Abstract
The ability to regulate the function of specific proteins using cell-permeable molecules can be a powerful method for interrogating biological systems. To bring this type of “chemical genetic” control to a wide range of proteins, we recently developed an experimental system in which the stability of a small protein domain expressed in mammalian cells depends on the presence of a high-affinity ligand. This ligand-dependent stability is conferred to any fused partner protein. The FK506- and rapamycin-binding protein (FKBP12) has been the subject of extensive biophysical analyses, including both kinetic and thermodynamic studies of the wild-type protein as well as dozens of mutants. The goal of this study was to determine if the thermodynamic stabilities (ΔΔGU-F) of various amino acid substitutions within a given protein are predictive for engineering additional ligand-dependent destabilizing domains. We used FKBP12 as a model system and found that in vitro thermodynamic stability correlates weakly with intracellular degradation rates of the mutants and that the ability of a given mutation to destabilize the protein is context dependent. We evaluated several new FKBP12 ligands for their ability to stabilize these mutants and found that a cell-permeable molecule called Shield-1 is the most effective stabilizing ligand. We then performed an unbiased microarray analysis of NIH3T3 cells treated with various concentrations of Shield-1. These studies show that Shield-1 does not elicit appreciable cellular responses.
Cell permeable small molecules have long been powerful tools for interrogating biology (1). They can be used to conditionally probe biological processes, often with high temporal resolution (2–4). Interest in using perturbants of this type has grown significantly in the past decade, but one of the pressing questions remains: How can one discover a cell permeable perturbant for any protein of interest? Nature was the source for many of the early examples, especially when the biologically relevant target of the perturbant was easily discerned. To accelerate the discovery process, many investigators are screening large libraries of small molecules either against purified proteins or against living cells using high-content imaging and phenotype-based scoring to evaluate library members (5). Irrespective of the discovery process used to identify the perturbant, the question of specificity remains critical for research biology (6). When a particular reagent is used to conditionally perturb a biological process, how confident can one be that the resulting phenotype can be ascribed to the putative molecular target of the perturbant?
We have chosen an alternate strategy to provide small molecule control of any protein of interest (7). Our approach relies upon one well characterized protein-ligand interaction that can be used to regulate many different proteins of interest. We have engineered small protein domains called destabilizing domains (DDs)1 that are constitutively degraded when expressed in mammalian cells. This instability is conferred to any other protein fused to the DD. A high-affinity ligand binds to the DD and prevents it from being degraded. This ligand-mediated stability allows the fused protein to perform its cellular function, provided it is fully functional in the context of a fusion protein. Specificity is provided by the genetic fusion of the DD to the protein of interest, and speed, reversibility, and tunability are provided by the stabilizing ligand.
One of the challenges to this approach is engineering the desired DDs. What is the most efficient procedure to take an otherwise stable protein-ligand pair and selectively perturb the protein structure to the point of instability in the absence of a ligand and stability in the ligand-bound state? In one approach, one can assume nothing and use an unbiased screening strategy to identify mutants of the protein that endow the structure with the desired ligand-dependent stability (7). Alternatively, one can rely upon thermodynamic analyses of the protein, if available, to rationally choose mutations that might confer ligand-dependent stability. Another challenge for this strategy is the desire to use stabilizing ligands that do not perturb the cellular environment by binding to off-target proteins. In this study, we evaluate the ability of directed amino acid substitutions to confer ligand-dependent stability to a small protein domain. We also investigate the specificity of ligands that stabilize these domains. This comparison of two possible discovery processes (unbiased screening versus biophysics-guided engineering) will likely inform future attempts to develop new destabilizing domains.
EXPERIMENTAL PROCEDURES
FKBP mutant generation
The retroviral vector pBMN FKBP-YFP-i-HcRed-tandem was assembled by ligating FKBP mutants into the pBMN YFP HA-i-HcRed-t vector. Quickchange Site-Directed Mutagenesis (Stratagene) was used to incorporate single point mutations into the FKBP-F36V gene. Double and triple mutants were obtained by sequential mutagenesis or PCR amplification. Transfection quality DNA was produced in E. coli.
