Abstract
The myosin superfamily is a versatile group of molecular motors involved in the transport of specific biomolecules, vesicles and organelles in eukaryotic cells. The processivity of myosins along an actin filament and transport of intracellular ‘cargo’ are achieved by generating physical force from chemical energy of ATP followed by appropriate conformational changes. The typical myosin has a head domain, which harbors an ATP binding site, an actin binding site, and a light-chain bound ‘lever arm’, followed often by a coiled coil domain and a cargo binding domain. Evolution of myosins started at the point of evolution of eukaryotes, S. cerevisiae being the simplest one known to contain these molecular motors. The coiled coil domain of the myosin classes II, V and VI in whole genomes of several model organisms display differences in the length and the strength of interactions at the coiled coil interface. Myosin II sequences have long-length coiled coil regions that are predicted to have a highly stable dimeric interface. These are interrupted, however, by regions that are predicted to be unstable, indicating possibilities of alternate conformations, associations to make thick filaments, and interactions with other molecules. Myosin V sequences retain intermittent regions of strong and weak interactions, whereas myosin VI sequences are relatively devoid of strong coiled coil motifs. Structural deviations at coiled coil regions could be important for carrying out normal biological function of these proteins.
Keywords: myosin structure, myosin domain architecture, coiled coil
Gene Organization and Evolutionary History
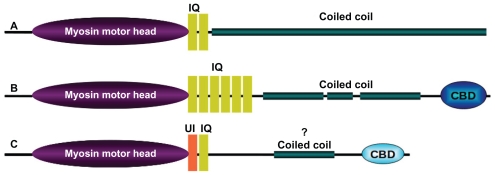
Myosin is a family of actin based molecular motors that hydrolyze ATP and generate physical force to move different cargoes inside the cell. This superfamily, divided into at least twenty four classes based on head domain sequence similarity and domain organization1,2 (Fig. 1), drive a large number of biological processes like cytokinesis,3,4 organelle transport,5,6 cell polarization and signal transduction7,8 in eukaryotes. They are typically 1000–2000 residues long and comprise of three domains: a conserved motor head with ATPase activity involved in actin binding and movement; a neck domain with bound regulatory proteins called myosin light chains; and a variable tail that often connects to various cargo associated proteins. The tail domain architectures vary considerably between the subtypes of myosin owing to their functional variation.9 In conventional myosin (myosin II) the myosin tail region forms a long coiled coil that facilitates dimerization and oligomerization.10 Coiled coils in many unconventional myosins terminate at a globular cargo binding domain, which associates with specific cargo to be localized in the cell (Fig. 2).
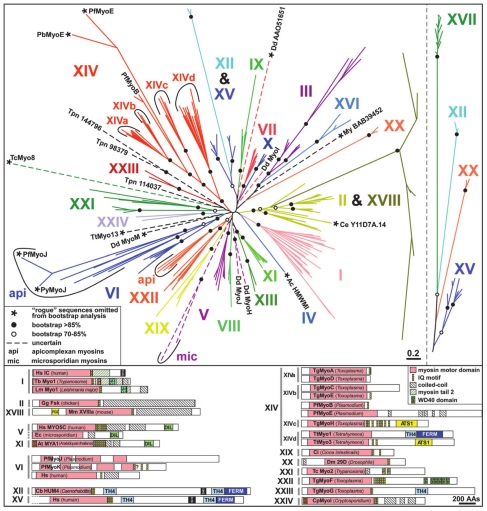
Figure 1.
Different classes of myosin superfamily: Phylogenetic tree of 114 myosin sequences showing 24 classes of myosins (Foth et al;65 Copyright (2006) National Academy of Sciences, USA.).
Figure 2.
Domain architecture. The overall length and domain organization vary considerably between the subclass members of myosin superfamily. Domain architectures of human (A) myosin II (NP_005954.3: 1939 amino acids) (B) myosin Va (NP_000250.3:1855 amino acids) and (C) myosin VI (NP_004990.3: 1285 amino acids). The length of predicted coiled coil domain using COILS program reflects the variation in length and architecture of the coiled coil domains. Lengths of coiled coil of human (A) myosin II (NP_005954.3 : residue 843–1931), (B) myosin Va (NP_000250.3: residue 911–1104; residue 1153–1234; residue 1339–1445) and (C) myosin VI (NP_004990.3: residue 864–1030) are shown in green.
