Using computational modeling and laser microsurgery, we establish that neither the centrosomal microtubule array nor the Golgi-derived array is solely sufficient for correct Golgi assembly. Only the concerted effort of both MT arrays results in the integral, polarized Golgi complex necessary for polarized trafficking and cell motility.
Abstract
Assembly of an integral Golgi complex is driven by microtubule (MT)-dependent transport. Conversely, the Golgi itself functions as an unconventional MT-organizing center (MTOC). This raises the question of whether Golgi assembly requires centrosomal MTs or can be self-organized, relying on its own MTOC activity. The computational model presented here predicts that each MT population is capable of gathering Golgi stacks but not of establishing Golgi complex integrity or polarity. In contrast, the concerted effort of two MT populations would assemble an integral, polarized Golgi complex. Indeed, while laser ablation of the centrosome did not alter already-formed Golgi complexes, acentrosomal cells fail to reassemble an integral complex upon nocodazole washout. Moreover, polarity of post-Golgi trafficking was compromised under these conditions, leading to strong deficiency in polarized cell migration. Our data indicate that centrosomal MTs complement Golgi self-organization for proper Golgi assembly and motile-cell polarization.
INTRODUCTION
The Golgi complex in the majority of interphase mammalian cells is present as a single organelle that supports integrated protein processing and sorting (Burkhardt, 1998; Thyberg and Moskalewski, 1999; Mironov and Beznoussenko, 2010; Lowe, 2011). When the cell enters mitosis, the Golgi breaks down and subsequent daughter cells generate this organelle anew. Golgi membranes fuse and stack into multiple ministacks throughout the cytoplasm. Thereafter these ministacks are transported by the molecular motor dynein along the newly emerging interphase microtubule (MT) network and assemble to form a single complex. Mechanisms responsible for proper formation and positioning of the Golgi complex are essential for the cell (Rios and Bornens, 2003; Lowe, 2011).
Several recent studies demonstrate that in mammalian cells the Golgi acts as a major MT-organizing center (MTOC) in addition to the centrosome (Chabin-Brion et al., 2001; Efimov et al., 2007; Rivero et al., 2009). Prior to this finding, it was assumed that Golgi assembly is driven by the radial MT array emanating from the centrosome (Thyberg and Moskalewski, 1999; Mironov and Beznoussenko, 2010). However, our recent analyses of Golgi-derived MTs indicated that this MT subpopulation is specifically essential for proper Golgi assembly and integrity (Miller et al., 2009). It still remains unknown whether Golgi assembly can be supported exclusively by its own MTOC activities or whether it requires centrosomal MTs. Intuitively, the radial array of MTs formed by the centrosome would provide an important spatial cue for the collection of scattered Golgi stacks in the cell center. However, it is also conceivable that Golgi self-assembly does not rely on the centrosome, as the Golgi can accumulate in the central cell area in the absence of the centrosome by a yet unclear mechanism (Tangemo et al., 2011). In this regard, noncentrosomal MTs can self-organize into radial arrays in a dynein-dependent manner (Mitchison, 1992; Nedelec et al., 1997; Surrey et al., 2001; Cytrynbaum et al., 2004). Furthermore, dynein activity combined with MT nucleation on the surface of melanin granules (melanosomes) is sufficient to aggregate the melanosomes in the center of fish melanophores (Malikov et al., 2005). A similar mechanism may be sufficient for the assembly of the Golgi. However, unlike melanosomes, single Golgi stacks are structurally and functionally asymmetric (Cole et al., 1996), and the Golgi complex is not a random collection of vesicles but rather a highly organized, continuous membrane organelle (Snapp et al., 2003; Mironov and Beznoussenko, 2010). Thus, even if the self-organization mechanisms are sufficient to aggregate Golgi stacks, it is not clear whether self-organization would provide for correct positioning and subsequent fusion of the Golgi elements.
In this study, we have developed a computational model that evaluates contributions of specific MT arrays in the Golgi assembly. The model supports our previous conclusion that both MT arrays are involved in the Golgi assembly (Miller et al., 2009), suggesting the concerted effort of both MT arrays bring coordination of the Golgi assembly to a new level, as neither MT array on its own provides polarity and integrity of the complex. More specifically, the model illustrates how decreased formation of Golgi-derived MTs modulates the Golgi organization and predicts an essential role of the radial centrosomal array for Golgi integrity and polarity.
Accordingly, we experimentally tested the role of the centrosomal MT array in Golgi organization and function. We used a highly focused laser beam to physically ablate the centrosome that prevents formation of the centrosomal MTs. In contrast to genetic manipulations, physical ablation of the centrosome does not affect MT nucleation at other locales (Khodjakov et al., 2000; Khodjakov and Rieder, 2001). Without the centrosome, Golgi-associated MT nucleation remains normal (Efimov et al., 2007). Unexpectedly, we find that the centrosome per se is not required for the maintenance and function of the already-assembled Golgi complex. However, the MT network in acentrosomal cells fails to correctly assemble the Golgi, which leads to inefficiency in polarized post-Golgi trafficking, as predicted by the model. Moreover, our work indicates that the role of the centrosome in the control of polarized cell migration is indirect. When the Golgi and/or Golgi-derived MT array is already assembled, ablation of the centrosome has no effect on the cell polarization and motility. However, when emerging MTs assemble the Golgi in the absence of the centrosome, the cell is incapable of polarized cell migration. These data strongly suggest that the centrosome modulates cell polarization and motility through Golgi organization and trafficking.
RESULTS
Efficient Golgi complex assembly requires two MT populations
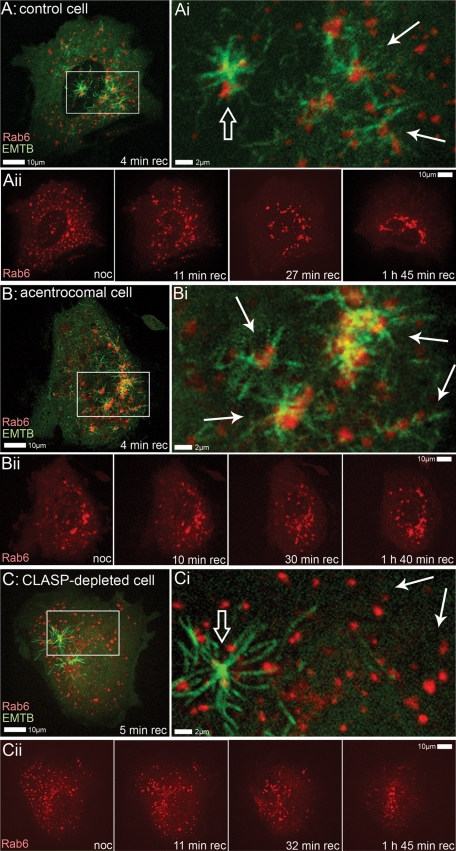
To visualize the process of MT-dependent Golgi assembly by emerging MT network in interphase cells, we applied nocodazole washout assay. Retinal pigment epithelium 1 (RPE1) cells, which demonstrate two prominent MT arrays (that is, emanating from the Golgi and the centrosome), were cotransfected with mCherry-Rab6 as a Golgi marker and 3GFP-EMTB (ensconsin MT-binding domain) as an MT marker. If MTs are depolymerized by nocodazole, the Golgi randomly disperses in the cytoplasm (Supplemental Figure S1); however, a fully functional Golgi complex assembles within 60 min upon nocodazole washout (Miller et al., 2009). After full MT depolymerization, nocodazole was removed, and the Golgi assembly was recorded (Figure 1 and Supplemental Movies S1–S6). Consistent with our previous finding (Miller et al., 2009), when two types of regrowing MTs are involved (Figure 1, A–Aii, and Movie S1), Golgi ministacks are clustered together throughout the cell and at the same time, these clusters are being relocated to the cell center, resulting in the formation of an integral Golgi complex (Movie S2).
