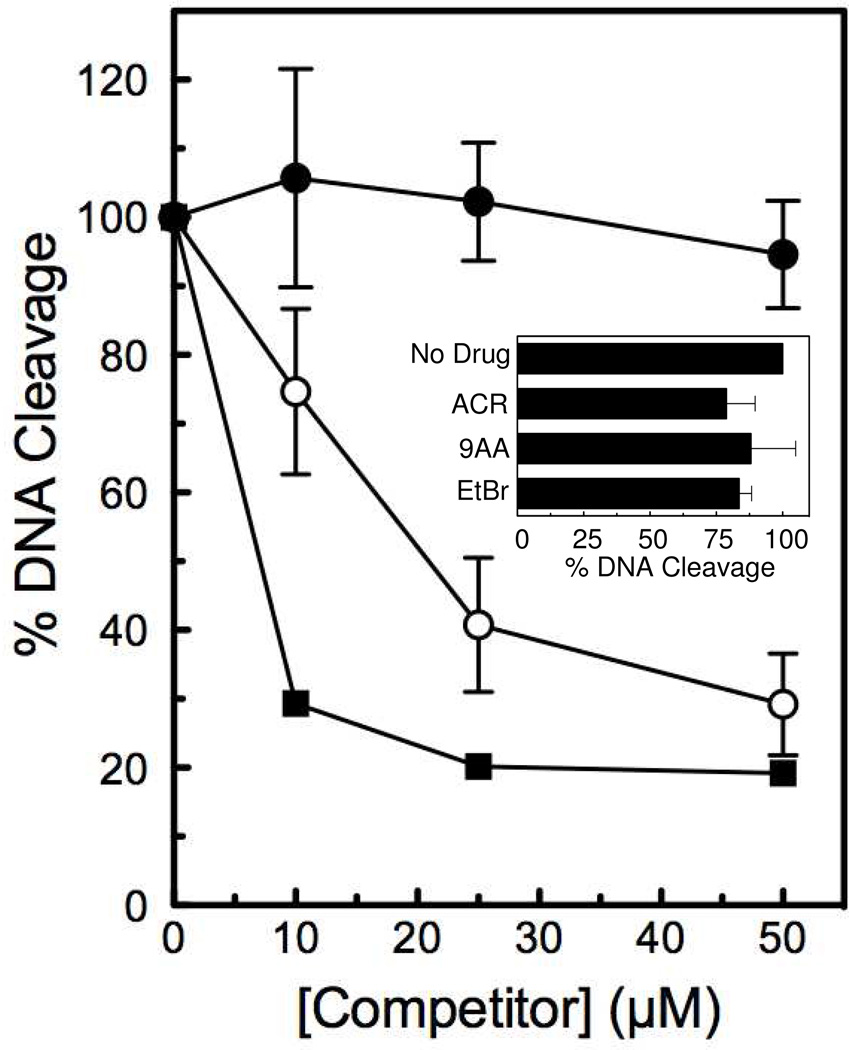
As seen in Figure 7, acridine, 9-aminoacridine, and ethidium bromide inhibited topoisomerase IIα-mediated DNA cleavage induced by 25 µM m-AMSA to an extent proportional to their DNA binding strengths. Acridine showed little inhibition of DNA cleavage at 50 µM. In contrast, 9-aminoacridine and ethidium bromide inhibited m-AMSA-induced DNA cleavage by 50% at ~21 and ~7 µM, respectively.
Figure 7.
Inhibition of m-AMSA-induced topoisomerase IIα-mediated DNA cleavage by intercalators. The abilities of 0–50 µM acridine (ACR, closed circles), 9-aminoacridine (9AA, open circles), and ethidium bromide (EtBr, squares) to inhibit DNA cleavage induced by 25 µM m-AMSA were determined. A singly end-labeled 50-mer oligonucleotide substrate was used as the cleavage substrate. The level of DNA cleavage in the presence of 25 µM m-AMSA and the absence of competitor was set to 100%. The inset shows the effects of 50 µM ACR, 9AA, and EtBr on DNA cleavage mediated by human topoisomerase IIα in the absence of m-AMSA. The baseline level of DNA cleavage in the absence of intercalators was set to 100%. Error bars represent the standard deviation of three independent experiments for both the figure and the inset.