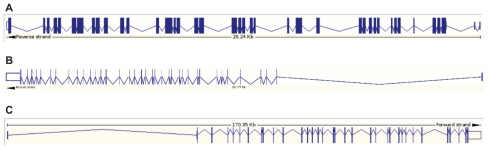
Myosin genes are highly interrupted with a large number of introns (Fig. 3). The number of exons in mammalian myosin can be as low as 12, as seen in myosins III and VII, whereas the exon number shoots up to 40 and higher in certain subclasses like myosin V. This variation is indicative of many isoforms with different cargo specificity and differences in localization. Most eukaryotes rely on myosins, except a few taxonomic groups, viz. red algae and diplomonad protists1 which are not known to possess genes with myosin head domain. No classical myosins have been found in prokaryotes. The ATPase domain of myosins along with that of the kinesin family of microtubule based motors is believed to have evolved from the core GTPase domain of G-proteins through deletion of strands 6 and 7 and addition of two N terminal strands.11,12 The wide distributions of myosins I and II across taxonomic levels ranging from yeast to human imply the early evolution of these two classes. Myosins V and XI, which diverged to animal and plant lineages respectively, are thought to have evolved from a common myosin II-like ancestor.1 The most recent hypothesis put forward the evolution of present day variability among myosins from three ancient myosin subfamilies (see Domain Architecture section for details): (i) the MSD subfamily standing for the general domain architecture of MYSc-SMC-DIL (MYSc-Myosin head domain, SMC-Structural Maintenance of Chromosomes is a chromosome segregation protein domain, DIL-DILute domain), comprising myosins V and XI; (ii) the subfamily of the architecture MYSc-MYTH4/FERM (MYTH4-Myosin Tail Homology 4, FERM-band 4.1/Ezrin/Radixin/Moesin-like domain) domain including myosin classes IV, VII, XII, and XV; and (iii) the subfamily containing a membrane binding TH1 tail domain comprising myosin I.1
Figure 3.
Gene organization of myosin superfamily. Myosin genes are highly interrupted and contain large number of exons. (A) human myosin II (NP_005954.3: 40 exons; Transcript length: 6,023 bps), (B) human myosin Va (NP_000250.3: 41 exons; Transcript length: 12,225 bps) and (C) human myosin VI (NP_004990.3: 35 exons; Transcript length: 8,662 bps).
Localization and Function
Myosins are localized to different organs or cell types based on their structural and functional peculiarities. Most of the non-muscle myosins are cytoplasmic. There are a number of myosins suggested to function during transcription within the nucleus by facilitating RNA polymerase movement, like myosin I,13 myosin II14 and myosin VI.15 Possible roles of myosins in transcription and other possible nuclear functions is in need of additional research. Nuclear organization and dynamics of that organization, clearly an exciting and important area of study, is in its infancy. Within the cytoplasm, myosins are localized to various specialized zones, viz. myosin II in the rear of polarized cells,16 myosin V in growth cones of neuronal cells,17 myosin VI at the base of stereocilia, and myosin VIII in plasmodesmata of plant cells.18
Myosin was first described in muscle cells, now known as muscle myosin II. Non-muscle myosin II is found in the cell division furrow of many unicellular and multicellular organisms,19 where it forms oligomeric thick filaments and facilitates cell division. Non-muscle myosin II isoforms are also localized to the leading edge of growth cones of rat dorsal root ganglion cells (myosin IIB isoform) and in the perinuclear region (myosin-IIA isoform).20 Myosin II association with Golgi-associated Rab6 resulting in fission of transport carriers was recently described.21
Myosin V has three isoforms and they are primarily involved in transport of various cellular organelles, such as secretory vesicles (reviewed in Desnos et al., 2007).22 Myosin V in melanocytes is involved in melanosome transport.23 In neurons and glial cells, abundant punctate staining of myosin V was found in the peri-nuclear region, along the cellular processes and in the distal tips of processes.24,25 Myosin V is also localized in the centrosome,26 where it is proposed to facilitate chromosomal segregation.27 The myosin Va isoform is also involved in acrosome formation and nuclear morphogenesis during spermatogenesis.28 Myosin VI and VII are present in the inner ear epithelial cells and hair cells.29 Myosin VI is also localized to the cortical region of the cell, which is the point of endocytosis30 and to specialized clathrin-coated invaginations at the base of the brush-border microvilli.31 Myosin VI is involved in two distinct steps of endocytosis in higher eukaryotes: the formation of clathrin-coated vesicles and the movement of nascent uncoated vesicles from the actin-rich cell periphery to the early endosome with the help of various adapter proteins.32 For a more extensive review of the roles of the myosin family of molecular motors in cells and tissues, see Hartman et al., 2011.33
General Structural Features
Myosins function as monomers,34 dimers35 or oligomers36 to bring about the diverse sub-cellular movements described above. In general, the myosin protomer consists of a long polypeptide known as the heavy chain comprising an ~80 kDa head domain followed by a variable length tail domain. The head domain is the most conserved and possesses the ATPase activity and actin binding ability.37,38 The C-terminal portion of the head is an α-helix with bound light chains that acts as a lever arm that amplifies smaller movements within the catalytic domain during the chemo-mechanical ATPase cycle. This light chain bound α-helix consists of a varying number of IQ motifs ([I,L,V]QxxxRGxxx[R,K]) where the light chains, calmodulin or calmodulin-like proteins, bind. A typical dimeric myosin consists of two heavy chains with a varying number of light chains, depending on the myosin type. In type II myosins, an essential light chain (ELC) and a regulatory light chain (RLC) bind at the IQ motifs.36
The C-terminal tail of unconventional myosins often, but not always, have a coiled coil region and a terminal globular cargo-binding domain. Conventional myosins (myosins II) possess long coiled coil tails and lack the C-terminal globular domain typical of the unconventional myosins.