Tissue Culture and Transfections
NIH3T3 cells were cultured in DMEM supplemented with 10% heat-inactivated donor bovine serum (Invitrogen), 2 mM glutamine, 100 U/mL penicillin, and 100 μg streptomycin. The ΦNX ecotropic packaging line was cultured with 10% heat-inactivated fetal bovine serum (Invitrogen), 2 mM glutamine, 100 U/mL penicillin, and 100 μg streptomycin. The ΦNX cell line was transfected using standard Lipofectamine 2000 protocols. Viral supernatants were harvested 48 hours post transfection, filtered and concentrated using an Amicon Ultra centrifugal filter (Millipore, 100-kDa cutoff). NIH3T3 cells were incubated with the concentrated viral supernatant supplemented with 4 μg/mL polybrene for 4 hr at 37 °C. The viral supernatant was then removed, and fresh media was added to the cells. Cells were cultured for 24 hours to allow for viral integration.
Flow Cytometry
Transduced NIH3T3 cells were plated at 1 × 105 cells per well of a 12-well plate, and treated as described twenty-four hours prior to analysis. Cells were removed from wells using trypsin-EDTA (Invitrogen) and quenched with 1 mL growth media. The media was removed and cells were resuspended in 300 μL PBS. Cells were analyzed at the Stanford Shared FACS Facility with 10,000 events represented.
Antibodies
Immunoblotting was performed using the following antibodies: FKBP (mouse, 554091, BD Biosciences); YFP, Aequorea victoria (JL-8, Clontech).
Microarray Analysis
NIH3T3 cells were cultured in media containing vehicle or 1 μM, 100 nM, or 10 nM Shield-1 for 24 hours. Total RNA was extracted using the RNeasy kit (Qiagen), labeled, and hybridized to the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) arrays. The hybridizations were performed by the Stanford Functional Genome Facility. After overnight hybridization at 65 °C, each array was washed and scanned using the GenePix 4000A microarray scanner (Axon Instruments). SAM was used to identify genes whose expression had significantly changed after treatment with Shield-1 (19). A complete description of the experiment including data filtering and analysis is included in the Supporting Information.
FKBP Ligands
Reagent requests should be directed to the corresponding author. Shield-1 will be distributed to all qualified investigators.
RESULTS
Human FKBP12 (hereafter called FKBP) has been the subject of numerous kinetic and thermodynamic analyses of protein folding and stability (8–13). In addition to the wild-type protein, the relative contributions of dozens of individual amino acid mutations to the stability of FKBP have been analyzed in vitro (8). These biophysical studies provide a rich source of data with which to guide the “rational design” strategy for engineering ligand-dependent stability. Mutations in different regions of the protein that confer similar levels of instability can be tested to investigate the context dependence of this design strategy.
Biologically active FKBP ligands (e.g., FK506 and rapamycin) exert their respective activities through FKBP and distinct partner proteins (2, 14, 15). However, ligands that bind to FKBP alone are generally not considered to elicit significant responses when administered to cultured mammalian cells. To ensure greater specificity between protein and ligand, we chose the FKBP F36V mutant and a series of ligands pioneered by Dennis Holt and colleagues that possess a synthetic “bump” that occupies the cavity created by the F36V mutation (16, 17). These bumped ligands bind approximately 1600-fold more tightly to the F36V mutant relative to wild-type FKBP. The F36V mutation is present in all proteins described in this study, and we identify mutants using only destabilizing amino acid changes (i.e., “V2A” denotes the V2A/F36V mutant).