It is notable that intercalators can decrease topoisomerase II-mediated cleavage by interfering with DNA binding or by altering the apparent topology of DNA (making the substrate appear to be positively supercoiled) in a closed topological system (15). To minimize the effects of intercalators on baseline levels of DNA cleavage mediated by human topoisomerase IIα, an oligonucleotide system was utilized for the competition experiments. Indeed, at 50 µM ethidium bromide, the highest concentration of the strongest intercalator employed (Table 1), baseline levels of enzyme-mediated DNA cleavage dropped only 16% (Figure 7, inset).
Taken together, the above findings support the conclusion that DNA intercalation plays an important role in the actions of m-AMSA as a topoisomerase II poison.
Activity of the m-AMSA Head Group
A number of topoisomerase II-targeted drugs contain a multi-ring system attached to a head group (1, 2). For example, the anticancer drug etoposide is comprised of a glycosylated polycyclic core (albeit non-intercalative) that is linked to a 3’,5’-dimethoxy-4’-hydroxyphenyl head group (E-ring) (1, 2, 24). A recent structure of a covalent human topoisomerase IIβ-DNA cleavage complex formed in the presence of etoposide indicates that the polycyclic drug core interacts primarily with DNA, while the head group is positioned at the interface between the enzyme and the cleaved scissile bond and interacts with both the protein and DNA (41). Saturation transfer difference NMR studies of the etoposide-topoisomerase IIα binary complex coupled with activity studies also suggest strong interactions between portions of the polycyclic core/glycosyl group and DNA, and between the head group and the enzyme (42–44).
Consistent with the studies on etoposide, the findings described above for m-AMSA imply that the acridine core and head group of the drug may play different and complementary functions in stabilizing the topoisomerase II-DNA complex. We propose that the acridine portion of m-AMSA is largely responsible for DNA binding, while the head group interacts with the enzyme and the scissile bond in the ternary complex.
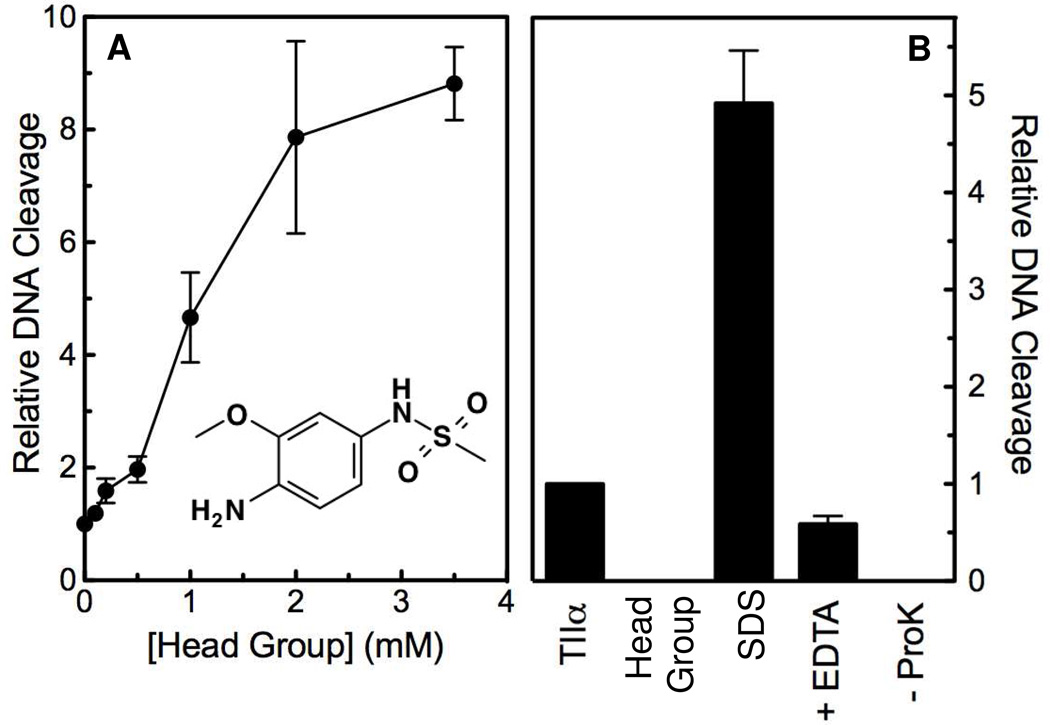
In order to address this hypothesis, the ability of the isolated m-AMSA head group to enhance DNA cleavage mediated by human topoisomerase IIα was assessed (unfortunately, the o-AMSA head group was unavailable for testing). As seen in Figure 8 (left panel), the detached head group stimulated DNA scission ~8– to 9–fold, which is comparable to levels observed with ~25 µM m-AMSA. In contrast to the parent drug, however, ~3.5 mM head group was required to induce this level of cleavage. Thus, the efficacy of the isolated head group is similar to that of the parent compound, but the potency is >2 orders of magnitude lower.