FIGURE 1:
Golgi assembly in RPE1 after nocodazole washout. (A–Aii) Control cell. (A and Ai) Centrosomal (open arrow) and Golgi-derived (solid arrows) MTs organized at initial stage of recovery after nocodazole washout (4 min). MTs are highlighted by 3GFP-EMTB marker and the Golgi is highlighted by mCherry-Rab6. Box from (A) is enlarged in (Ai). (Aii) Selected frames from live-cell imaging sequence of the same cell. After complete MT depolymerization by nocodazole, Golgi marked by mCherry-Rab6 is dispersed throughout the cell (noc). After nocodazole washout, Golgi ministacks cluster all over the cell (at 11 and 27 min recovery) and at the same time are being collected in the cell center (at 27 min and 1 h, 45 min recovery). (B–Bii) Acentrosomal cell, treated as in (A). (Bi and Bii) In the absence of the centrosome, all MTs organized at initial stage of recovery (4 min) are formed at the Golgi (solid arrows). Box from (B) is enlarged in (Bi). (Bii) Selected frames from live-cell imaging sequence. After nocodazole washout, Golgi ministacks are being collected in the cell center but integral Golgi ribbon is not formed. (C–Cii) CLASP-depleted cell, treated as in (A). (Ci and Cii) MTs organized at initial stage of recovery (5 min) are predominantly centrosomal (open arrow). Golgi-derived MTs are significantly diminished (solid arrows). Box from (C) is enlarged in (Ci). (Cii) Selected frames from live-cell imaging sequence. After nocodazole washout, Golgi ministacks are being collected in the cell center without significant clustering.
To test the role of the centrosomal MT array, we laser-ablated the centrosome in cultured RPE1 cells that stably express centrin1-GFP (Efimov et al., 2007; Uetake and Sluder, 2007; Figure S2). We have previously demonstrated that the Golgi remains fully capable of MT nucleation and organization, while centrosome-derived MTs do not form after centrosome ablation (Efimov et al., 2007). Immunofluorescence analyses of cells fixed at different time points after centrosome ablation confirmed that the MT network and the Golgi complex remained similar to control cells that contained centrosomes (Figure S2). When the centrosome was ablated in nocodazole-treated cells coexpressing mCherry-Rab6 and 3GFP-EMTB, and the drug was subsequently washed out (Figure 1, B–Bii), MTs were readily formed at dispersed Golgi stacks (Movie S3) and drove Golgi stacks together. Subsequently, the Golgi was self-organized in the perinuclear area in the absence of the centrosome (Movie S4). However, assembly appeared incomplete and morphology of the complex was aberrant, suggestive of a significant role for the centrosome in the Golgi assembly process.
The same assay was applied to cells in which Golgi-derived MTs were suppressed by CLIP-associated protein (CLASP) depletion (Figure S3) to test the involvement of the Golgi-derived MTs in Golgi assembly (Figure 1, C–Cii). In this case, MTs were formed predominantly at the centrosomes (Movie S5), and resulting Golgi integrity was diminished (Figure 1Cii and Movie S6), indicating that the centrosomal array on its own is insufficient for correct Golgi assembly. This result is consistent with our previous finding that Golgi-derived MTs are needed for Golgi assembly (Miller et al., 2009).
Combined, these data suggest that de novo Golgi complex assembly requires contributions from both centrosomal and Golgi-derived MTs.
Computational model supports necessity of two geometrically distinct MT populations for the assembly of integral Golgi complex
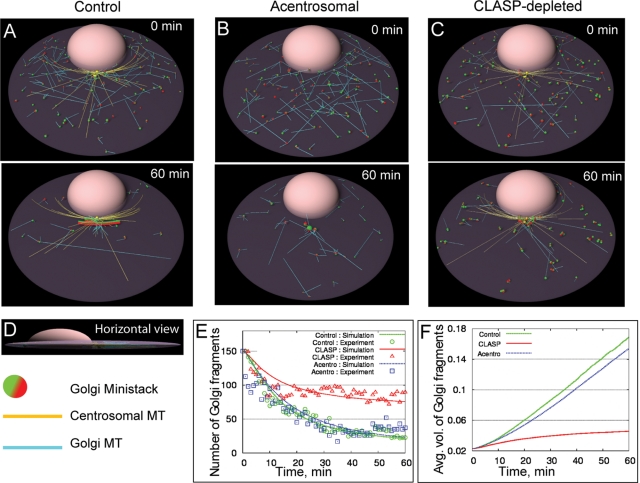
To evaluate relative significance of each type of MT in Golgi assembly, we modeled this process in silico. We used agent-based, three-dimensional simulations (Alberts and Odell, 2004; Paul et al., 2009) of the assembly process. The geometry and dimensions in the simulations are close to those observed. The model simulations seen in Movies S7–S10 (snapshots are depicted in Figure 2, A–C) revealed that the Golgi assembly occurs in three stages in control cells: 1) Ministacks in close proximity interact among themselves and form intermediate fragments. 2) As a result, these fragments are more frequently captured and transported by centrosomal MTs and accumulate around the centrosome. 3) In the vicinity of the centrosome, fragments interact once again to form bigger Golgi stacks. The predicted time series for the fragment numbers are in excellent agreement with the experimental data (Figure 2E).
FIGURE 2:
In silico, both MT arrays are required for the proper Golgi assembly. (A–C) Snapshots from the simulations. Top, initial configuration; bottom, predicted state of the system after assembly for 60 min. Centrosome- and Golgi-nucleated MTs are shown in yellow and cyan, respectively. In the control and acentrosomal cases, each initial Golgi fragment nucleates ∼ 4 MTs, whereas in the CLASP-depleted case, each initial Golgi fragment nucleates ∼1–2 MTs. Red/green ministack surfaces correspond to trans/cis sides, respectively. (A) In control, the majority of multiple Golgi fragments assemble within 60 min into the integral Golgi ribbon in the vicinity of the centrosome. It is oriented with trans (MT-nucleating) surface away from the nucleus. (B) Without centrosomal MTs, the majority of fragments self-assemble, albeit to a random location near the nucleus. (C) When CLASP is depleted, ministacks collected in the vicinity of the centrosome remain unfused and are randomly oriented. Many fragments remain scattered away from the centrosome after 60 min. (D) Side view of the control simulation snapshot. (E) Average number of Golgi fragments in control, CLASP-depleted, and acentrosomal cells plotted as a function of time. Solid curves: simulations; symbols: experimental data obtained from images like Figure 1, as described in the Supplemental Material. (F) Average predicted number of Golgi fragments in control, CLASP-depleted, and acentrosomal cells plotted as a function of time.
In acentrosomal cells, assembly is almost as rapid as in control cells (Figure 2, E and F). However, acentrosomal assembly is not able to make a single complex: it creates Golgi fragments very dispersed in size (Figures 2B and S4B), unlike the uniformly large stacks that emerge in control cells (Figures 2A and S4A), so that the average radius of Golgi fragments at each time point is decreased and the spanning cluster radius is increased (Figure S5, A–C). Some smaller fragments, most notably those “behind” the nucleus, remain uncaptured in the absence of the centrosomal MT array, because the fragments at opposite sides of the nucleus do not “see” each other. Accordingly, the compactness of the resulting complex is decreased in acentrosomal cells (Figure S5D).
In the CLASP-depleted cell, the centrosomal MTs bring many fragments to the center, but many other ministacks remain uncaptured (Figure 2C). Thus one significant function of Golgi-derived MTs in Golgi assembly is the premerging of ministacks into larger fragments that are more easily captured by the centrosomal radial array, thereby significantly accelerating the assembly (Figures 2, E and F, and S5, A and B). Moreover, the best fitting to experimental data was achieved upon assumption that centrosomal and Golgi-derived MTs were equally good at capturing, but only Golgi-derived MTs were effective at fusing, Golgi fragments. Owing to insufficient clustering and fusion, assembly in CLASP-depleted cells is inefficient (Figure S5C), and the radius of Golgi fragments remains significantly lower than in control cells. In addition, compactness of the assembly (see Supplemental Material) worsens in the CLASP-depleted cells (Figure S5D). Predicted Golgi organization reaches its steady state at 1 h of assembly and does not change after 3 h of assembly (Figure S6).
Centrosome is required for the assembly of the integral Golgi complex in nocodazole washout assay
Dynamics of the Golgi assembly were in excellent agreement with the model predictions (Figure 2E). Moreover, model-predicted defects of the Golgi shape and integrity in CLASP-depleted cells was supported by our previous experimental findings (Miller et al., 2009).