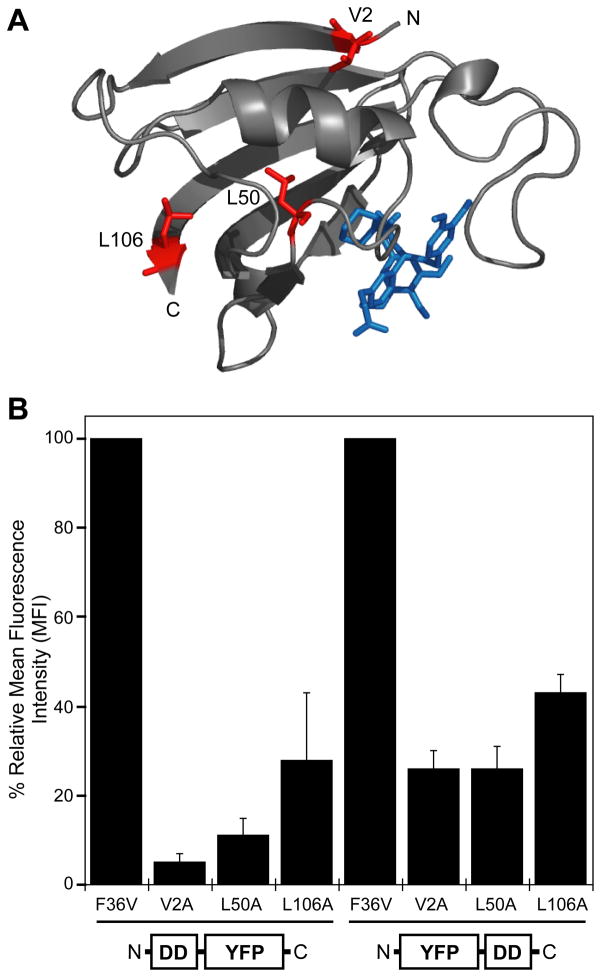
In order to test the stability of various mutants expressed in cultured cells, the FKBP F36V gene was cloned at either the N- or C-terminus of yellow fluorescent protein (YFP), and site-specific mutations were introduced into the FKBP sequence. These chimeric genes were transduced into NIH3T3 cells using an MMLV-derived retroviral expression system to create clonally mixed populations of stably transduced cells. Analytical flow cytometry was used to quantitate the expression levels of the fusion proteins. We first examined the V2A, L50A, and L106A mutants that destabilize FKBP by 2.43, 2.57, and 2.32 kcal/mol, respectively (8). These three mutants confer similar degrees of thermodynamic instability to the FKBP protein, although they are located in different environments (Fig. 1A). Expression levels of the parent F36V protein fused to YFP were similar to YFP alone, however the V2A, L50A, and L106A FKBP mutants were all destabilizing when fused to either the N- or C-terminus of FKBP (Fig. 1B). FKBP mutants fused to the N-terminus of YFP were more destabilizing than those fused at the C-terminus.
Figure 1. FKBP mutants destabilize YFP expression.
A, The structure of FKBP F36V is shown as a ribbon diagram (PyMol) complexed with the SLF* ligand (PDB ID 1BL4). Three representative mutations are highlighted in red, and the ligand is shown in blue.
B, Fluorescence of FKBP-YFP and YFP-FKBP fusions expressed in NIH3T3 cells in the absence of stabilizing ligand as determined by analytical flow cytometry. Data are presented as the average mean fluorescence intensity (MFI) ± SEM standardized to the fusions between F36V and YFP.
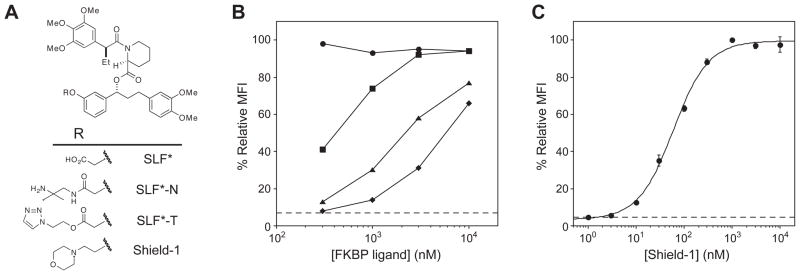
During our original screen for FKBP-derived destabilizing domains we used a ligand called SLF* to stabilize FKBP (Fig. 2A) (7). In an attempt to improve the FKBP-stabilizing activity of SLF* we synthesized several variants by attaching solubilizing functional groups to a region of the ligand that does not affect binding to FKBP. These SLF* variants were then tested for their ability to stabilize the most destabilized of the fusion proteins, the V2A mutant fused to the N-terminus of YFP (V2A-YFP). Cells expressing the V2A-YFP fusion protein were dosed with various concentrations of ligand 24 hours prior to analysis by analytical flow cytometry. The original SLF* compound showed the lowest level of stabilization (Fig. 2B). The functional groups appended to SLF* all resulted in improved stabilizing activity, and the FKBP ligand possessing a morpholine group attached through an ether linkage proved to be most effective in rescuing the fusion protein. This ligand was named Shield-1, and further characterization of the interaction between Shield-1 and the V2A-YFP fusion protein revealed an EC50 of ~60 nM (Fig. 2C).
Figure 2. Synthetic FKBP ligands stabilize the V2A-YFP fusion protein.
A, Chemical structures of FKBP ligands.