Figure 8.
Enhancement of topoisomerase IIα-mediated DNA cleavage by the detached m-AMSA head group. Panel A shows the effects of the isolated m-AMSA head group (structure shown as inset) on the cleavage of negatively supercoiled plasmid DNA by human topoisomerase IIα. determined. Panel B shows a series of control experiments that confirm that DNA cleavage induced by the isolated head group is mediated by human topoisomerase IIα. Reactions contained DNA and enzyme in the absence of m-AMSA head group (TIIα), DNA and 3 mM head group in the absence of enzyme, or complete reaction mixtures treated with SDS prior to adding EDTA (SDS). The reversibility of DNA cleavage induced by 3 mM head group was determined by incubating reactions with EDTA prior to trapping cleavage complexes with SDS (EDTA). To determine whether DNA cleavage induced by 3 mM head group was protein-linked, proteinase K treatment was omitted (−ProK). Error bars for both panels represent standard deviations for three independent experiments.
To ensure that the detached head group was inducing DNA cleavage through an effect on topoisomerase IIα, several control experiments were carried out (Figure 8, right panel). No DNA cleavage was observed in the absence of enzyme. In addition, DNA scission was reversed by the addition of EDTA, which chelates the essential divalent cation (45), and no free linear DNA was seen in reactions that were not treated with proteinase K to digest the topoisomerase II. Thus, the isolated head group induces DNA cleavage through its effects on the type II enzyme.
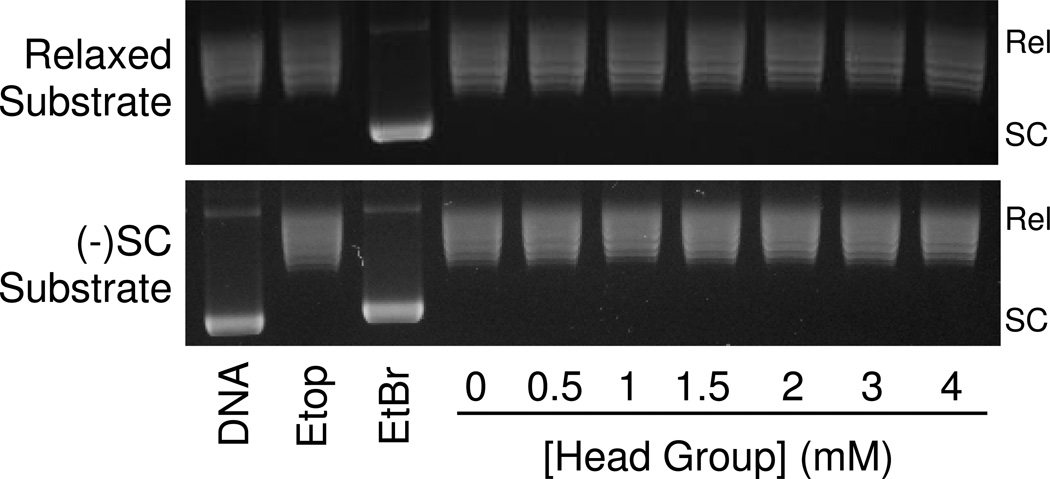
Studies described above predict that the intercalation of m-AMSA is mediated by the acridine portion of the drug. If this is the case, the isolated head group would not be expected to intercalate. Indeed, even at concentrations as high as 4 mM, no intercalation by the head group was observed (Figure 9).
Figure 9.
The isolated m-AMSA head group does not intercalate in DNA. Representative topoisomerase I-based DNA intercalation assay gels are shown for the isolated m-AMSA head group. Assays starting with either relaxed or negatively supercoiled DNA plasmid substrates are included. Etoposide (Etop, 100 µM) and ethidium bromide (EtBr, 10 µM) are included as positive and negative controls, respectively. DNA standards (DNA) also are shown. Results are representative of three independent experiments.
