Abstract
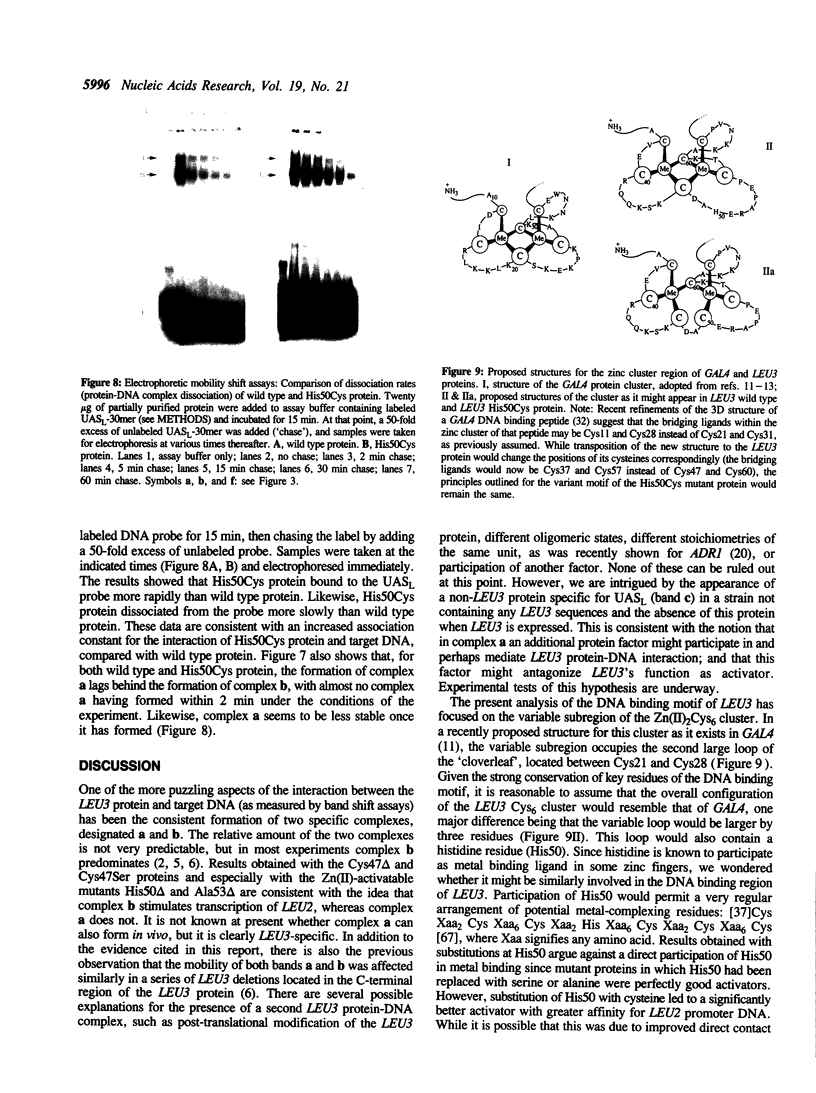
The transcriptional activator LEU3 of Saccharomyces cerevisiae belongs to a family of lower eukaryotic DNA binding proteins with a well-conserved DNA binding motif known as the Zn(II)2Cys6 binuclear cluster. We have constructed mutations in LEU3 that affect either one of the conserved cysteines (Cys47) or one of several amino acids located within a variable subregion of the DNA binding motif. LEU3 proteins with a mutation at Cys47 were very poor activators which could not be rescued by supplying Zn(II) to the growth medium. Mutations within the variable subregion were generally well-tolerated. Only two of seven mutations in this region generated poor activators, and both could be reactivated by Zn(II) supplements. Three of the other five mutations gave rise to activators that were better than wild type. One of these, His50Cys, exhibited a 1.5 fold increase in in vivo target gene activation and a notable increase in the affinity for target DNA. The properties of the His50Cys mutant are discussed in terms of a variant structure of the DNA binding motif. During the course of this work, evidence was obtained suggesting that only one of the two LEU3 protein-DNA complexes routinely seen actually activates transcription. The other (which may contain an additional protein factor) does not.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990 Jan;220(2):269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- Balzi E., Chen W., Ulaszewski S., Capieaux E., Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987 Dec 15;262(35):16871–16879. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brisco P. R., Kohlhaw G. B. Regulation of yeast LEU2. Total deletion of regulatory gene LEU3 unmasks GCN4-dependent basal level expression of LEU2. J Biol Chem. 1990 Jul 15;265(20):11667–11675. [PubMed] [Google Scholar]
- Friden P., Reynolds C., Schimmel P. A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol Cell Biol. 1989 Sep;9(9):4056–4060. doi: 10.1128/mcb.9.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol Cell Biol. 1988 Jul;8(7):2690–2697. doi: 10.1128/mcb.8.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever R. F., Huiet L., Baum J. A., Tyler B. M., Patel V. B., Rutledge B. J., Case M. E., Giles N. H. DNA sequence, organization and regulation of the qa gene cluster of Neurospora crassa. J Mol Biol. 1989 May 5;207(1):15–34. doi: 10.1016/0022-2836(89)90438-5. [DOI] [PubMed] [Google Scholar]