B, NIH3T3 cells stably expressing the V2A-YFP fusion protein were treated with 3-fold dilutions of each FKBP ligand (10 μM to 300 nM). YFP expression levels were measured upon treatment with SLF* acid (diamonds), SLF*-N (squares), SLF*-T (triangles) and Shield-1 (circles) by flow cytometry.
C, NIH3T3 cells stably expressing the V2A-YFP fusion protein were treated with 3-fold dilutions of Shield-1 (10 μM to 1 nM) and assayed by flow cytometry. Data are presented as the average MFI ± SEM relative to that of the F36V FKBP protein. The dotted line represents the YFP expression level for V2A-YFP in the absence of Shield-1.
In order to probe the basis for the improved activity of the SLF variants, we measured the dissociation constant between each ligand and the F36V protein [see supporting information (SI) Fig. 1]. A fluorescence polarization-based competition binding assay revealed that the affinities of the four ligands for the F36V protein were experimentally indistinguishable (SI, Fig. 1) (18). Fluorescence polarization was also used to measure the affinities of Shield-1 for the F36V parent protein as well as for the three mutants (SI, Figs. 2 and 3). The dissociation constants of Shield-1 for the four proteins varied between 2 and 4 nM, suggesting that the different potencies of stabilization observed for the analogs are the result of differences in cellular permeability rather than differences in affinity for FKBP.
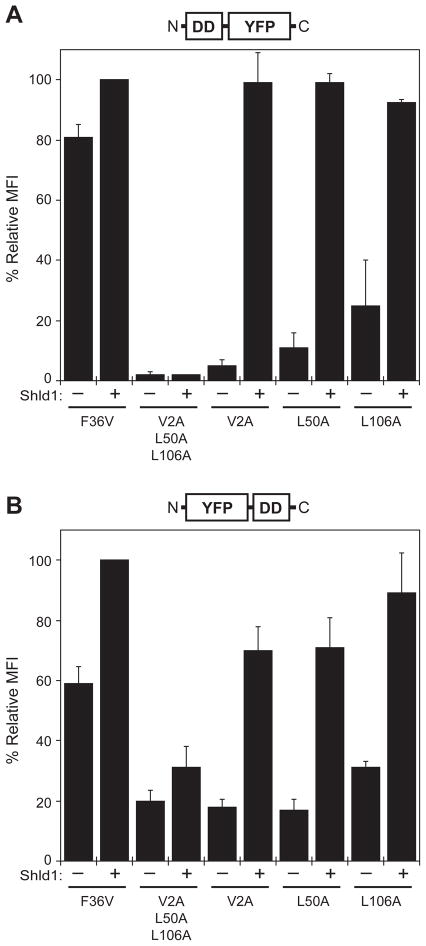
We next tested all three FKBP mutants using Shield-1 as the stabilizing ligand. With the FKBP mutants fused to the N-terminus of YFP, a 1 μM dose of Shield-1 fully stabilized YFP expression levels (Fig. 3A). A triple mutant incorporating all three destabilizing mutations showed very low expression with no detectable Shield-1 dependence (Fig. 3A). The FKBP mutants are not as destabilizing when they are fused to the C-terminus of YFP (Fig. 3B). Shield-1 stabilizes the C-terminal fusions, although the expression levels do not quite reach those of the parent F36V protein. Treatment of the F36V parent fusion proteins with Shield-1 causes a modest rise in expression levels, suggesting that the binding of the ligand to the FKBP domain reduces the rate of degradation. When fused to the C-terminus of YFP, the triple mutant (V2A/L50A/L106A) is not significantly more destabilizing then the V2A or L50A mutants, although stabilization of the triple mutant by Shield-1 is attenuated. Transduced cells were treated with either vehicle or Shield-1, and lysates were immunoblotted with antibodies against either FKBP12 or YFP (SI, Fig. 4A). As expected, protein levels by Western blot correlate well with YFP levels measured by analytical flow cytometry, and no evidence of partial degradation was observed. Inhibition of the proteasome with lactacystin prevented degradation of the V2A-YFP fusion protein (SI, Fig. 4B), which is consistent with our previous findings (7).
Figure 3.