Both the head group and the intercalative acridine moiety are required for the high potency of m-AMSA as a topoisomerase II poison. Because intercalation alters the structure of DNA, acridine may increase the potency of m-AMSA by a specific or a general mechanism. In the first case, intercalation by the acridine moiety of an individual m-AMSA molecule increases the affinity of the attached head group for the enzyme and the scissile bond. In the second case, intercalation by m-AMSA molecules outside of the topoisomerase II active site alters the local DNA structure in a manner that increases the affinity of a different m-AMSA head group for the scissile bond. To distinguish between these two possibilities, we characterized the importance of the linkage between the acridine moiety and the head group for drug function. As seen in Figure 10, the activity of a 1:1 mixture of detached head group:acridine was lower than that of the head group alone, and the potency of the mixture was considerably lower than that of m-AMSA (in which the head group and acridine moiety are linked). This finding provides strong evidence that the linkage between the acridine moiety and the head group is critical for the potent activity of m-AMSA.
Figure 10.
Covalent linkage of the head group to the acridine moiety is necessary for the high potency of m-AMSA as a topoisomerase II poison. The ability of the detached head group (HG, closed circles) or a 1:1 mixture of head group + acridine (HG + ACR, open circles) to stimulate topoisomerase IIα-mediated DNA cleavage is shown. A control experiment assessing the effects of acridine alone on the DNA cleavage activity of topoisomerase IIα (ACR, squares) also is shown. Error bars represent the standard deviation of three independent experiments.
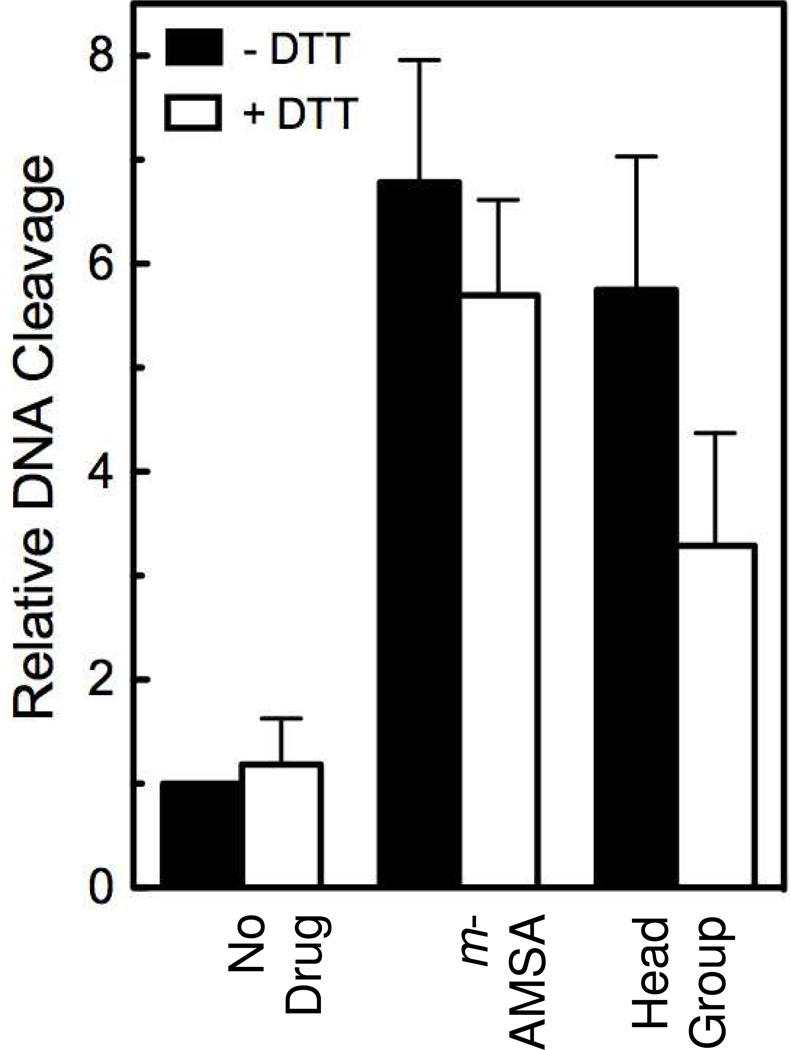
Because the m-AMSA head group has para amino substituents, it has the potential to undergo redox cycling with concomitant amino-imino tautomerization. Previous studies indicate that quinone-based compounds can poison topoisomerase II by a mechanism that differs from that of interfacial poisons such as m-AMSA (46–49). This redox-dependent mechanism involves covalent attachment of the drug to the enzyme (1, 47–50). It is not known whether imino-based compounds can also act as redox poisons of topoisomerase II. Therefore, to determine if a proportion of the activity of the m-AMSA head group against human topoisomerase IIα reflects a redox-dependent mechanism, the reducing agent DTT was added to DNA cleavage reactions. DTT blocks redox cycling, and reduces quinones to the corresponding hydroquinones and imines to the corresponding amines. Thus, it abrogates the effects of redox-dependent poisons on topoisomerase II.