Specific Domain Architectures
To date, the most extensively studied myosin head is that of myosin II, with numerous crystal structures, the first one being the motor domain of chicken skeletal muscle myosin.39 In the recent past, the myosin V motor domain was crystallized,40 as well as that of myosin VI.41 In all cases, the myosin head has an upper 50 kDa domain and a lower 50 kDa domain, connected by a loop. It has a converter domain, and a P-loop, switch I and switch II near the active site. Myosin II and a few other myosin classes possess an N-terminal SH3 domain. In myosin VI, there is a unique insert just after the converter domain that enables this motor to reverse its direction of movement toward the minus end of an actin filament. In addition, there are a number of domains, such as the PH domain, SH3 domain, and Kinase domain, which when combined with the myosin head make a plethora of domain architectures.
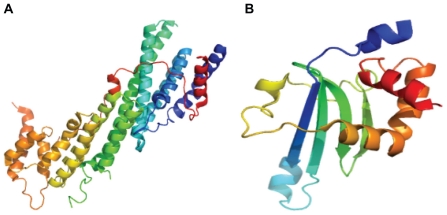
At the C-terminus of many non-muscle myosins is a globular cargo binding domain (CBD) that provides specificity for particular myosins binding to particular cargo. The CBD of myosins are relatively poorly conserved within the superfamily. Structures of only two CBDs, belonging to myosin V and VI, are known (Fig. 4). Myo2p is an essential myosin V in Saccharomyces cerevisiae involved in multiple functions through binding to multiple cargoes. Cargo binding domain of yeast M yosin V is an all-alpha fold containing 15 amphipathic α-helices connected by short and long loops organized into two five-helical bundles. These helical bundles correspond to sub-domains I and II originally defined by mild proteolysis and corresponds to two binding sites (one for vacuole binding and the other for secretory vesicle binding) that are oppositely placed at a 180° angle.42 The myosin V CBD is also known as the DIL domain. The myosin VI CBD forms a new fold that contains four β strands (βA–βD) and six α helices (αA–αF).43 The four βstrands form an antiparallel β sheet, creating the core of the domain. One side of the βsheet is covered by three helices (αD–αF) that are oriented perpendicular to each other. The other side of the sheet is largely exposed to the solvent with a short αC helix capped at one of its edges. The mouse myosin VI CBD binds to a fragment of the clathrin-coated vesicle adaptor Dab2 with high affinity.44
Figure 4.
Crystal structures of Cargo Binding Domain (CBD). (A) crystal structure of Myosin V CBD from Saccharomyces cerevisiae (PDB ID: 2F6H), (B) NMR structure of Myosin VI CBD from Mus musculus (PDB ID: 2KIA).
Between the head domain and the C-terminus of many myosins is a coiled coil structural motif that facilitates dimerization or oligomerization. The first myosin coiled coil crystal structure was from scallop myosin.45 The length of the coiled coil region in myosin varies from class to class. Myosins II have a very long heptad repeat, whereas myosin VI has essentially no predicted coiled coil regions (see below) and myosin V possesses a coiled coil of intermediate length (Fig. 2).
Coiled Coil Interaction Strength
In spite of the prominence of the coiled coil as an important motif in the myosin superfamily, structural and functional details of these coiled coils are severely lacking, due to the general absence of crystal structures. Though most of the myosin coiled coils have typical heptad repeats, the strength of the interactions between the two α-helices forming the various segments of the coiled coil are likely to vary. Very recently, the participation of the coiled coil motif in thick filament assembly of yeast myosin II has been suggested to involve two pathways. These investigators have employed deletion mutants and in vivo assays to demonstrate the role of the coiled coil, and an evolutionarily conserved structural kink within, in forming cleavage furrow ingression at the division during cytokinesis.46 In the literature, demarcation of coiled coil regions into weak and strong regions, protein-protein interaction sites and the recognition of structural kinks are just emerging.
There are many prediction programs that are available to predict the rough positions of coiled coils given the amino acid sequence of a protein.47 There are also structural analysis programs that enable identification of the coiled coil boundaries following the ridges-grooves arrangement of amino acid side chains.48 However, most of the predictive programs assume a uniform and ideal strength to the predicted regions, albeit being sensitive to sequence breaks like stutters and stammers. The sequence signatures at the coiled coils have evolved to such an extent that one can hardly find any conserved motifs across the subtypes. Yet many conserved (at the amino acid level) motifs can be found in the predicted coiled coil tail (please see a multiple sequence alignment of myosin VI in Supplementary Fig. 1).