Shield-1 stabilizes the FKBP-YFP fusion proteins. NIH3T3 cells stably expressing the indicated fusion proteins were split into two populations that were either mock-treated (−) or treated with 1 μM Shield-1 (+) for 24 hours. FKBP mutants were fused to the N-terminus (panel A) or the C-terminus (panel B) of YFP, and expression levels were measured by flow cytometry and normalized to cells expressing the parent F36V-YFP protein treated with 1 μM Shield-1. Data present the average of three experiments ± SEM.
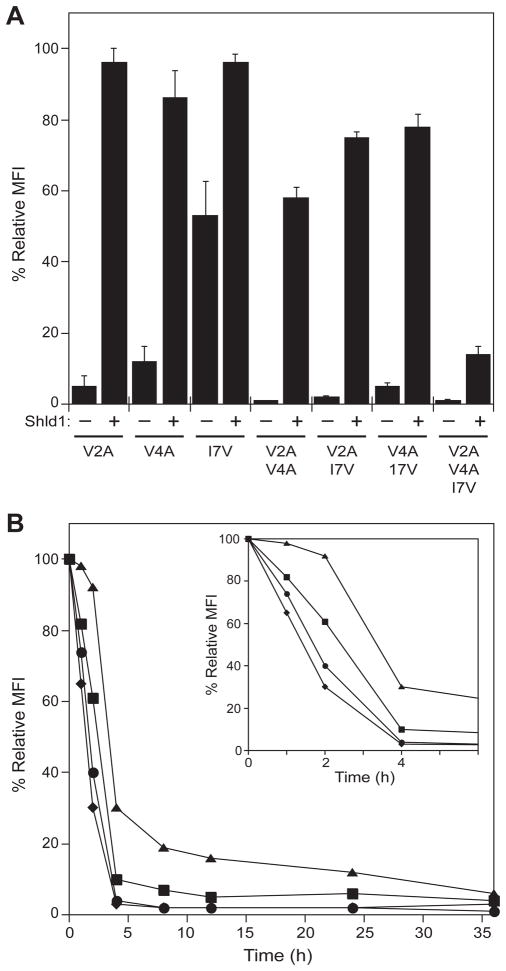
To determine if additional destabilizing mutations would result in an additive effect on FKBP stability, we returned to the biophysical analyses and made further mutations at the N-terminus of FKBP. The V4A and I7V mutations are destabilizing by 2.78 and 0.92 kcal/mol (8), respectively, so we incorporated these into the FKBP F36V background in all possible combinations with the V2A mutation. These single-, double-, and triple-mutants were fused to the N-terminus of YFP, individually transduced into NIH3T3 cells, and YFP expression levels were measured in the absence and presence of 1 μM Shield-1 (Fig. 4A). Treatment with Shield-1 stabilized the expression levels of all seven mutants, and the most efficiently rescued mutants were the single point mutants, V2A, V4A, and the double mutants, V2A/I7V and V4A/I7V. These four mutants were further characterized to determine the rates of protein degradation upon removal of Shield-1 (Fig. 4B). The mutants that displayed the lowest expression levels in the absence of Shield-1 showed the fastest rate of protein degradation, with both double mutants returning to basal levels after four hours.
Figure 4. Characterization of additional FKBP mutants.
A, FKBP-YFP fusions were either mock-treated (−) or treated with 1 μM Shield-1 (+) and YFP expression levels were monitored by flow cytometry. Data are presented as the average MFI ± SEM relative to that of the parent F36V FKBP.
B, NIH3T3 cells stably expressing FKBP-YFP fusion proteins were treated with 1 μM Shield-1 for 24 hr. The cells were then washed with media to remove Shield-1, and decreases in fluorescence intensity were monitored by flow cytometry: V2A-YFP (squares), V4A-YFP (triangles), V2A/I7V-YFP (diamonds) and V4A/I7V-YFP (circles).
As a simple test of the potential generality of this destabilizing activity, we fused the V2A mutant to the N-terminus of firefly luciferase. Firefly luciferase is stabilized by Shield-1 in a dose-responsive manner (SI, Fig. 5A). As observed with the YFP reporter, Shield-1 treatment provides modest stabilization to the fusion of F36V to luciferase. In contrast, fusion of V2A to luciferase results in destabilization of the fusion protein, with expression levels, and in turn luminescence, rescued upon addition of Shield-1 (SI, Fig. 5B). A kinetic analysis showed that Shield-1 treatment causes maximum stabilization in 2–3 hours, and further, that removal of Shield-1 from stabilized cells results in loss of luciferase expression in 1–2 hours (SI, Figs. 5C and 5D).