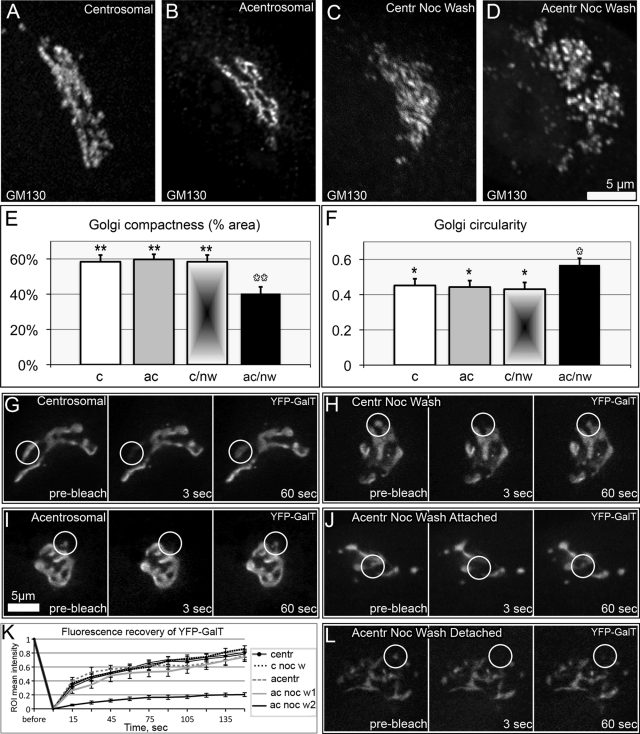
To test the model predictions for the acentrosomal Golgi assembly, we assessed the morphological features of a Golgi complex assembled in the absence of centrosomal MTs. The Golgi was assembled upon nocodazole washout in acentrosomal cells, as described for Figure 1B. Two hours after nocodazole washout of acentrosomal cells, morphology of the Golgi complex was compared with control conditions, including untreated centrosomal cells, untreated cells lacking centrosomes, and centrosomal cells 2 h after nocodazole washout. Distribution of cis-Golgi (GM130, Figure 3, A–D) as well as trans-Golgi network (TGN; GCC185, unpublished data) Golgi markers was detected by immunofluorescence analysis. The structure of the Golgi complex assembled without the centrosome was abnormal, though the Golgi stacks were collected in the cell center (Figure 3D). Circularity and compactness parameters (see Supplemental Material) were used to quantitatively assess the structural complexity and integrity of the Golgi complex. In accordance with the model predictions, high circularity indicated randomization and low compactness reflected insufficient integrity of the Golgi ribbon assembled without centrosomal input. In contrast, morphology and quantitative parameters of the Golgi complex assembled after nocodazole washout in cells containing centrosomes (Figure 3C) were indistinguishable from the Golgi in nontreated cells (Figure 3A).
FIGURE 3:
Golgi complex organization in centrosomal and acentrosomal cells. (A–D) The Golgi revealed by immunostaining for GM130. (A) Nontreated centrosomal cell; (B) acentrosomal cell; (C) centrosomal cell fixed 2 h after nocodazole washout; and (D) acentrosomal cell fixed 2 h after nocodazole washout. (E) Compactness of Golgi complex (percentage of space within the Golgi area occupied by GM130) in the four types of cells corresponding to (A–D). (F) Circularity [4π*(area/perimeter2)] of the Golgi complex in the four types of cells corresponding to (A–D). Data from more than three independent experiments; n = 10 for each condition. *, p < 0.05; **, p < 0.01 in unpaired Student's t test between bars marked with black and white asterisks. Error bars represent SEM. (G–L) FRAP analyses of Golgi ribbon continuity. (G–J and L) frames from live-cell imaging sequences in which YFP-GalT was used as an integral Golgi marker. (G) Nontreated centrosomal cell; (H) centrosomal cell 2 h after nocodazole washout; (I) acentrosomal cell; (J) and (L) acentrosomal cell 2 h after nocodazole washout. For each example, a frame prior to bleaching (prebleach), after bleaching (3 s), and upon fluorescence recovery (60 s) is shown. White circles indicate bleached regions of interest. In contrast to fast recovery in (G–J), detached Golgi fragment in (L) does not recover, indicating that connection with the Golgi ribbon is lost. See also solid black line in (K). (K) Quantification of FRAP as in (G–J) and (L). Error bars represent SEM.
We next assessed the role of the centrosomal array in integrity of the Golgi complex by fluorescence recovery after photobleaching (FRAP) analyses of yellow fluorescent protein (YFP)-GalT dynamics (Figure 3, G–L; as in Snapp et al., 2003; Miller et al., 2009). Our model predicts that the Golgi complex assembled in the absence of the centrosome consists of large clusters within which the Golgi fragments have sufficient conditions for fusion and smaller fragments that stay unassociated with large clusters. Indeed, rates of fluorescence recovery in acentrosomal cells fell into two distinct groups (Figure 3, J–L). The first group (Figure 3, J and K) did not significantly differ from the recovery rates in control centrosomal cells, either untreated or recovered after nocodazole treatment. The second group was observed in smaller Golgi fragments and was characterized by very slow recovery rates (Figure 3, K and L). Thus the large Golgi clusters presented integral Golgi ribbons, but the small fragments did not establish functional contact with the major Golgi mass. These data indicate that the centrosome supports the proper assembly of the integral Golgi complex.
Alternative mechanisms of collecting Golgi stacks in the cell center substitute for role of the centrosome in the Golgi complex integrity
Interestingly, the morphology of the existing Golgi complex was not affected by the centrosome ablation per se (Figure 3, A and B). In both centrosomal and acentrosomal cells, a typical Golgi ribbon morphology with similar circularity and compactness parameters was observed (Figure 3, E and F). Moreover, FRAP rates for acentrosomal cells with preexisting Golgi complex did not differ from control centrosomal cells (Figure 3, G–I and K), revealing that the continuity of the existing Golgi ribbon was unaltered by centrosome ablation. This result suggests the centrosome is not required for the maintenance of already-assembled Golgi complexes. According to our model, the centrosome provides the radial signal for Golgi assembly, and it is possible that Golgi-derived MTs are sufficient to support Golgi integrity when it is already collected in the cell center.
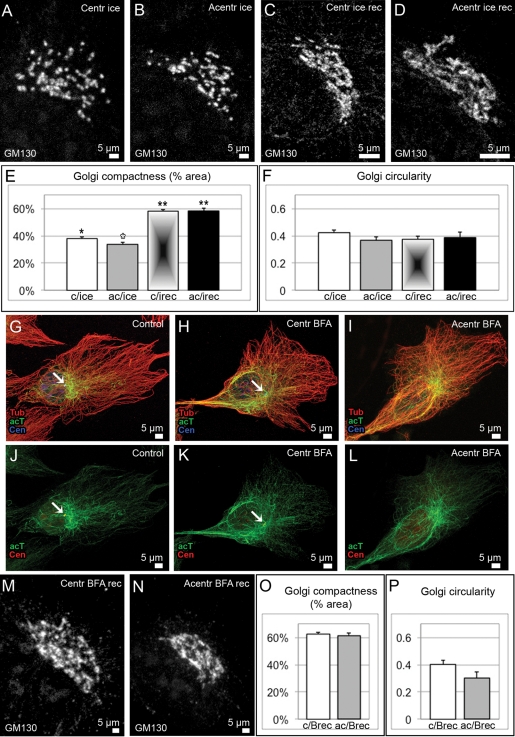
To test this possibility, we applied cold treatment that results in MT depolymerization without Golgi scattering throughout the cell (Figure S7, B and C). Cell were put on ice for 40 min and analyzed thereafter. As expected, the Golgi remained in the cell center both in centrosomal and acentrosomal cells and maintained an overall polarized shape with low circularity index (Figure 4, A–D and F). However, the Golgi ribbon was fragmented without MTs, regardless of the presence of the centrosome, indicating that Golgi integrity is MT-dependent (Figures 4E and S7C). Indeed, initial Golgi integrity was recovered upon MT regrowth both in centrosomal and acentrosomal cells. This result indicates that Golgi-derived MTs are necessary and sufficient to support Golgi ribbon continuity, as long as Golgi ministacks are collected in advance through centrosomal guidance. This result is in accordance with our computational model, which predicts that the role of the centrosome in Golgi assembly is restricted to organizing a radial array of MTs, which brings Golgi clusters to the cell center.