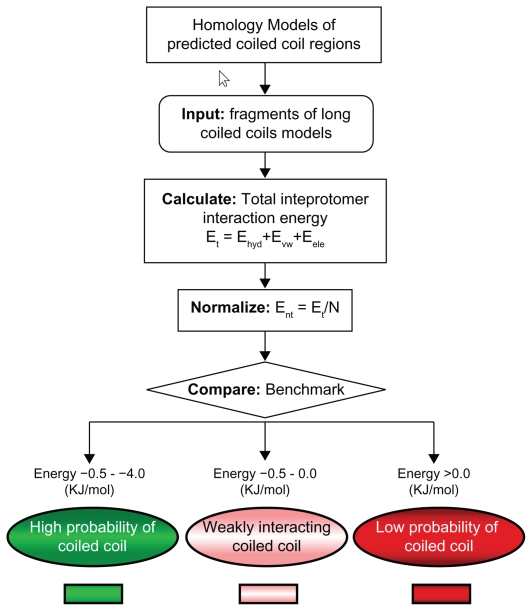
We have calculated the strength of interactions between the coiled coil α-helices within dimeric myosins through the pseudo-energy function inscribed in COILCHECK.49 For this we applied an algorithm (Fig. 5) that computationally ana lyzes the interaction strength of myosin at the coiled coil motif by three-dimensional modeling using MODELLER50 followed by retrospective calculations of interaction energies at the coiled coil interface using COILCHECK. Unlike general protein-protein interfaces51,52 that are largely dominated by hydrophobic interactions, the coiled coil interfaces are special, elaborate and retain alternating hydrophobic interactions and salt bridges built around a simple system of a pair of helices. Van der Waals and electrostatic interactions are measured all along the coiled coils across the dimer (see Fig. 5 for Methods). Further, interactions such as hydrogen bonds and salt bridges also contribute to the stability of these domains. This method has been applied to 26 protein structural entries that contain coiled coil. The binding strength values correlate well with the structure and stability of coiled coils. Indeed, COILCHECK results are sensitive to demarcate DNA-binding proteins that structurally deviate from ideal coiled coils (Fig. 6A). The guanine nucleotide exchange factor (GEF) domain of sec2p protein is a 22-nm long coiled coil and has a weakly interacting N-terminal region and a strongly intertwined middle region as evident from the crystal structure.53 Calculation of the interaction energies at different regions of the GEF domain using COILCHECK are consistent with the experimental results (Fig. 6B).
Figure 5.
Coiled coils interaction strength analysis protocol. Molecular models of myosin coiled coils were made using tropomyosin as template. The models were generated using MODELLER program.50 The long models were split into penta-heptad sized fragments and interaction energy values were calculated using COILCHECK program.
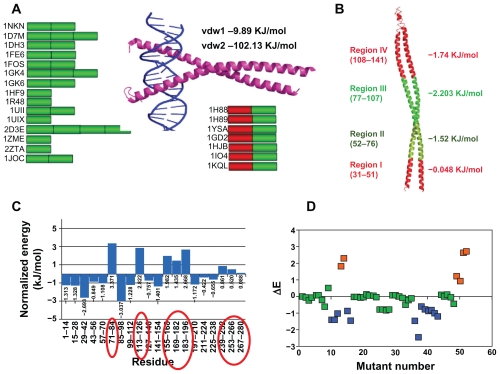
Figure 6.
COILCHECK based inter-protomer interactions. (A) Left: Pictorial representation of parallel coiled coil crystal structures depicting the interaction energy. Each block corresponds to a 35 residue window. Green blocks are regions of low inter-protomer interaction energy in COILCHECK calculations. Right top: PyMOL representation of coiled coil of BZIP transcription factor bound to DNA (PDB ID: 1IO4). Vdw1, vdw2 are the van der Waals energies for the first and second penta-heptad fragments of the coiled coil respectively. Van der Waals contribution in the first half that interacts with DNA is very less compared to the second half. The total normalized energy per residue for this region is also positive (shown on Right bottom). Right bottom: Pictorial representation of parallel coiled coils complexed with DNA. Red block represents the high calculated interaction energy that indicates the absence purely dimeric coiled coil interaction. This high interaction energy reflects either a poorly interacting dimer, a higher order assembly or interaction with other proteins. (B) Region II and III of GEF domain of sec2p (shown in green) has low COILCHECK calculated interaction energies as compared to the N terminal segment (shown in green) that is not a coiled coil and the C terminal segment (shown in red) that is weakly interacting. (C) Normalized energy for 14 residue long continuous fragments of tropomyosin (1C1G) structure. The positive energy regions (circled in red) correspond either to alanin repeats or breaks in hydrophobic cores.66 Alanine repeats and sequence periods are quasi-equivalent to actin binding sites.67 (D) ΔE is the difference in normalized energy values of wild type GCN4 and single point in silico mutants. A single residue was mutated at a time in a,d, e and g positions of an ideal heptad of GCN4. When positions a and d (Valine and Leucine) were mutated to hydrophobic residues the mutants showed least ΔE (shown in light green) and when mutated to charged residues ΔE was higher and destabilizing (shown in orange). Position g when mutated to any other residue did not show much deviation (light green). Position e when mutated to any other residue we found a stabilizing effect (dark green). The color scale from red to green is shown on the left side of the graph. The numbers (in the negative side) corresponding to each bar in the graph are the normalized stabilization energy for that fragment. The two lines of numbers (in the positive side) corresponds to the total electrostatic (first line from top) and Van der Waals energies in kJ/mol (second line from top).