The ideal system to conditionally regulate protein expression levels using small molecules would utilize a stabilizing ligand that does not exhibit any off-target effects in a biological environment. In order to assess the ability of Shield-1 to perturb gene expression patterns, we treated NIH3T3 cells with vehicle or one of three concentrations of Shield-1 (10, 100, and 1000 nM). Each set of conditions was performed in triplicate, and following 24 hours of Shield-1 treatment RNA was isolated, processed, and used to probe Mouse Exonic Evidence Based Oligonucleotide (MEEBO) arrays. We used Significance Analysis of Microarray (SAM) to identify genes that were differentially expressed between the treated and mock-treated samples (19).
The microarrays were evaluated using a relatively high false discovery rate (FDR) to identify as many perturbations to mRNA levels as possible. Using a FDR ≤ 27.5%, SAM identified four genes whose transcript levels were significantly altered in cells treated with 1 μM Shield-1 (Table 1 and SI Figure 6A). The mRNA levels increased less than threefold for three of the four genes and decreased (11-fold) for the other gene. Using the same threshold, no transcript levels were significantly altered upon treatment with 100 nM Shield-1 (SI Figure 6B). For cells treated with 10 nM Shield-1, SAM predicted five genes as significantly changed. The mRNA levels for all five genes decreased expression (< 3-fold) compared with control. Notably, none of the five genes identified in 10 nM treated cells were perturbed in cells treated with higher concentrations of Shield-1.
Table 1.
Effects of Shield-1 on Gene Expression in NIH3T3 Cells
| Upregulated Genes | |||||
|---|---|---|---|---|---|
| Clone ID | Gene Symbol | Gene description | Fold Change (FDR*) | ||
| 10 nM | 100 nM | 1 μM | |||
| mSQ030922 | Epb4.1l2 | Erythrocyte protein band 4.1-like 2 | 2.2 (<0.1%) | ||
| mSQ001430 | Csnk1e | Casein kinase 1, epsilon | 2.7 (<0.1%) | ||
| mSQ002857 | Tcfe3 | Transcription factor E3 | 1.9 (27.5%) | ||
| Downregulated Genes | |||||
|---|---|---|---|---|---|
| Clone ID | Gene Symbol | Gene description | Fold Change (FDR*) | ||
| 10 nM | 100 nM | 1 μM | |||
| mSQ036549 | MtCrVgp3 | Murine type C retrovirus | −11.3 (<0.1%) | ||
| mSQ009285 | 4930422M22Rik | RIKEN cDNA 4930422M22 gene | −2.2 (<0.1%) | ||
| mSQ007437 | A430110N23 | hypothetical protein A430110N23 | −2.1 (<0.1%) | ||
| mSQ014243 | Ptpn1 | Protein tyrosine phosphatase, non- receptor type 1 | −1.7 (<0.1%) | ||
| mSQ001772 | Npas4 | Neuronal PAS domain protein 4 | −1.7 (22.7%) | ||
| mSQ028092 | Shkbp1 | Sh3kbp1 binding protein 1 | −2.3 (22.7%) | ||
NIH3T3 cells in triplicate were cultured in media and mock-treated or treated with 10 nM, 100 nM or 1 μM Shield-1 for 24 hours. Total RNA from each of the twelve samples was extracted and hybridized to the MEEBO oligonucleotide array. The false discovery rate (FDR) was obtained using unpaired two-class Significance Analysis of Microarrays (SAM), and genes whose mRNA levels changed significantly upon Shield-1 treatment were identified.
DISCUSSION
The ability to conditionally perturb the function of specific proteins using cell-permeable small molecules is a powerful technique with which to probe biological processes. We recently used an unbiased screening process to identify destabilizing domains based on FKBP (7). For the purposes of engineering new protein-ligand combinations that behave as DDs, we were interested in knowing the most efficient method to identify mutants of a protein that would display the desired ligand-dependent stability. The purpose of these studies was to determine if biophysics-based design is a competitive discovery method for engineering conditional instability, so we chose three mutants of the 107-residue FKBP protein that destabilize the protein fold to the same approximate degree. To test the context dependence of these mutations, we chose residues at the N-terminus, in the middle, and at the C-terminus of the protein. We further fused the FKBP mutants to either the N- or the C-terminus of the YFP reporter protein. If there was no context dependence to the induced instability, we might expect that all mutants would exhibit similar behavior, regardless of the placement of the destabilizing mutation within the structure of FKBP.