No significant decrease in the activity of m-AMSA was seen in the presence of DTT, but incubation of the head group with the reducing agent did lower drug activity (Figure 11). However, because the head group retained the majority of its activity in the presence of DTT, we conclude that at least part of its activity toward topoisomerase IIα reflects a redox-independent mechanism like that of the parent drug.
Figure 11.
Partial redox-dependence of the isolated m-AMSA head group as a topoisomerase II poison. The effects of 25 µM m-AMSA or 3 mM m-AMSA head group on the cleavage of negatively supercoiled plasmid DNA by human topoisomerase IIα in the absence (closed bars) or presence (open bars) of 3 mM dithiothreitol (DTT) are shown. A control experiment carried out in the absence of drugs also is shown (No Drug). DNA cleavage levels are relative to those induced by the enzyme in the absence of drug or DTT. Error bars represent the standard deviation of three independent experiments.
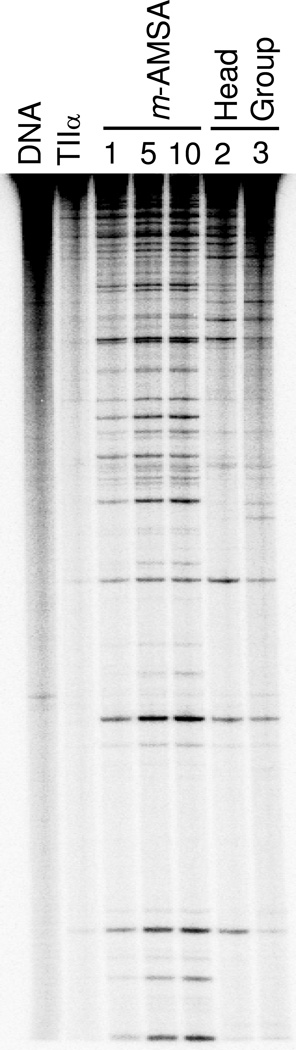
To further explore the properties of the head group, sites of topoisomerase IIα-mediated DNA cleavage induced by the compound were determined. Sites cleaved in the presence of the head group represented a subset of those induced by m-AMSA (Figure 12). Therefore, the head group appears to be responsible for much of the specificity of the drug.
Figure 12.
DNA cleavage site specificity and utilization by human topoisomerase IIα in the presence of the m-AMSA head group. A singly end-labeled linear 4332 bp fragment of pBR322 was used as the cleavage substrate. An autoradiogram of a polyacrylamide gel is shown. DNA cleavage reactions were carried out in the absence of drug (TIIα), or in the presence of 1, 5, or 10 µM m-AMSA or 2 or 3 mM head group. A DNA standard (DNA) also is shown. Results are representative of three independent experiments.
Conclusions
Although m-AMSA was the first compound shown to poison eukaryotic topoisomerase II, the specific functions of the individual components of the drug are still undefined. Taken together, our findings suggest that the activity and specificity of m-AMSA reside largely in the head group. Both the 3’-methoxy and 1’-methanesulfonamide substituents contribute positively to drug efficacy. Finally, the linkage between the head group and the intercalative acridine moiety provides a strong DNA anchor for the drug and, consequently, dramatically increases the affinity of m-AMSA for the topoisomerase II-DNA cleavage complex.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Katie J. Aldred and Dr. Steven L. Pitts for critical reading of the manuscript.
Footnotes
This work was supported by National Institutes of Health research grant GM33944 (to N.O.), The Auckland Division of the Cancer Society of New Zealand (to W.A.D.), and Department of Defense Breast Cancer Research Program grant BC095831P1 (to D.E.G.). A.C.K. was a trainee under the Chemistry-Biology Interface training grant T32 GM065086 from the National Institutes of Health.