FIGURE 4:
Golgi complex organization in centrosomal and acentrosomal cells influenced by cold or BFA. (A–D), (M), and (N) the Golgi revealed by immunostaining for GM130. (A) centrosomal cell fixed after 40 min on ice; (B) acentrosomal cell fixed after 40 min on ice; (C) centrosomal cell fixed 2 h after cold treatment; and (D) acentrosomal cell fixed 2 h after cold treatment. (E) Compactness of Golgi complex (percentage of space within the Golgi area occupied by GM130) in the four types of cells corresponding to (A–D). (F) Circularity [4π*(area/perimeter2)] of the Golgi complex in the four types of cells corresponding to (A–D). Data from three independent experiments. p < 0.01 in unpaired Student's t test between bars marked with single and double asterisks; p < 0.05 between white and black asterisks. Error bars represent SEM. (G–L) MT organization in nontreated (G) and (J), BFA-treated centrosomal (H) and (K), and BFA-treated acentrosomal (I) and (L) cells. Immunostaining: (G–I) Tubulin, red; acetylated tubulin, green (far red, pseudocolored); centrin-GFP, blue (pseudocolored). (J–L) Acetylated tubulin, green (far red, pseudocolored); centrin-GFP, red (pseudocolored). Arrow, centrosome. (M) Centrosomal cell fixed 2 h after BFA washout; (D) acentrosomal cell fixed 2 h after BFA washout. (O) Compactness and (P) circularity of the Golgi complex in cells corresponding to (M–N). Data from five independent experiments. p > 0.05 in unpaired Student's t test. Error bars represent SEM.
We were able to further test whether the radial array of MTs can perform the function of the centrosome in Golgi assembly. After 1–2 h of breffeldin A (BFA) treatment, the Golgi can no longer be detected in RPE cells (Figure S7, D and E), while the MT array is similar to those of untreated cells (Figure S7). We and others have shown previously that noncentrosomal MTs in this case can still be formed at random cytosolic sites (Efimov et al., 2007; Rivero et al., 2009). Interestingly, centrosome ablation, which completely abolishes new formation of centrosomal MTs (Khodjakov et al., 2000; Khodjakov and Rieder, 2001), only slightly disorganizes the MT network, which remains primarily radial (Figure 4, G–I). Such organization is likely due to the high stability of MT parts in the cell center, which is evident from high tubulin acetylation levels in this area (Figure 4, G–L).
We took advantage of this experimental condition to investigate efficiency of Golgi assembly in the presence of the radial MT array, but in the absence of the centrosome. Golgi complexes assembled 2 h after BFA washout demonstrate compactness and circularity parameters indistinguishable from those assembled in the presence of the centrosome in nocodazole washout experiments (Figure 4, M–P). We assume that, once stabilized, MTs in the cell center persist throughout the BFA treatment and fulfill the role of the radial MT population during Golgi assembly, while the nonradial MT population, which forms at emerging Golgi stacks, allows for ministack clustering.
Computational model predicts that collaboration of two MT populations is required for the establishment of overall Golgi polarity
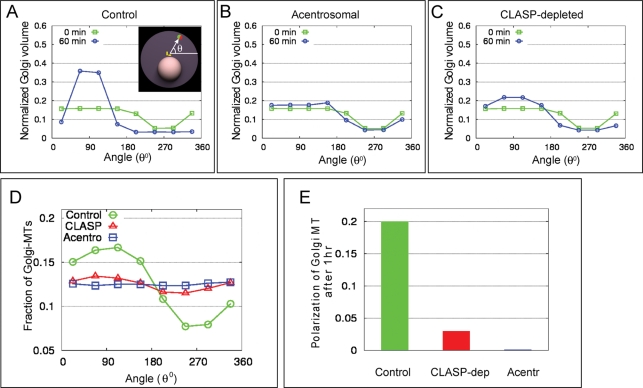
Computational simulations of the Golgi assembly revealed that participation of both MT arrays in the process is critical for the resulting complex integrity and compactness. Additionally, a requirement of both arrays for overall Golgi asymmetry was revealed. In particular, the control assembly driven by collaboration of centrosomal and Golgi-derived MTs focuses the stacks in a narrow angular space, placing them exactly in front of the centrosome. In CLASP-depleted cells, this focusing is significantly impaired, and in the acentrosomal case, it is nonexistent (Figure 5, A–C), in agreement with experimental data (Figure S8). Moreover, summarized cis–trans orientation of stacks within the Golgi assembled by both MT populations is polarized, while it is random in acentrosomal or CLASP-depleted cells (Figure 2, A–C). Since Golgi-derived MTs in the model form at the trans side only, angular distribution of Golgi-derived MTs reflects polarization of the Golgi complex and is highly directional in control cells, but not when the Golgi is assembled by solely Golgi-derived or centrosomal MTs (Figure 5, D and E). Thus our model predicts that Golgi complex assembled by concerted effort of two MT arrays acquires an important property, which makes it qualitatively different from Golgi complexes assembled by either MT population alone.
FIGURE 5:
Both centrosomal and Golgi-derived MTs are required for the proper polarization of the Golgi and MT array in silico. (A–C) Simulation results for the angular distribution of Golgi fragments before (0 min) and after (60 min) the assembly. Initially, Golgi fragments are distributed uniformly across the cell. The minimum at 270° is due to the excluded volume of the nucleus. In the control case (A), significant focusing of the Golgi density to 90° is predicted. In the acentrosomal case (B), no organization in the angular space is predicted. In the CLASP-depleted case (C), little focusing to 90° is predicted. (D) Computed angular distribution of the Golgi-derived MTs after 60 min of assembly. Ninety degrees corresponds to outward-pointing normal to the nuclear surface. Significant polarization of MTs in angular space takes place in control cell, but not in CLASP-depleted and acentrosomal cells. (E) Predicted quantitative measure of the MT array polarization calculated as defined in the Supplemental Material on modeling Golgi assembly.
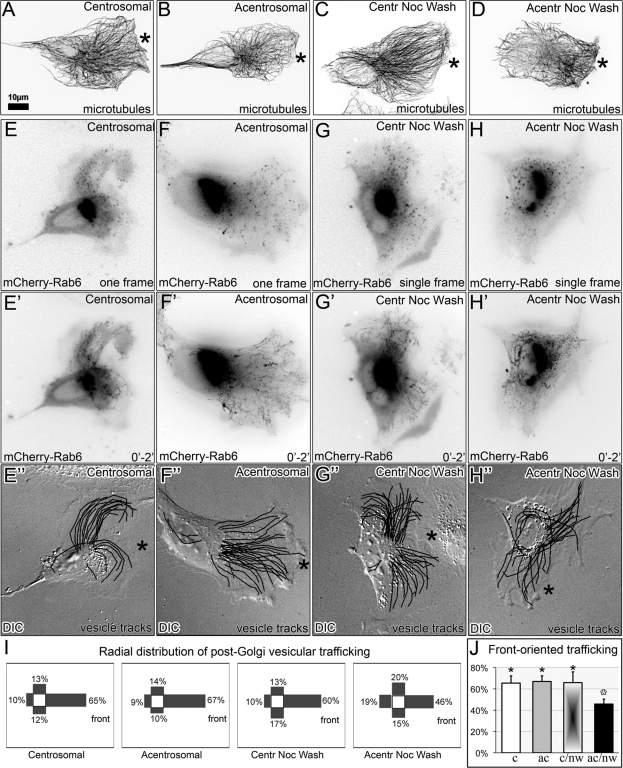
Directionality of post-Golgi vesicular trafficking is disorganized in cells in which the Golgi is assembled in the absence of radial MTs
Overall polarity of the Golgi complex and directionality of MT arrays is essential for the directionality of post-Golgi trafficking, which in motile cells is necessary for directional cell migration. Our previous data (Miller et al., 2009) indicate that the model predictions for CLASP-depleted cells are correct, and polarity of both MT array and post-Golgi trafficking is lost. To evaluate model predictions for acentrosomal cells, we compared RPE1 cells in which the Golgi was assembled in the absence of the centrosome with control conditions, including untreated centrosomal cells, untreated cells lacking centrosomes, and centrosomal cells 2 h after nocodazole washout. In accordance with the model predictions, the distribution of MTs in acentrosomal cells after nocodazole washout was less polarized than in control cells or acentrosomal cells with preexisting Golgi complex (Figure 6, A–D). As post-Golgi vesicular trafficking is considered the major functional readout of the MT array directionality, we analyzed the movements of exocytic vesicles labeled with mCherry-Rab6 (Grigoriev et al., 2007). Transport of individual Rab6-marked vesicles from the Golgi toward the plasma membrane was tracked (Figure 6, E–H), and the directionality of vesicle tracks was quantified for the “forward,” “rear,” “left,” and “right” quadrants (Figure 6I). As expected, nontreated centrosomal cells and centrosomal cells after nocodazole washout exhibited polarized trafficking toward the leading edge (Figure 6, E″ and G″). No changes were observed in the trafficking pattern of untreated acentrosomal cells (Figure 6F″). In contrast, polarity of post-Golgi trafficking in acentrosomal cells after nocodazole washout was significantly disturbed (Figure 6H″). In these cells, the trafficking pattern was more random, with fewer front-directed tracks (Figure 6J). Thus alteration in the trafficking directionality correlates with abnormal Golgi complex organization (Figure 3) and changes in MT distribution but not directly with the presence of the centrosome. These results are in agreement with the theoretical predictions (Figure 5).