The data shown in Figure 3 suggest that there is a strong context dependence with respect to both the sites of mutation as well as the orientation of the fusion protein. FKBP mutants fused at the N-terminus of YFP are more strongly destabilizing than C-terminal fusions. The higher efficiency with which the N-terminal fusions are degraded may reflect mechanistic differences in the quality control machinery monitoring protein stability. The nature of the amino substitution is also important. When fused at the N-terminus of YFP, the L106P mutant is strongly destabilizing (1–2% expression in the absence of Shield-1) (7). However, the L106A mutation is only moderately destabilizing (25–30% expression in the absence of Shield-1).
Within a given fusion protein orientation, (i.e., FKBP fused to the N-terminus of YFP) we observed a positive, but weak, correlation between in vitro thermodynamic stability and intracellular stability in the context of a fusion protein. All of the mutants are destabilizing, but the extent of destabilization differs. The V2A, L50A, and L106A mutations are similarly destabilizing in terms of ΔΔGU-F, however the expression levels of their YFP fusion proteins are 5%, 11%, and 25%, respectively, in the absence of Shield-1.
Mammalian cells must be able to unfold proteins for degradation by ATP-dependent proteases as well as for translocation across some membranes (20). When analyzing the susceptibility of proteins to protease-mediated degradation, Matouschek and coworkers have shown that the local structure of the substrate is more important than overall protein stability (21). They have also demonstrated that efficient proteasome-mediated degradation requires an unstructured region within the target protein to serve as an initiation site (22). Similar context-dependent effects have been observed by Baker and Sauer (23) as well as Robinson and Dobson (24). In light of these studies it is possible that the mutations of the single-domain FKBP protein regulate the different intracellular rates of degradation through specific local effects rather than global effects on protein stability.
The effects of our destabilizing mutations appear to be additive. When we made additional mutations to the FKBP domain of the V2A-YFP fusion protein, we observed that a greater degree of instability was conferred relative to V2A alone (Fig. 4A). As we have observed previously, mutants that are strongly destabilizing exhibit faster degradation rates when Shield-1 is withdrawn (7). Upon addition of Shield-1, the rates of synthesis for the mutants are indistinguishable. In this respect, these FKBP-derived DDs function as ligand-dependent degrons that modulate the rate of degradation. It is the ratio of the rates of synthesis and degradation that govern the expression level of a given protein.
Several new FKBP ligands were synthesized and tested as stabilizing ligands, and we observed that their potencies differed widely (Fig. 2B). The affinity of each ligand for the F36V protein was measured, and the dissociation constants are very similar (SI, Fig. 1). This observation suggests that the range of stabilizing potencies results from different cellular permeabilities rather than underlying biophysical differences. Shield-1 binds equally well to the V2A, L50A, and L106A mutants, as expected.
It is difficult to prove that Shield-1 is not binding to other proteins and causing cellular responses that are unrelated to FKBP fusion proteins. To probe the specificity of the interaction between Shield-1 and other cellular constituents, we used an unbiased microarray analysis to look for perturbations in gene expression patterns in NIH3T3 cells treated with different concentrations of Shield-1. We were particularly interested in genes whose expression exhibited dose-dependent changes in response to different concentrations of Shield-1. Using fairly liberal thresholds for significance, we found only a very small number of genes whose RNA levels changed appreciably relative to mock-treated control cells. Importantly, none of the gene expression changes were observed in all three concentrations of Shield-1 that were tested. These data support the notion that Shield-1 is relatively free of off-target perturbations to the cellular environment. Our findings are consistent with those of the Ariad group that tested a structurally similar dimeric FKBP ligand in a Phase I human clinical trail (25).
The ability to conditionally regulate a single protein of interest will likely provide many new insights into complex biological systems. However, many investigations would benefit from the ability to independently regulate the levels of two or more different proteins. For example, a method to separately control the intracellular expression levels of GSK-3α and GSK-3β would be useful for probing the potentially overlapping functions of these two kinases. The existence of two or more DD systems that are regulated by orthogonal small molecules would be useful, so what is the best discovery method to identify new DDs?