When COILCHECK was applied to the tropomyosin structure (PDB code: 1C1G), actin-interacting zones were observed to acquire relatively poor energies for coiled coil formation (Fig. 6C). Furthermore, in order to probe the sensitivity of COILCHECK in detecting local structural perturbation due to the introduction of mis-sense mutations, we performed a series of virtual mutations (Fig. 6D). Interestingly, COILCHECK energy differences between the wild type and ‘single-site mutants’ were not very high when permitted amino acid exchanges, such as Leu to Val, were performed at either ‘a’ or ‘d’ positions. On the other hand, if drastic amino acid changes, such as Leu to Asp or Val to Lys, were introduced in the coiled coil, COILCHECK energy differences were quite high (Fig. 6D), suggesting that the method is sensitive to predict the effect of small sequence changes on the stability at the interface.
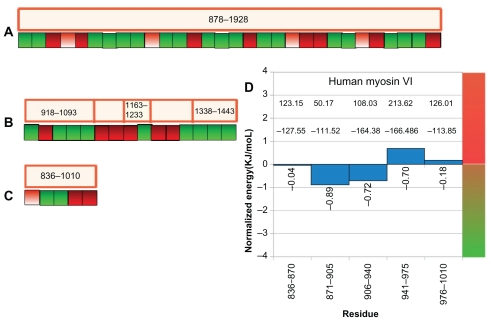
Using this approach, we found that the coiled coil regions of myosins V are generally interrupted by poorly interacting dimeric zones (Fig. 7B). Myosins VI have only a couple of short predicted strong interactions in its α-helical tail (Fig. 7C and D). These myosin VI results are consistent with experimental findings that the putative coiled coil region ( ie region predicted as coiled coil by COILS or PAIRCOIL algorithms) of myosin VI, are actually not coiled coil but stable single α-helices.54 Coiled coils of myosins II are predicted to have many segments of weak interactions that may have roles in oligomerization or protein-protein interactions (Fig. 7A).
Figure 7.
Inter-protomer interaction strength. Non-uniform inter-protomer interaction strength in predicted coiled coil regions are color coded and represented as boxes. Each box corresponds to 35 amino acids. Strongly interacting regions are shown in green and non interacting regions are shown in red. An intermediate level of interaction is also possible and shown in lighter shade of red. (A) human myosinII NP_001070654.1. (B) human myosin Va NP_000250.3. (C) human myosin VI (NP_004990.3). (D) Human myosin VI normalized energy per residue plotted against the pentaheptads. The first line of numbers in the positive side of the graph corresponds to the total electrostatic interaction energy and the second line corresponds to the total Van der Waals interaction energy.
Molecular Mechanisms
The myosin motor proteins convert chemical energy stored in ATP into mechanical movement by way of conformational changes within the head that are amplified by the neck domains. Myosins work by a swinging cross-bridge mechanism,55 the cross-bridge being the head domain.56 Following the first crystal structure determination of the myosin II head domain (see Rayment et al, 1993),39 the swinging cross-bridge hypothesis was refined to the swinging lever arm hypothesis. In the swinging lever arm hypothesis, the light chain binding region acts as a lever arm and generates at least three conformational isomers of the myosin during the ATPase cycle.
Myosin II has a step size of 5–15 nm, as evident from single molecule measurements.57 The ATPase site of the head domain has a nucleotide-binding site, which is a phosphate-binding loop (P-loop) that is closely associated with switch-I and switch-II helices. The actin-binding site and the P-loop communicate the presence or absence of Pi through switch-I and switch-II helix movements.