SUPPORTING INFORMATION
Representative DNA intercalation gels for many of the compounds included in Table 1 are shown in Supplemental Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009;37:738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Cancer Institute. Clinical Trials. 2011 http://www.cancer.gov/clinicaltrials/search/results?protocolsearchid=9234167.
- 4.Jehn U, Heinemann V. New drugs in the treatment of acute and chronic leukemia with some emphasis on m-AMSA. Anticancer Res. 1991;11:705–711. [PubMed] [Google Scholar]
- 5.Kell J. Treatment of relapsed acute myeloid leukaemia. Rev. Recent Clin. Trials. 2006;1:103–111. doi: 10.2174/157488706776876445. [DOI] [PubMed] [Google Scholar]
- 6.Verma D, Kantarjian H, Faderl S, O'Brien S, Pierce S, Vu K, Freireich E, Keating M, Cortes J, Ravandi F. Late relapses in acute myeloid leukemia: analysis of characteristics and outcome. Leuk. Lymphoma. 2010;51:778–782. doi: 10.3109/10428191003661852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson EM, Tewey KM, Liu LF. Mechanism of antitumor drug action: poisoning of mammalian DNA topoisomerase II on DNA by 4'-(9-acridinylamino)-methanesulfon-m-anisidide. Proc. Natl. Acad. Sci. USA. 1984;81:1361–1365. doi: 10.1073/pnas.81.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson MJ, Osheroff N. Stabilization of the topoisomerase II-DNA cleavage complex by antineoplastic drugs: inhibition of enzyme-mediated DNA religation by 4'-(9-acridinylamino)methanesulfon-m-anisidide. Biochemistry. 1990;29:2511–2515. doi: 10.1021/bi00462a012. [DOI] [PubMed] [Google Scholar]
- 9.Robinson MJ, Osheroff N. Effects of antineoplastic drugs on the post-strand-passage DNA cleavage/religation equilibrium of topoisomerase II. Biochemistry. 1991;30:1807–1813. doi: 10.1021/bi00221a012. [DOI] [PubMed] [Google Scholar]
- 10.Sorensen BS, Sinding J, Andersen AH, Alsner J, Jensen PB, Westergaard O. Mode of action of topoisomerase II-targeting agents at a specific DNA sequence. Uncoupling the DNA binding, cleavage and religation events. J. Mol. Biol. 1992;228:778–786. doi: 10.1016/0022-2836(92)90863-f. [DOI] [PubMed] [Google Scholar]
- 11.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat. Res. 2007;623:83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deweese JE, Burgin AB, Osheroff N. Using 3'-bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase IIα. Biochemistry. 2008;47:4129–4140. doi: 10.1021/bi702194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh KL, Willmore E, Tinelli S, Cornarotti M, Meczes EL, Capranico G, Fisher LM, Austin CA. Amsacrine-promoted DNA cleavage site determinants for the two human DNA topoisomerase II isoforms alpha and beta. Biochem. Pharmacol. 1996;52:1675–1685. doi: 10.1016/s0006-2952(96)00516-3. [DOI] [PubMed] [Google Scholar]
- 15.McClendon AK, Osheroff N. The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs. Biochemistry. 2006;45:3040–3050. doi: 10.1021/bi051987q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Withoff S, de Vries EG, Keith WN, Nienhuis EF, van der Graaf WT, Uges DR, Mulder NH. Differential expression of DNA topoisomerase II alpha and - beta in P-gp and MRP-negative VM26, m-AMSA and mitoxantrone-resistant sublines of the human SCLC cell line GLC4. Br. J. Cancer. 1996;74:1869–1876. doi: 10.1038/bjc.1996.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dereuddre S, Delaporte C, Jacquemin-Sablon A. Role of topoisomerase II beta in the resistance of 9-OH-ellipticine-resistant Chinese hamster fibroblasts to topoisomerase II inhibitors. Cancer Res. 1997;57:4301–4308. [PubMed] [Google Scholar]
- 18.Herzog CE, Holmes KA, Tuschong LM, Ganapathi R, Zwelling LA. Absence of topoisomerase IIβ in an amsacrine-resistant human leukemia cell line with mutant topoisomerase IIα. Cancer Res. 1998;58:5298–5300. [PubMed] [Google Scholar]