FIGURE 6:
Polarity of MTs and trafficking requires presence of the Golgi complex assembled by both MT arrays. (A and E–E″) Nontreated centrosomal cell; (B and F–F″) acentrosomal cell; (C and G–G″) centrosomal cell 2 h after nocodazole washout; (D and H–H″) acentrosomal cell 2 h after nocodazole washout. (A–D) MT organization revealed by tubulin immunostaining. Asterisks indicate leading edges. For clarity, fluorescence images are presented in inverted contrast. (E–H″) Post-Golgi vesicular trafficking analyzed in live mCherry-Rab6–expressing cells. (E–H) First frames from the time-lapse movies; (E′–H′) time projections of 2-min movies recorded at 1-s intervals. (E″–H″) Post-Golgi vesicle tracks superimposed on DIC images. Front of cell (asterisks) is designated according to the leading-edge protrusion. (I) quadrant distribution of vesicle trajectories in the experimental condition types corresponding to (E–H). (J) Percentage of front-oriented trafficking in the four experimental conditions corresponding to (E–H). n = 10, at least three independent experiments for each condition.
Directionality of migration is decreased in cells in which the Golgi is assembled in the absence of radial MTs
Our demonstration that directionality of post-Golgi vesicular trafficking is affected only in cells in which the Golgi is assembled in the absence of the centrosomes allowed us to address a long-standing question of whether the centrosome is directly required for directional cell migration (Schliwa et al., 1999; Etienne-Manneville, 2004; Wakida et al., 2010) or whether its role is mediated by the downstream effects on Golgi organization and transport (Miller et al., 2009; Yadav et al., 2009; Hurtado et al., 2011).
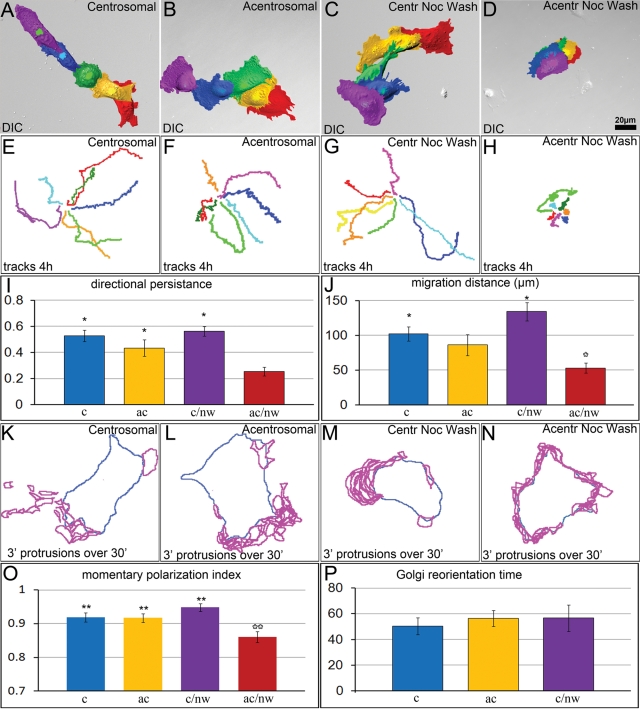
We monitored single-cell migration for centrosomal and acentrosomal cells over a 4-h period in untreated cells or in cells after nocodazole recovery, cold treatment, or BFA treatment. Total migration distance and directional persistence of migration (total migration distance divided by the length of cell migration track) were analyzed (Figure S9A). Both parameters were indistinguishable between the untreated centrosomal and acentrosomal cells, revealing that the centrosome is not required for directional migration in the presence of a functional Golgi complex (Figure 7, A and B, E and F, I, and J). The Golgi complex reorientation toward the leading edge, which is often referred to as a typical feature of directionally moving cells (Schliwa et al., 1999; Etienne-Manneville, 2004) was not altered by centrosome ablation (Figure S10). Motility of centrosomal cells released from nocodazole was similar to that of untreated controls (Figure 7, C, G, I, and J). In contrast, both the total migration distance and directional persistence of acentrosomal cells released from nocodazole were strongly decreased (Figure 7, D, H, I, and J). Thus abnormal Golgi organization and randomized post-Golgi trafficking correlate with the deficiency in directional migration.
FIGURE 7:
Migration and polarization ability of centrosomal and acentrosomal cells. (A–D) Random single-cell migration of: (A) nontreated centrosomal cell; (B) acentrosomal cell; (C) centrosomal cell starting 2 h after nocodazole washout; and (D) acentrosomal cell starting 2 h after nocodazole washout. Color-coded images depict positions of each cell at 1 h intervals. (E–H) Examples of migration trajectories (center of the nucleus) at 4-h observation periods in the four experimental conditions corresponding to (A–D). (I) Total cell migration distance and (J) directional persistence of migration in the four experimental conditions corresponding to (A–D). *, p < 0.05 in unpaired Student's t test between bars marked with black and white asterisks. Error bars represent SEM. (K–N) Distribution of protrusions during 30-min time-lapse imaging in experimental conditions corresponding to (A–D). Blue, contours of cells at the first frame of the time-lapse sequence; magenta, cell protrusions formed every 3 min throughout the time-lapse sequence. (O) Momentary polarization index in the four experimental conditions corresponding to (A–D). **, p < 0.01 in unpaired Student's t test between bars marked with black and white asterisks. (P) Time taken to restore the Golgi orientation from the back to the front of migrating cells after a cell turn has turned; shown for three cellular conditions. p > 0.05 in unpaired Student's t test.
To additionally test the motile properties of acentrosomal cells in which the Golgi was assembled in the presence of an alternative radial stimulus, we monitored the motility of cells recovered after cold or BFA treatments (Figure S11). Overall, cell motility parameters were similar to those of centrosomal cells under the same conditions. However, directional persistence of acentrosomal cell migration after cold treatment was slightly lower than in centrosomal cells (0.75 times), though significantly higher than in cells recovered after nocodazole (1.85 times). This result suggests the centrosomal MTs likely provide an additional, though minor, polarization signal that does not depend on the Golgi, or the Golgi complex recovered after cold treatment has defects in its organization that could not be detected by our analyses.
The inability of cells with an abnormal Golgi to migrate directionally may reflect defects in polarization of a cell at each particular time point (momentary polarization) or this inability may arise from frequent changes in the direction of polarized migration. To discriminate between these two possibilities, we monitored formation of the leading-edge protrusions over a 30-min period at 3-min temporal resolution and analyzed polarity of the protrusion (Figure S9B). In the directionally migrating cells with intact Golgi, protrusions formed predominantly at the leading edge (Figure 7, K–M and O). In contrast, after nocodazole washout, acentrosomal cells exhibited a random distribution of protrusions, indicating these cells lacked the ability to achieve morphological polarization (Figure 7, N and O). Thus proper organization of the Golgi and directionality of polarized trafficking is associated with momentary motile-cell polarity (Figure 7P).