Myosin V and VI follow variants of the swinging lever arm mechanism, and achieve distinct step sizes, processivity and directionality.58,59 The unique long neck of myosin V with six calmodulin binding sites (IQ motifs) contribute to the long step size of 36 nm. Electron micrographs of myosin VI bound to actin revealed that the light chain binding region is oriented toward the pointed end of the actin unlike other myosins.41,60 Albeit having a short neck similar to myosin II, myosin VI step sizes are comparable to that of Myosin V. This large step size of ~36 nm owes to a mechanism of a 180° twist of its lever arm59,61 and an unusual tail domain that allows the 36-nm stretch.62,63 The reverse directionality of myosin VI comes from a unique insert just following the converter region.41,61 Diverse domain combinations of the myosin superfamily members result in imparting a wide variety of motility associated functions in the cell. The general myosin head-coiled coil-cargo binding domain architecture of many unconventional myosins is designed in such a way that vesicles or organelles are attached at the cargo binding domain of the dimer and the ATPase hydrolysis results in the walking motion of the dimer along the actin filament. Myosin VI is the only known retrograde myosin, which walks toward the minus end of the actin filament. Myosin VI dimerization appears to occur upon cargo binding.64
Conclusion
On the basis of available literature and our own analysis, we have narrated the molecular features of the myosin superfamily. The subfamily types described here are derived based on sequence similarity relationships of the motor domain. Further investigations on sequence, structural and functional diversity of the tail domain will provide the basis for a deeper understanding about this superfamily and new classification schemes.
We wish to draw attention to the putative coiled coil regions of the myosins because these are relatively understudied. Indeed, as shown for myosin VI, the strongly predicted coiled coil region appears not to be a coiled coil at all. Interestingly, COILCHECK, unlike COILS or PAIRCOILS, suggests very weak coiled coil interactions in myosin VI. COILCHECK is now being applied to all the myosin coiled coils to better understand which regions are likely to be strong interacting domains and which are weak. These data together with experimental data on the functions of these different domains are likely to shed new light on the roles that these myosin tail domains play in myosin structure and function.
Supplementary Figure
A multiple sequence alignment of Myosin VI predicted coiled coil region.
Acknowledgements
DPS’s stay at NCBS is supported by NCBS and Indian Council of Agricultural Research. This work is supported by Human Frontier Science Program. We also thank NCBS (TIFR) for infrastructural support.
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukayotes. Nature. 2005;436:1113–8. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 2.Goodson HV, Spudich JA. Molecular evolution of myosin family:relationships derived from comparisons of amino acid sequences. Proc Natl Acad Sci U S A. 1993;90:659–3. doi: 10.1073/pnas.90.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–91. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 4.Knecht DA, Loomis WF. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science. 1987;236:1081–5. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- 5.Tuxworth RI, Titus MA. Unconventional myosins: anchors in the membrane traffic relay. Traffic. 2000;1:11–8. doi: 10.1034/j.1600-0854.2000.010103.x. [DOI] [PubMed] [Google Scholar]
- 6.Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiol. 2005;20:239–51. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 7.Breshears LM, Wessels D, Soll DR, Titus MA. An unconventional myosin required for cell polarization and chemotaxis. PNAS. 2010;107(15):6918–23. doi: 10.1073/pnas.0909796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahler M. Are class III and Class IX molecules motorised signalling molecules. Biochim Biophys Acta. 2000;1496:52–9. doi: 10.1016/s0167-4889(00)00008-2. [DOI] [PubMed] [Google Scholar]
- 9.Thompson RF, Langford GM. Myosin superfamily evolutionary history. Anat Rec. 2002;268:276–89. doi: 10.1002/ar.10160. [DOI] [PubMed] [Google Scholar]
- 10.O’Halloran TJ, Ravid S, Spudich JA. Expression of Dictyostelium myosin tail segments in Escherichia coli: domains required for assembly and phosphorylation. The Journal of Cell Biology. 1990;110:63–70. doi: 10.1083/jcb.110.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kull FJ, Vale RD, Fletterick RJ. The case for a common ancestor: Kinesin and myosin motor proteins and G proteins. J Muscle Res Cell Motil. 1998;19:877–86. doi: 10.1023/a:1005489907021. [DOI] [PubMed] [Google Scholar]