- 19.Errington F, Willmore E, Tilby MJ, Li L, Li G, Li W, Baguley BC, Austin CA. Murine transgenic cells lacking DNA topoisomerase II® are resistant to acridines and mitoxantrone: analysis of cytotoxicity and cleavable complex formation. Mol. Pharmacol. 1999;56:1309–1316. doi: 10.1124/mol.56.6.1309. [DOI] [PubMed] [Google Scholar]
- 20.Zwelling LA, Michaels S, Erickson LC, Ungerleider RS, Nichols M, Kohn KW. Protein-associated deoxyribonucleic acid strand breaks in L1210 cells treated with the deoxyribonucleic acid intercalating agents 4'-(9-acridinylamino) methanesulfon-m-anisidide and adriamycin. Biochemistry. 1981;20:6553–6563. doi: 10.1021/bi00526a006. [DOI] [PubMed] [Google Scholar]
- 21.Yang L, Rowe TC, Nelson EM, Liu LF. In vivo mapping of DNA topoisomerase II-specific cleavage sites on SV40 chromatin. Cell. 1985;41:127–132. doi: 10.1016/0092-8674(85)90067-4. [DOI] [PubMed] [Google Scholar]
- 22.Ross W, Rowe T, Glisson B, Yalowich J, Liu L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984;44:5857–5860. [PubMed] [Google Scholar]
- 23.Chow KC, Macdonald TL, Ross WE. DNA binding by epipodophyllotoxins and N-acyl anthracyclines: Implications for mechanism of topoisomerase II inhibition. Mol. Pharmacol. 1988;34:467–473. [PubMed] [Google Scholar]
- 24.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr. Med. Chem. Anti-Cancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 25.Cain BF, Seelye RN, Atwell GJ. Potential antitumor agents. 14. Acridylmethanesulfonanilides. J. Med. Chem. 1974;17:922–930. doi: 10.1021/jm00255a003. [DOI] [PubMed] [Google Scholar]
- 26.Cain BF, Atwell GJ, Denny WA. Potential antitumor agents. 16. 4'-(Acridin9-ylamino)methanesulfonanilides. J. Med. Chem. 1975;18:1110–1117. doi: 10.1021/jm00245a013. [DOI] [PubMed] [Google Scholar]
- 27.Waring MJ. DNA-binding characteristics of acridinylmethanesulphonanilide drugs: comparison with antitumour properties. Eur. J. Cancer. 1976;12:995–1001. doi: 10.1016/0014-2964(76)90066-9. [DOI] [PubMed] [Google Scholar]
- 28.Elmore RH, Wadkins RM, Graves DE. Cooperative binding of m-AMSA to nucleic acids. Nucleic Acids Res. 1988;16:9707–9719. doi: 10.1093/nar/16.20.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wadkins RM, Graves DE. Thermodynamics of the interactions of m-AMSA and o-AMSA with nucleic acids: Influence of ionic strength and DNA base composition. Nucleic Acids Res. 1989;17:9933–9946. doi: 10.1093/nar/17.23.9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wadkins RM, Graves DE. Interactions of anilinoacridines with nucleic acids: effects of substituent modifications on DNA-binding properties. Biochemistry. 1991;30:4277–4283. doi: 10.1021/bi00231a025. [DOI] [PubMed] [Google Scholar]
- 31.Austin CA, Marsh KL, Wasserman RA, Willmore E, Sayer PJ, Wang JC, Fisher LM. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J. Biol. Chem. 1995;270:15739–15746. doi: 10.1074/jbc.270.26.15739. [DOI] [PubMed] [Google Scholar]
- 32.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 33.Kingma PS, Greider CA, Osheroff N. Spontaneous DNA lesions poison human topoisomerase IIα stimulate cleavage proximal to leukemic 11q23 chromosomal breakpoints. Biochemistry. 1997;36:5934–5939. doi: 10.1021/bi970507v. [DOI] [PubMed] [Google Scholar]
- 34.Fortune JM, Osheroff N. Merbarone inhibits the catalytic activity of human topoisomerase IIα by blocking DNA cleavage. J. Biol. Chem. 1998;273:17643–17650. doi: 10.1074/jbc.273.28.17643. [DOI] [PubMed] [Google Scholar]
- 35.Baldwin EL, Byl JA, Osheroff N. Cobalt enhances DNA cleavage mediated by human topoisomerase IIα in vitro and in cultured cells. Biochemistry. 2004;43:728–735. doi: 10.1021/bi035472f. [DOI] [PubMed] [Google Scholar]