DISCUSSION
Golgi assembly from the scattered ministacks is an essential process that occurs every time a cell exits mitotic division. Previously we demonstrated that Golgi-derived MTs are indispensable for the Golgi complex assembly (Miller et al., 2009); this was further confirmed by a recent study (Hurtado et al., 2011). Our current results reveal that the two MT arrays act in concert and are both essential for Golgi integrity and polarity.
We have shown previously that these two MT arrays are characterized by distinct geometries (Efimov et al., 2007). In particular, the centrosome produces a strictly radial MT array (Bergen et al., 1980; Salaycik et al., 2005), Golgi-derived MTs intrinsically grow tangentially to a stack of origin, and their directions are random when ministacks are dispersed throughout the cell (Miller et al., 2009). An intriguing question is whether this distinct geometry or another distinction of the two MT populations is a critical factor for their specific functions.
Our data indicate that the radial centrosomal MT array is necessary for arranging the Golgi complex organization in the course of assembly from scattered ministacks. At the same time, Golgi-derived MTs are critical for efficient Golgi fragment merging and Golgi ribbon continuity (Miller et al., 2009; Hurtado et al., 2011). Unexpectedly, the computation simulation presented here predicts that even aberrant Golgi assembly observed in CLASP-depleted cells (Miller et al., 2009) partially relies on Golgi-derived MTs formed due to incomplete CLASP depletion. The model also predicts that Golgi-derived MTs support fusion of ministacks upon clustering much more efficiently than centrosomal MTs. Indeed, Golgi-derived MTs are likely biochemically different from centrosomal ones, as indicated by the enrichment of CLASPs specifically at Golgi-derived MTs (Efimov et al., 2007). This property may be important for efficient ministack coalescence at an MT and subsequent fusion. However, when only Golgi-derived MTs are present, scattered ministacks cannot be properly collected into a single integral complex. The strong centripetal signal provided by the radial centrosomal array is likely necessary to properly coalesce scattered Golgi clusters and finalize Golgi assembly. This function of the centrosomal array can be substituted by prior positioning of ministacks in the cell center (cold recovery) or by a radial array of stable MTs (BFA recovery), indicating that the centripetal signal, and not other properties of the centrosomal array, is important for this function. We conclude that most likely both geometric and biochemical properties of Golgi-derived MTs, but only geometric organization of the radial centrosomal array, are important for Golgi integrity.
We further demonstrate that the concerted effort of two MT arrays is necessary and sufficient for Golgi complex polarity. MTs assemble the Golgi complex by bringing together single Golgi stacks. cis–trans polarity of these stacks does not require MTs, as single polar stacks exist in a nocodazole-treated cell (Cole et al., 1996), can be reconstituted in a tube (Tang et al., 2008), and can emerge in the absence of the centrosome (Tangemo et al., 2011). However, our data indicate that Golgi complex collected and assembled in the absence of radial MTs lacks its overall polarity, as illustrated by observed randomization of the Golgi shape (increased circularity) and of post-Golgi trafficking directionality. Similarly, the Golgi assembled solely by centrosomal MTs lack polarity (Miller et al., 2009; Hurtado et al., 2011). Lack of polarity in a complex composed of polar stacks may arise from insufficient complex integrity and randomized mutual positioning of polar Golgi fragments within the complex. Indeed, in silico simulations predict that lack of either one of the MT arrays leads to the loss of complex polarity associated with integrity disturbance and shape randomization. Thus our results strongly indicate that neither MT subpopulation alone is capable of substituting for the concerted effort of centrosomal and Golgi-derived MTs in the Golgi assembly. We propose that mutual stack positioning requires both strong radial signal and tangential forces driving single stacks together. Interestingly, similar collaboration of centrosomal and noncentrosomal MTOCs is important for efficient mitotic spindle formation (O’Connell and Khodjakov, 2007).
Improperly assembled Golgi complex produces nondirectional post-Golgi trafficking. This may occur through loss of MT network polarity (Musch, 2004; Hoogenraad and Bradke, 2009). Decreased trafficking polarity in the absence of the centrosome correlates with the decrease in the polarity of the MT network, which in this case forms as a whole at the Golgi (Efimov et al., 2007). Golgi-derived MTs are initiated at one distinct side of a Golgi stack (at the TGN, according to our data [ Efimov et al., 2007], or cis-Golgi, as proposed by others [ Rivero et al., 2009]). We propose that these MT nucleation sites are located randomly within the fragmented Golgi complex formed without a centrosome, leading to randomization of a normally polar Golgi-derived MT array, and subsequently, post-Golgi trafficking. Of note, endoplasmic reticulum-to-Golgi trafficking apparently can be supported by Golgi-derived MTs only, since already-assembled Golgi is properly maintained in acentrosomal cells.
Interestingly, our computational model also predicts that correct assembly of the fully functional Golgi complex requires two MT arrays but not a previously proposed direct centrosome–Golgi interaction (Rios et al., 2004; Bornens, 2008; Hurtado et al., 2011). We also show that the Golgi correctly assembles without the centrosome in acentrosomal cells recovering after cold treatment. This argues that the role of the centrosome in Golgi organization is likely restricted to the role of dynein-dependent centripetal transport of scattered ministacks during the new Golgi assembly. Also, already-assembled Golgi is capable of maintaining polarization and integrity without the centrosome. Thus, while multifunctional Golgi matrix protein GM130 was shown to regulate the centrosomal organization and function (Kodani and Sutterlin, 2008; Kodani et al., 2009), there is likely no direct functional feedback from the centrosome to the Golgi. Our findings argue that the centrosome is not needed for Golgi organization and function when Golgi translocation to the cell center is complete.
The role of the centrosome in polarized cell motility has been intensively investigated in the last decade (Zmuda and Rivas, 1998; Etienne-Manneville, 2004; de Anda et al., 2005; Li and Gundersen, 2008). More recent study implicates mutual positioning of the centrosome and the Golgi in motile-cell polarity (Hurtado et al., 2011). Surprisingly, our results demonstrate that directional migration of cells with an alternative source of polarized MT array, such as RPEs, does not require the centrosome per se. Acentrosomal cells become depolarized and seize migration only upon nocodazole washout. Thus certain polarity cues established by the centrosome are lost during nocodazole treatment. Our data implicate proper Golgi organization as the major contributor to this mechanism, which is consistent with the fact that nonspecific Golgi dispersal prevents directional motility (Prigozhina and Waterman-Storer, 2004; Yadav et al., 2009). We propose that the centrosome supports polarized cell migration indirectly, via organizing integral Golgi complex in an MT-dependent manner. Golgi polarity, in turn, provides asymmetry of the noncentrosomal (Golgi-derived) MT array, which supports trafficking polarity as well as other asymmetric MT-dependent processes (focal adhesion disassembly, mRNA delivery, etc.). It is interesting that polarity of migration is also abolished in cell types in which MT asymmetry is lost after the centrosome ablation (Wakida et al., 2010).
Our data also indicate that additional, yet minor, Golgi-independent polarity features exist that are stable upon centrosome ablation but lost during the MT-depolymerization period. These features appear to depend directly on the centrosome, because acentrosomal cells move slightly less directionally after cold treatment than do centrosomal cells, despite their proper Golgi organization. This minor polarity regulation may be provided directly by the centrosomal MTs or through a centrosome–Golgi connection that has been proposed to play a role in cell polarity (Hurtado et al., 2011). Our data also argue that this centrosome–Golgi connection is essential only temporally during the Golgi assembly phase.
To conclude, we establish in this study that the centrosomal array alone or the Golgi-derived array alone is insufficient for correct Golgi assembly from scattered ministacks. Only the concerted effort of both MT arrays results in an integral Golgi complex possessing the overall polarity necessary for polarized trafficking and cell motility.