- 12.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 13.Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, et al. A Myosin I isoform in the nucleus. Science. 2000;290(5490):337–41. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Sarna SK. Nuclear myosin II regulates the assembly of preinitiation complex for ICAM-1 gene transcription. Gastroenterology. 2009;137(3):1051–60. doi: 10.1053/j.gastro.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vreugde S, Ferrai C, Miluzio A, et al. Nuclear Myosin VI enhances RNA Polymerase II-dependent transcription. Molecular Cell. 2006;23(5):749–55. doi: 10.1016/j.molcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Wong K, Van Keymeulen A, Bourne HR. PDZRhoGEF and myosin II localize RhoA activity to the back of polarizing neutrophil-like cells. J Cell Biol. 2007;179(6):1141–8. doi: 10.1083/jcb.200706167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bridgman PC, Dave S, Asnes CF, Tullio AN, Adelstein RS. Myosin IIB is required for growth cone motility. The Journal of Neuroscience. 2001;21(16):6159–69. doi: 10.1523/JNEUROSCI.21-16-06159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radford JE, White RG. Localization of a myosin-like protein to plasmodesmata. Plant J. 1998;14(6):743–50. doi: 10.1046/j.1365-313x.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- 19.Maupin P, Phillips CL, Adelstein RS, Pollard TD. Differential localization of myosin-II isozymes in human cultured cells and blood cells. Journal of Cell Science. 1994;107:3077–90. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- 20.Cheng TP, Murakami N, Elzinga M. Localization of myosin-IIB at the leading-edge of growth cones from rat dorsal-root ganglionic cells. FEBS Lett. 1992;311:91–4. doi: 10.1016/0014-5793(92)81374-u. [DOI] [PubMed] [Google Scholar]
- 21.Valente C, Polishchuk R, De Matteis MA. Rab6 and myosin II at the cutting edge of membrane fission. Nature Cell Biology. 2010;12:635–8. doi: 10.1038/ncb0710-635. [DOI] [PubMed] [Google Scholar]
- 22.Desnos C, Huet S, Darchen F. ‘Should I stay or should I go?’: myosin V function in organelle trafficking. Biol Cell. 2007;99:411–23. doi: 10.1042/BC20070021. [DOI] [PubMed] [Google Scholar]
- 23.Huang JD, Cope MJ, Mermall V, et al. Molecular genetic dissection of mouse unconventional myosin-Va: Head region mutations. Genetics. 1998;148:1951–61. doi: 10.1093/genetics/148.4.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espreafico EM, Cheney RE, Matteoli M, et al. Primary structure and cellular localization of chicken brain myosin V, an unconventional myosin with calmodulin light chains. J Cell Biol. 1992;119:1541–58. doi: 10.1083/jcb.119.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coling DE, Espreafico EM, Kachar B. Cellular distribution of myosin-V in the guinea pig cochlea. J Neurocytol. 1997;26:113–20. doi: 10.1023/a:1018523827852. [DOI] [PubMed] [Google Scholar]
- 26.Espreafico EM, Coling DE, Tsakraklides V, et al. Localization of myosin-V in the centrosome. Proc Natl Acad Sci U S A. 1998;95:8636–41. doi: 10.1073/pnas.95.15.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin H, Pruyne D, Huffaker TC, Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406(6799):1013–5. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- 28.Sun X, He Y, Hou L, Yang WX. Myosin Va Participates in Acrosomal Formation and Nuclear Morphogenesis during Spermatogenesis of Chinese Mitten Crab Eriocheir sinensis. PLoS One. 2010;5(9):e12738. doi: 10.1371/journal.pone.0012738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coffin AB, Dabdoub A, Kelley MW, Popper AN. Myosin VI and VIIa distribution among inner ear epithelia in diverse fishes. Hear Res. 2007;224(1–2):15–30. doi: 10.1016/j.heares.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biemesderfer D, Mentone SA, Mooseker M, Hasson T. Expression of myosin-VI within the endocytic pathway in the adult and developing proximal tubule. Am J Physiol Renal Physiol. 2002;282:f785–94. doi: 10.1152/ajprenal.00287.2001. [DOI] [PubMed] [Google Scholar]
- 31.Yang LE, Maunsbach AB, Leong PK, McDonough AA. Redistribution of Myosin VI from Top to Base of Proximal Tubule Microvilli during Acute Hypertension. J Am Soc Nephrol. 2005;16:2890–6. doi: 10.1681/ASN.2005040366. [DOI] [PubMed] [Google Scholar]
- 32.Hasson T. Myosin VI: two distinct roles in endocytosis. Journal of Cell Science. 2003;116:3453–61. doi: 10.1242/jcs.00669. [DOI] [PubMed] [Google Scholar]
- 33.Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu Rev Cell Dev Biol. 2011;27:133–55. doi: 10.1146/annurev-cellbio-100809-151502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn BD, Sakamoto T, Hong MS, Sellers JR, Takizawa PA. Myo4p is a monomeric myosin with motility uniquely adapted to transport mRNA. J Cell Biol. 2007;178(7):1193–206. doi: 10.1083/jcb.200707080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrington WF, Burke M. Geometry of the Myosin Dimer in High-Salt Media. I. Association Behavior of Rod Segments from Myosin. Biochemistry. 1972;11(8):1448–55. doi: 10.1021/bi00758a019. [DOI] [PubMed] [Google Scholar]
- 36.Saitoh T, Takemura S, Ueda K, et al. Differential localization of non-muscle myosin II isoforms and phosphorylated regulatory light chains in human MRC-5 fibroblasts. FEBS Lett. 2001;509:365–9. doi: 10.1016/s0014-5793(01)03186-6. [DOI] [PubMed] [Google Scholar]
- 37.Warrick HM, Spudich JA. Myosin structure and function in cell motility. Annu Rev Cell Biol. 1987;3:379–421. doi: 10.1146/annurev.cb.03.110187.002115. [DOI] [PubMed] [Google Scholar]