- Halvorsen Y. C., Nandabalan K., Dickson R. C. LAC9 DNA-binding domain coordinates two zinc atoms per monomer and contacts DNA as a dimer. J Biol Chem. 1990 Aug 5;265(22):13283–13289. [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature. 1987 Jul 23;328(6128):353–355. doi: 10.1038/328353a0. [DOI] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Kim J., Michels C. A. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr Genet. 1988 Oct;14(4):319–323. doi: 10.1007/BF00419988. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczak J. E., Brandriss M. C. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol Cell Biol. 1991 May;11(5):2609–2619. doi: 10.1128/mcb.11.5.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. GAL4 transcription factor is not a "zinc finger" but forms a Zn(II)2Cys6 binuclear cluster. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2077–2081. doi: 10.1073/pnas.87.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Coleman J. E. Sequential assignments of the 1H NMR resonances of Zn(II)2 and 113Cd(II)2 derivatives of the DNA-binding domain of the GAL4 transcription factor reveal a novel structural motif for specific DNA recognition. Biochemistry. 1991 Apr 30;30(17):4212–4222. doi: 10.1021/bi00231a016. [DOI] [PubMed] [Google Scholar]
- Peters M. H., Beltzer J. P., Kohlhaw G. B. Expression of the yeast LEU4 gene is subject to four different modes of control. Arch Biochem Biophys. 1990 Jan;276(1):294–298. doi: 10.1016/0003-9861(90)90041-v. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Eisen A., Young E. T. Two monomers of yeast transcription factor ADR1 bind a palindromic sequence symmetrically to activate ADH2 expression. Mol Cell Biol. 1991 Mar;11(3):1566–1577. doi: 10.1128/mcb.11.3.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L., Coleman J. E., Auld D. S. Zinc fingers, zinc clusters, and zinc twists in DNA-binding protein domains. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):999–1003. doi: 10.1073/pnas.88.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. M., Bai Y. L., Kohlhaw G. B. Yeast regulatory protein LEU3: a structure-function analysis. Nucleic Acids Res. 1990 Jan 25;18(2):291–298. doi: 10.1093/nar/18.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K. M., Kohlhaw G. B. Transcriptional activator LEU3 of yeast. Mapping of the transcriptional activation function and significance of activation domain tryptophans. J Biol Chem. 1990 Oct 15;265(29):17409–17412. [PubMed] [Google Scholar]
- Zhou K., Brisco P. R., Hinkkanen A. E., Kohlhaw G. B. Structure of yeast regulatory gene LEU3 and evidence that LEU3 itself is under general amino acid control. Nucleic Acids Res. 1987 Jul 10;15(13):5261–5273. doi: 10.1093/nar/15.13.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]