MATERIALS AND METHODS
Computational modeling
To simulate the Golgi assembly model, we implemented time-dependent, explicit, agent-based simulations (Alberts and Odell, 2004; Paul et al., 2009). The Golgi assembly process is modeled in an oblate-spheroidal cell (Figure 2, A–D) with a spherical nucleus located nonconcentrically and a centrosome near the nuclear surface facing distal cell boundary. In the beginning of each simulation, two classes of objects—ministacks and MTs—were constructed, and their positions and sizes were then changed in the three-dimensional space according to the computational rules described below. Initially, 150–200 spherical Golgi fragments, each consisting of two hemispheres 0.2-µm in radius representing trans and cis sides, are uniformly and randomly distributed within the cell. The trans surface of each ministack nucleates 2–4 MTs in control and acentrosomal cells and 0–2 MTs in CLASP-depleted cells. Fitting of the simulation results to the experimental data (Figure 2E) illustrates that the dynamics of Golgi assembly in CLASP-depleted cells still involves a small number of Golgi-derived MTs, consistent with the fact that CLASP depletion was below 100% (Efimov et al., 2007; Miller et al., 2009). In addition, 250–300 MTs are nucleated from the centrosome in control and CLASP-depleted cells. MTs undergo dynamic instability; when a growing MT plus end encounters a Golgi fragment, a capture event takes place. If a centrosomal MT makes the capture, the fragment is transported by dynein motors toward the centrosome, but if it is a Golgi-derived MT, two fragments—captor and captured—glide toward each other with rates graded by their sizes. The fragments brought together by a Golgi-nucleated MT merge upon colocalization after a certain delay time. The cis–trans orientation of the merged fragment is equal to the geometric average of the initial orientations of two merged fragments. The surface area of the resulting fragment is equal to the sum of the areas of the merged fragments. A stack that is bigger than a threshold size starts to flatten against the nucleus. A constant average number of MTs is nucleated per unit surface area of the trans side of the resulting fragment. Additional model assumptions, variations, parameters, and simulation details can be found in the Supplemental Material.
Cells
Immortalized human pigment epithelial cells hTert-RPE1 (Clontech, Mountain View, CA) that stably express centrin1-GFP (see Uetake et al., 2007) were maintained in DMEM/F12 with 10% fetal bovine serum at 37°C in 5% CO2. Cells were plated on glass coverslips coated with 5 μg/ml fibronectin 24 h prior to experiments. For laser microsurgery and live-cell observations, these coverslips were mounted in Rose chambers (Khodjakov and Rieder, 2001). Alternatively, glass bottom dishes (MatTek, Ashland, MA) were used in some experiments. In all live-cell experiments, cells were maintained on the microscope stage at 37°C.
Small interfering RNA depletion
Two different combinations of mixed small interfering RNA (siRNA) oligonucleotides against CLASP1 and CLASP2 were transfected into cells using HiPerFect (Qiagen, Valencia, CA), as described previously (Miller et al., 2009). Experiments were conducted 72 h after transfection, as at this time minimal protein levels were detected. Nontargeting siRNA (Dharmacon, Chicago, IL) was used for controls.
Treatments
Nocodazole (2.5 μg/ml) or BFA (5 μg/ml) was added to the culture media for 2 h. For washout, cells were rinsed five times and transferred to fresh, warm (37°C) medium for 2 h.
For cold treatment, cells were placed on ice for 40 min. For recovery, cells were placed to 37°C for 2 h.
In cell migration assays, time-lapse sequences were initiated 2 h after drug washout or temperature release, and cells were monitored for 4–8 h.
Transfection and immunostaining
3GFP-EMTB, ensconsin MT-binding domain (a gift from J. C. Bulinski, Columbia University, New York, NY) and/or mCherry-Rab6 (a gift from A. Akhmanova, Rotterdam, The Netherlands), or YFPGalT (Clontech) were transiently expressed in RPE1 cells. Fugene6 (Roche, Indianapolis, IN) was used for transfection according to the manufacturer's protocols.
The following antibodies were used: mouse monoclonal anti-GM130 (Transduction Laboratories, San Jose, CA), guinea pig polyclonal anti–GCC185 VU-140 (Efimov et al., 2007), rabbit polyclonal acti–CLASPs VU-83 (Efimov et al., 2007), mouse monoclonal anti–α-tubulin DM1a (Sigma-Aldrich, St. Louis, MO), rabbit polyclonal anti-tubulin (Abcam, Cambridge, MA), mouse monoclonal anti–Ac tubulin (Sigma-Aldrich). Alexa Fluor 488– and Alexa Fluor 568–conjugated highly cross-absorbed goat anti–mouse immunoglobulin G (IgG) antibodies, Alexa Fluor 568–and Alexa Fluor 633–conjugated highly cross-absorbed goat anti–rabbit IgG antibodies, and Alexa Fluor 568 anti–guinea pig IgG (Molecular Probes, Eugene, OR) were used as secondary antibodies. For immunostaining, cells were fixed for 15 min (room temp.) in 2% paraformaldehyde, 0.1% glutaraldehyde, and 0.5% saponin in cytoskeleton buffer (10 mM 2-(N-morpholino)ethanesulfonic acid, 150 mM NaCL, 5 mM ethylene glycol tetraacetic acid, 5 mM glucose, and 5 mM MgCL2, pH 6.1).
Immunofluorescence and live-cell imaging
Confocal stacks were taken with Leica TCS SP5 laser-scanning confocal system using a 100×, 1.47 numerical aperture (NA) Plan Apo lens, or with a Yokogawa QLC-100/CSU-10 spinning-disk head (Visitec assembled by Vashaw, Norcross, GA) attached to a Nikon TE2000E microscope using a 100×, 1.4 NA Plan Apo lens and a back-illuminated EMCCD camera (Cascade 512B; Photometrics, Tucson, AZ) driven by IPLab software (Scanalytics, Rockville, MD). For cell motility assays, differential interference contrast differential interference contrast (DIC) and two-color fluorescence images were recorded using 20×, 0.75 NA Plan Apo lens and an interline CCD camera (CoolSnap HQ; Photometrics).
Centrosome ablation and FRAP
The detailed layout of our laser microablation system is described elsewhere (Magidson et al., 2007). Briefly, 532-nm laser pulses generated by a Q-switched Nd:YAG laser were focused on the specimen with a 100×, 1.4 NA Plan Apo lens. Centrosome ablation was achieved by a series of two to four laser pulses (∼0.5 each) aimed sequentially at the two centrin-GFP–labeled centrioles. For each cell, success of the ablation was confirmed by collecting an image stack ∼30 min after the operation. Owing to dynamic exchange of centrin-GFP, lack of localized centrin-GFP fluorescence proves that the centrosome was destroyed (Magidson et al., 2007).
The unique morphology of centrin-GFP–labeled centrosomes (two diffraction-limited spots) allowed us to unambiguously delineate this organelle, even when cells coexpressed centrin-GFP and green-labeled Golgi markers. Further, centrin-GFP–labeled centrosomes remained clearly delineated in cells expressing 3GFP-EMTB when MTs were depolymerized with nocodazole.
The same microscopy workstation was utilized in the FRAP experiments, except a continuous-wave 488-nm laser beam was used in the latter case. Golgi stacks were photobleached with 1.5-s exposure to the beam. Recovery of the Golgi-associated fluorescence was measured on a single focal plan recorded at 3-s intervals.
In both laser microsurgery and FRAP experiments, images were recorded with a back-illuminated EM-CCD camera (Cascade 512B; Photometrics, Tuscon, AZ) in spinning-disk confocal mode (GSU-10; Yokogawa). All light sources were shuttered by either fast mechanical shutters (Vincent Associates, Falmouth, MA) or acousto-optic tunable filters (Solamere Technology Group, Salt Lake City, UT), such that cells were exposed to light only during actual image acquisition.