Ligand-dependent stability can be engineered into FKBP using either biophysical data as detailed in this manuscript or through screening an unbiased library of FKBP mutants (7). The biophysics-based design strategy delivers mutants that display the desired ligand-dependent stability, although the extent of destabilization conferred by the various mutants is not uniform or predictable. Additionally, the biophysical data required for this design strategy is not as widely available for other proteins as might be needed. Conversely, one can manipulate various parameters in a screening process (e.g., concentration of stabilizing ligand, incubation periods), to deliver mutants with specific characteristics. From this perspective, the unbiased screening strategy appears to be a more reliable strategy for engineering new destabilizing domains.
Supplementary Material
Acknowledgments
This work was supported by the NIH (GM 073046). We thank the Crabtree lab for the luciferase constructs and Janos Demeter, John Coller, and Jason Myers for helpful discussions related to the microarray analyses.
Footnotes
The following abbreviations are used: FDR, false discovery fate; FKBP, human 12-kDa FK506- and rapamycin-binding protein; HcRed, red fluorescent protein from Heteractis crispa; MEEBO, mouse exonic evidence based oligonucleotide; MMLV, Moloney Murine Leukemia Virus; SAM, significance analysis of microarrays; SLF, synthetic ligand for FKBP; YFP, yellow fluorescent protein.
References
- 1.Weisenberg RC, Borisy GG, Taylor WE. Biochemistry. 1968;7:4466–4478. doi: 10.1021/bi00852a043. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Farmer JD, Lane WS, Friedman J, Weissman I, Schreiber SL. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 3.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 4.Taunton J, Hassig CA, Schreiber SL. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 5.Yeh JRJ, Crews CM. Dev Cell. 2003;5:11–19. doi: 10.1016/s1534-5807(03)00200-4. [DOI] [PubMed] [Google Scholar]
- 6.Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, Gutbrod H, Salassidis K, Stein-Gerlach M, Missio A, Cotten M, Daub H. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AGL, Wandless TJ. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Main ERG, Fulton KF, Jackson SE. Biochemistry. 1998;17:6145–6153. doi: 10.1021/bi973111s. [DOI] [PubMed] [Google Scholar]
- 9.Main ERG, Fulton KF, Jackson SE. J Mol Biol. 1999;291:29–444. doi: 10.1006/jmbi.1999.2941. [DOI] [PubMed] [Google Scholar]
- 10.Fulton KF, Main ERG, Daggett V, Jackson SE. J Mol Biol. 1999;291:445–461. doi: 10.1006/jmbi.1999.2942. [DOI] [PubMed] [Google Scholar]
- 11.Main ERG, Jackson SE. Nat Struct Biol. 1999;6:831–835. doi: 10.1038/12287. [DOI] [PubMed] [Google Scholar]
- 12.Main ERG, Fulton KF, Daggett V, Jackson SE. J Biol Phys. 2001;27:99–117. doi: 10.1023/A:1013137924581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korepanova A, Douglas C, Leyngold I, Logan TM. Protein Sci. 2001;10:1905–1910. doi: 10.1110/ps.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 15.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 16.Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, Stevenson LF, Magari SR, Wood SA, Courage NL, Lu X, Cerasoli F, Gilman M, Holt DA. Proc Natl Acad Sci USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang W, Rozamus LW, Narula S, Rollins CT, Yuan R, Andrade LJ, Ram MK, Phillips TB, van Schravenkijk MR, Dalgarno D, Clackson T, Holt DA. J Med Chem. 2000;43:1135–1142. doi: 10.1021/jm9904396. [DOI] [PubMed] [Google Scholar]
- 18.Braun PD, Wandless TJ. Biochemistry. 2004;43:5406–5413. doi: 10.1021/bi035839g. [DOI] [PubMed] [Google Scholar]
- 19.Tusher VG, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prakash S, Matouschek A. Trends Biochem Sci. 2004;29:593–600. doi: 10.1016/j.tibs.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 22.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 23.Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 24.Canet D, Last AM, Tito P, Sunde M, Spencer A, Archer DB, Redfield C, Robinson CV, Dobson CM. Nat Struct Biol. 2002;9:308–315. doi: 10.1038/nsb768. [DOI] [PubMed] [Google Scholar]
- 25.Iuliucci JD, Oliver SD, Morley S, Ward C, Ward J, Dalgarno D, Clackson T, Berger HJ. J Clin Pharmacol. 2001;41:870–879. doi: 10.1177/00912700122010771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.