- 38.Vibert P, Cohen C. Domains, motions and regulation in the myosin head. J Muscle Res Cell Motil. 1988;9:296–305. doi: 10.1007/BF01773873. [DOI] [PubMed] [Google Scholar]
- 39.Rayment I, Rypniewski WR, Schmidt-Bäse K, et al. The three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–8. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442(7099):208–11. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 41.Ménétrey J, Bahloul A, Wells AL, et al. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435(7043):779–85. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 2006;25(4):693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu C, Feng W, Wei Z, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138(3):537–48. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 44.Morris SM, Arden SD, Roberts RC, et al. Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic. 2002;3(5):331–41. doi: 10.1034/j.1600-0854.2002.30503.x. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Brown JH, Reshetnikova L, et al. Visualization of an unstable coiled coil from the scallop myosin rod. Nature. 2003;424(6946):341–5. doi: 10.1038/nature01801. [DOI] [PubMed] [Google Scholar]
- 46.Fang X, Luo J, Nishihama R, et al. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191(7):1333–50. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lupas A, Van Dyke M, Stock J. Predicting Coled Coils from Protein Sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 48.Walshaw J, Woolfson DN. Socket: a program for identifying and analysing coiled coil motifs within protein structures. J Mol Biol. 2001;307(5):1427–50. doi: 10.1006/jmbi.2001.4545. [DOI] [PubMed] [Google Scholar]
- 49.Alva V, Syamala Devi DP, Sowdhamini R. COILCHECK: An Interactive Server for the Analysis of Interface Regions in Coiled Coils. Protein and Peptide Letters. 2008;15(1):33–8. doi: 10.2174/092986608783330314. [DOI] [PubMed] [Google Scholar]
- 50.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 51.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janin J, Chothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990;265(27):16027–30. [PubMed] [Google Scholar]
- 53.Sato Y, Shirakawa R, Horiuchi H, Dohmae N, Fukai S, Nureki O. Asymmetric Coiled coil Structure with Guanine Nucleotide Exchange Activity. Structure. 2007;15(2):245–52. doi: 10.1016/j.str.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 54.Spink BJ, Sivaramakrishnan S, Lipfert J, Doniach S, Spudich JA. Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI. Nature: Structural and Molecular Biology. 2008;15(6):591–97. doi: 10.1038/nsmb.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holmes KC. The swinging lever-arm hypothesis of muscle contraction. Current Biology. 1997;7(2):112–8. doi: 10.1016/s0960-9822(06)00051-0. [DOI] [PubMed] [Google Scholar]
- 56.Toyoshima YY, Kron SJ, McNally EM, Niebling KR, Toyoshima C, Spudich JA. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987;328:536–9. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 57.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–9. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 58.Purcell TJ, Morris C, Spudich JA, Sweeney HL. Role of the lever arm in the processive stepping of myosin V. PNAS. 2002;99(22):14159–64. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spudich JA, Sivaramakrishnan S. Myosin VI an innovative motor that challenged the swinging lever arm hypothesis. Nature Reviews Molecular Cell Biology. 2010;11:128–37. doi: 10.1038/nrm2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wells AL, Lin AW, Chen LQ, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–8. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 61.Reifenberger JG, Toprak E, Kim H, Safer D, Sweeney HL, Selvin PR. Myosin VI undergoes a 180° power stroke implying an uncoupling of the front lever arm. PNAS. 2009;106(43):18255–60. doi: 10.1073/pnas.0900005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okten Z, Churchman LS, Rock RS, Spudich JA. Myosin VI walks hand-over-hand along actin. Nature Structural & Molecular Biology. 2004;11(9):884–7. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 63.Mukherjea M, Llinas P, Kim H, et al. Myosin VI dimerization triggers an unfolding of a 3-helix bundle in order to extend its reach. Mol Cell. 2009;35(3):305–15. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phichith D, Travaglia M, Yang Z, et al. Cargo binding induces dimerization of myosin VI. PNAS. 2009;106:17320–4. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. PNAS. 2006;103(10):3681–6. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Minakata S, Maeda K, Oda N, Wakabayashi K, Nitanai Y, Maéda Y. Two-Crystal Structures of Tropomyosin C-Terminal Fragment 176–273. Exposure of the Hydrophobic Core to the Solvent Destabilizes the Tropomyosin Molecule. Biophysical Journal. 2008;95:710–9. doi: 10.1529/biophysj.107.126144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh A, Hitchcock-DeGregori SE. Tropomyosin’s periods are quasi-equivalent for actin binding but have specific regulatory functions. Biochemistry. 2007;46:14917–27. doi: 10.1021/bi701570b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A multiple sequence alignment of Myosin VI predicted coiled coil region.