- 36.O'Reilly EK, Kreuzer KN. A unique type II topoisomerase mutant that is hypersensitive to a broad range of cleavage-inducing antitumor agents. Biochemistry. 2002;41:7989–7997. doi: 10.1021/bi025897m. [DOI] [PubMed] [Google Scholar]
- 37.Fortune JM, Velea L, Graves DE, Osheroff N. DNA topoisomerases as targets for the anticancer drug TAS-103: DNA interactions and topoisomerase catalytic inhibition. Biochemistry. 1999;38:15580–15586. doi: 10.1021/bi991792g. [DOI] [PubMed] [Google Scholar]
- 38.Fortune JM, Dickey JS, Lavrukhin OV, Van Etten JL, Lloyd RS, Osheroff N. Site-specific DNA cleavage by Chlorella virus topoisomerase II. Biochemistry. 2002;41:11761–11769. doi: 10.1021/bi025802g. [DOI] [PubMed] [Google Scholar]
- 39.Shieh TL, Hoyos P, Kolodziej E, Stowell JG, Baird WM, Byrn SR. Properties of the nucleic acid photoaffinity labeling agent 3-azidoamsacrine. J. Med. Chem. 1990;33:1225–1230. doi: 10.1021/jm00166a022. [DOI] [PubMed] [Google Scholar]
- 40.Freudenreich CH, Kreuzer KN. Localization of an aminoacridine antitumor agent in a type II topoisomerase-DNA complex. Proc. Natl. Acad. Sci. USA. 1994;91:11007–11011. doi: 10.1073/pnas.91.23.11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 42.Wilstermann AM, Bender RP, Godfrey M, Choi S, Anklin C, Berkowitz DB, Osheroff N, Graves DE. Topoisomerase II - drug interaction domains: identification of substituents on etoposide that interact with the enzyme. Biochemistry. 2007;46:8217–8225. doi: 10.1021/bi700272u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bender RP, Jablonksy MJ, Shadid M, Romaine I, Dunlap N, Anklin C, Graves DE, Osheroff N. Substituents on etoposide that interact with human topoisomerase IIα in the binary enzyme-drug complex: Contributions to etoposide binding and activity. Biochemistry. 2008;47:4501–4509. doi: 10.1021/bi702019z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitts SL, Jablonksy MJ, Duca M, Dauzonne D, Monneret C, Arimondo PB, Anklin C, Graves DE, Osheroff N. Contributions of the D-ring to the activity of etoposide against human topoisomerase IIα: Potential interactions with DNA in the ternary enzyme-drug-DNA complex. Biochemistry. 2011;50:5058–5066. doi: 10.1021/bi200531q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osheroff N, Zechiedrich EL. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: Trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987;26:4303–4309. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- 46.Bender RP, Lindsey RH, Jr, Burden DA, Osheroff N. N-acetyl-p-benzoquinone imine, the toxic metabolite of acetaminophen, is a topoisomerase II poison. Biochemistry. 2004;43:3731–3739. doi: 10.1021/bi036107r. [DOI] [PubMed] [Google Scholar]
- 47.Lindsey RH, Bender RP, Osheroff N. Stimulation of topoisomerase II-mediated DNA cleavage by benzene metabolites. Chem. Biol. Interact. 2005;153–154:197–205. doi: 10.1016/j.cbi.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 48.Bender RP, Lehmler HJ, Robertson LW, Ludewig G, Osheroff N. Polychlorinated biphenyl quinone metabolites poison human topoisomerase IIα: Altering enzyme function by blocking the N-terminal protein gate. Biochemistry. 2006;45:10140–10152. doi: 10.1021/bi0524666. [DOI] [PubMed] [Google Scholar]
- 49.Bandele OJ, Osheroff N. (−)-Epigallocatechin gallate, a major constituent of green tea, poisons human type II topoisomerases. Chem. Res. Toxicol. 2008;21:936–943. doi: 10.1021/tx700434v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bender RP, Ham AJ, Osheroff N. Quinone-induced enhancement of DNA cleavage by human topoisomerase IIα: aAdduction of cysteine residues 392 and 405. Biochemistry. 2007;46:2856–2864. doi: 10.1021/bi062017l. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.