Image acquisition and processing
Figure 1: Maximum-intensity projection of confocal stacks (0.40-µm spacing, over thickness of 1.2 µm). Figure 3, A–D: Maximum-intensity projection of confocal stacks (0.20-µm spacing, over whole cell); Figure 3, G–J, L: Single plane confocal video frame. Figure 4, A–D and G–N: Maximum-intensity projection of confocal stacks (0.20-µm spacing, over whole cell); Figure 6, A–D: Wide-field fluorescence, inverted images; Figure 6, A′–D′: Maximum-intensity projection over time of time-lapse wild-field fluorescence, inverted images, 2-min recording with 1 s between frames and 300-ms exposure time; Figure 6, A″–D″: DIC images with superimposed tracks of post-Golgi vesicle movement (see “Post-Golgi vesicle tracking” in the Supplemental Material). Figure 6, G–J: Inverted maximum-intensity projection of confocal stacks (0.20-µm spacing, over whole cell). Figure 7, A–D: False-colored DIC video frames superimposed on one image, 1 h between frames, 100-ms exposure time. Figure S2: Maximum-intensity projection of confocal stacks (0.20-µm spacing, over whole cell). Figure S3, B and C: Wide-field fluorescence images. Figure S7: Maximum-intensity projection of confocal stacks (0.20-µm spacing, over whole cell). Figure S10: DIC (A–L) or phase (M–R) merged with wide-field fluorescence video frames, 30 min between frames, 100-ms exposure time for DIC and 200-ms exposure time for red fluorescent channel (pseudocolored green in the images). Figure S11, A–D: Pseudocolored DIC or phase-contrast video frames superimposed on one image, 1 h between frames, 100-ms exposure time.
Statistical analysis
Statistical significance was determined by Student's t test (two-tailed, unpaired).
Supplementary Material
Acknowledgments
We thank Nadia Efimova for technical assistance and advice. This work was supported by National Institutes of Health grants R01-GM078373 (to I.K.), RO1-GM59363 (to A.K.), and RO1-GM068952 (to A.M.); American Heart Association Grant-in-Aid 10GRNT4230026 (to I.K.); and predoctoral fellowship 09PRE2260729 (to P.M.M.). R.P. thanks DST India for financial support (grant SR/S2/CMP-0107/2010).
Abbreviations used:
- BFA
breffeldin A
- CLASP
CLIP-associated protein
- DIC
differential interference contrast
- EMTB
ensconsin MT-binding domain
- FRAP
fluorescence recovery after photobleaching
- Ig
immunoglobulin
- MT
microtubule
- MTOC
MT-organizing center
- NA
numerical aperture
- RPE1
retinal pigment epithelium 1
- siRNA
small interfering RNA
- TGN
trans-Golgi network
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0550) on January 19, 2012.
REFERENCES
- Alberts JB, Odell GM. In silico reconstitution of Listeria propulsion exhibits nano-saltation. PLoS Biol. 2004;2:e412. doi: 10.1371/journal.pbio.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen LG, Kuriyama R, Borisy GG. Polarity of microtubules nucleated by centrosomes and chromosomes of Chinese hamster ovary cells in vitro. J Cell Biol. 1980;84:151–159. doi: 10.1083/jcb.84.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK. The role of microtubule-based motor proteins in maintaining the structure and function of the Golgi complex. Biochim Biophys Acta. 1998;1404:113–126. doi: 10.1016/s0167-4889(98)00052-4. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cytrynbaum EN, Rodionov V, Mogilner A. Computational model of dynein-dependent self-organization of microtubule asters. J Cell Sci. 2004;117:1381–1397. doi: 10.1242/jcs.00919. [DOI] [PubMed] [Google Scholar]
- de Anda FC, Pollarolo G, Da Silva JS, Camoletto PG, Feiguin F, Dotti CG. Centrosome localization determines neuronal polarity. Nature. 2005;436:704–708. doi: 10.1038/nature03811. [DOI] [PubMed] [Google Scholar]
- Efimov A, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic. 2004;5:470–477. doi: 10.1111/j.1600-0854.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- Grigoriev I, et al. Rab6 regulates transport and targeting of exocytotic carriers. Dev Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Bradke F. Control of neuronal polarity and plasticity–a renaissance for microtubules? Trends Cell Biol. 2009;19:669–676. doi: 10.1016/j.tcb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Hurtado L, Caballero C, Gavilan MP, Cardenas J, Bornens M, Rios RM. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J Cell Biol. 2011;193:917–933. doi: 10.1083/jcb.201011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Kristensen I, Huang L, Sutterlin C. GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol Biol Cell. 2009;20:1192–1200. doi: 10.1091/mbc.E08-08-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Sutterlin C. The Golgi protein GM130 regulates centrosome morphology and function. Mol Biol Cell. 2008;19:745–753. doi: 10.1091/mbc.E07-08-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- Lowe M. Structural organization of the Golgi apparatus. Curr Opin Cell Biol. 2011;23:85–93. doi: 10.1016/j.ceb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Magidson V, Loncarek J, Hergert P, Rieder CL, Khodjakov A. Laser microsurgery in the GFP era: a cell biologist's perspective. Methods Cell Biol. 2007;82:239–266. doi: 10.1016/S0091-679X(06)82007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikov V, Cytrynbaum EN, Kashina A, Mogilner A, Rodionov V. Centering of a radial microtubule array by translocation along microtubules spontaneously nucleated in the cytoplasm. Nat Cell Biol. 2005;7:1213–1218. doi: 10.1038/ncb1332. [DOI] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Beznoussenko GV. Molecular mechanisms responsible for formation of Golgi ribbon. Histol Histopathol. 2010;26:117–133. doi: 10.14670/HH-26.117. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Self-organization of polymer-motor systems in the cytoskeleton. Philos Trans R Soc Lond B Biol Sci. 1992;336:99–106. doi: 10.1098/rstb.1992.0049. [DOI] [PubMed] [Google Scholar]
- Musch A. Microtubule organization and function in epithelial cells. Traffic. 2004;5:1–9. doi: 10.1111/j.1600-0854.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- Nedelec FJ, Surrey T, Maggs AC, Leibler S. Self-organization of microtubules and motors. Nature. 1997;389:305–308. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner A. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc Natl Acad Sci USA. 2009;106:15708–15713. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina NL, Waterman-Storer CM. Protein kinase D-mediated anterograde membrane trafficking is required for fibroblast motility. Curr Biol. 2004;14:88–98. doi: 10.1016/j.cub.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Rios RM, Bornens M. The Golgi apparatus at the cell centre. Curr Opin Cell Biol. 2003;15:60–66. doi: 10.1016/s0955-0674(02)00013-3. [DOI] [PubMed] [Google Scholar]
- Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits γ-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaycik KJ, Fagerstrom CJ, Murthy K, Tulu US, Wadsworth P. Quantification of microtubule nucleation, growth and dynamics in wound-edge cells. J Cell Sci. 2005;118:4113–4122. doi: 10.1242/jcs.02531. [DOI] [PubMed] [Google Scholar]
- Schliwa M, Euteneuer U, Graf R, Ueda M. Centrosomes, microtubules and cell migration. Biochem Soc Symp. 1999;65:223–231. [PubMed] [Google Scholar]
- Snapp EL, Altan N, Lippincott-Schwartz J. Measuring protein mobility by photobleaching GFP chimeras in living cells. Curr Protoc Cell Biol. 2003;19:Chapter 21–Unit 21.1. doi: 10.1002/0471143030.cb2101s19. [DOI] [PubMed] [Google Scholar]
- Surrey T, Nedelec F, Leibler S, Karsenti E. Physical properties determining self-organization of motors and microtubules. Science. 2001;292:1167–1171. doi: 10.1126/science.1059758. [DOI] [PubMed] [Google Scholar]
- Tang D, Mar K, Warren G, Wang Y. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangemo C, Ronchi P, Colombelli J, Haselmann U, Simpson JC, Antony C, Stelzer EH, Pepperkok R, Reynaud EG. A novel laser nanosurgery approach supports de novo Golgi biogenesis in mammalian cells. J Cell Sci. 2011;124:978–987. doi: 10.1242/jcs.079640. [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell-cycle progression without an intact microtuble cytoskeleton. Curr Biol. 2007;17:2081–2086. doi: 10.1016/j.cub.2007.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakida NM, Botvinick EL, Lin J, Berns MW. An intact centrosome is required for the maintenance of polarization during directional cell migration. PLoS One. 2010;5:e15462. doi: 10.1371/journal.pone.0015462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmuda JF, Rivas RJ. The Golgi apparatus and the centrosome are localized to the sites of newly emerging axons in cerebellar granule neurons in vitro. Cell Motil Cytoskeleton. 1998;41:18–38. doi: 10.1002/(SICI)1097-0169(1998)41:1<18::AID-CM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.