Nuclear pore complex assembly and kinetochore function depend on the NUP107 subcomplex, but the roles of each of its nine constituents are unknown. NUP107 itself is shown to be dispensable for NPC assembly but needed for proper localization of kinetochore protein NUF2 and Aurora B kinase. Moreover, a novel interaction is found with SAC protein MAD1.
Abstract
Nuclear pore complexes consist of several subcomplexes. The NUP107 complex is important for nucleocytoplasmic transport, nuclear envelope assembly, and kinetochore function. However, the underlying molecular mechanisms and the roles of individual complex members remain elusive. We report the first description of a genetic disruption of NUP107 in a metazoan. Caenorhabditis elegans NUP107/npp-5 mutants display temperature-dependent lethality. Surprisingly, NPP-5 is dispensable for incorporation of most nucleoporins into nuclear pores and for nuclear protein import. In contrast, NPP-5 is essential for proper kinetochore localization of NUP133/NPP-15, another NUP107 complex member, whereas recruitment of NUP96/NPP-10C and ELYS/MEL-28 is NPP-5 independent. We found that kinetochore protein NUF2/HIM-10 and Aurora B/AIR-2 kinase are less abundant on mitotic chromatin upon NPP-5 depletion. npp-5 mutants are hypersensitive to anoxia, suggesting that the spindle assembly checkpoint (SAC) is compromised. Indeed, NPP-5 interacts genetically and physically with SAC protein MAD1/MDF-1, whose nuclear envelope accumulation requires NPP-5. Thus our results strengthen the emerging connection between nuclear pore proteins and chromosome segregation.
INTRODUCTION
The nuclear pore complex (NPC) is a macrostructure within the nuclear envelope (NE) composed of multiple copies of ∼30 different proteins called nucleoporins (Nups; Hetzer and Wente, 2009). NPCs are estimated to have a total molecular weight of ∼50–60 MDa (Rout et al., 2000; Cronshaw et al., 2002; Alber et al., 2007), and they serve as the only known transport route of RNA and protein between the nucleus and the cytoplasm. In addition, it is becoming increasingly clear that Nups have a determinant role in regulation of gene expression (Capelson et al., 2010; Kalverda et al., 2010; Strambio-De-Castillia et al., 2010). Nups interact with each other to form subcomplexes in some cases, such as the NUP93/205/188 complex that regulates the NPC permeability barrier and passage of membrane proteins through the NPC (Grandi et al., 1997; Galy et al., 2003; Theerthagiri et al., 2010). Another example is the NUP107 complex, which is composed of nine different Nups (NUP37, NUP43, NUP85, NUP96, NUP107, NUP133, NUP160, SEH1, and SEC13) in vertebrates. The NUP107 complex is biochemically stable, and its purification from the yeast Saccharomyces cerevisiae revealed that it adopts a Y-shaped structure (Siniossoglou et al., 2000). Within this Y-shaped structure Nup84p, the yeast orthologue of NUP107, interacts with Nup133p at the bottom tip of the Y and with Nup145Cp/NUP96 + Sec13p further up the stalk (Lutzmann et al., 2002). Whereas several members of the yeast Nup84p complex, including Nup84p itself (Siniossoglou et al., 1996), are dispensable for growth, studies in vertebrates demonstrated that the NUP107 complex is critical for postmitotic and interphase NPC formation (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003; D’Angelo et al., 2006). Moreover, depletion of the NUP107 complex from Xenopus laevis egg extracts prevents efficient spindle assembly (Orjalo et al., 2006). On entry into mitosis the entire subcomplex localizes to kinetochores (Belgareh et al., 2001; Loiodice et al., 2004). Kinetochores are conserved structures composed of >80 proteins and link spindle microtubules to chromosomes to segregate DNA during anaphase (Cheeseman and Desai, 2008). Kinetochores are composed of three main layers: the inner kinetochore, which interacts with centromeric DNA; the outer kinetochore, which constitutes the surface of interaction with spindle microtubules; and the central kinetochore, which is the region between the inner and outer kinetochore. It was recently demonstrated that the localization of the NUP107 complex to kinetochores is dependent on NDC80 and CENP-F, two outer kinetochore components, and that NUP133 interacts directly with CENP-F (Zuccolo et al., 2007).
The spindle assembly checkpoint (SAC) is a mitotic control mechanism that ensures that chromosomes do not segregate until they are properly bioriented and attached to microtubules (Tanaka, 2010). If a kinetochore is not attached to spindle microtubules, the SAC delays anaphase onset by sequestering CDC20, an activator of the anaphase-promoting complex/cyclosome (APC/C; Fang et al., 1998). The APC/C is an E3 ubiquitin ligase that targets key mitotic substrates for degradation, such as cyclin B and securin. The SAC is also involved in detection of lack of stretch-induced tension between kinetochores on sister chromatids (Maresca and Salmon, 2009; Uchida et al., 2009; Wan et al., 2009). In the absence of tension, Aurora B, a member of the chromosomal passenger complex (CPC) that localizes at the inner centromere, phosphorylates NDC80 and other kinetochore proteins. This reduces the stability of microtubule–kinetochore interactions, thus signaling to the SAC due to an unbound kinetochore situation (Lampson and Cheeseman, 2011). Of interest, it has been shown that SEH1, a member of the NUP107 complex, regulates the centromeric localization of Aurora B and other CPC proteins (Platani et al., 2009). When proper attachment of spindle microtubules to kinetochore occurs, provoking tension between sister chromatids, the SAC is satisfied and mitosis can progress.
In this article, we characterize a NUP107-null mutation in Caenorhabditis elegans. Targeting of single NUP107 subcomplex members by RNA interference (RNAi) in mammalian cells or by immunodepletion from cell-free extracts frequently causes an efficient codepletion of most other subcomplex members (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003), which has prevented assignation of specific functions to individual proteins. However, we find that expression of all C. elegans Nups tested, except for NUP133/NPP-15, is unaffected by the removal of NUP107/NPP-5, making the nematode an attractive model with which to study the function of NUP107 complex components (Table 1). Unexpectedly, we observe that NPCs assemble and function in the absence of NPP-5, whereas NPP-5 localization is highly sensitive to depletion of other NUP107 complex members. We also show that kinetochore assembly is partially inhibited in npp-5 mutants and that NPP-5 binds to and regulates NE accumulation of MAD1/MDF-1, a member of the SAC. Depletion of MDF-1 in npp-5 mutants leads to high synthetic embryonic lethality, suggesting that the SAC is required for cell viability in the absence of NPP-5.
TABLE 1:
Overview of genes involved in this study.
| C. elegans protein | Human orthologue |
|---|---|
| AIR-2 | Aurora B |
| BUB-3 | BUB3 |
| CYB-3 | Cyclin B3 |
| HIM-10 | NUF2 |
| MDF-1 | MAD1 |
| MDF-2 | MAD2 |
| MEL-28 | ELYS |
| MIS-12 | MIS12 |
| NPP-2 | NUP85 |
| NPP-5 | NUP107 |
| NPP-6 | NUP160 |
| NPP-7 | NUP153 |
| NPP-8 | NUP155 |
| NPP-10N | NUP98 |
| NPP-10C | NUP96 |
| NPP-15 | NUP133 |
| NPP-18 | SEH1 |
| NPP-19 | NUP35 |
| NPP-23 | NUP43 |
| SAN-1 | BUBR1 |
RESULTS
NUP107/NPP-5 is required for proper development
To evaluate the implication of NUP107 in animal development, we characterized two mutant alleles of the C. elegans orthologue of NUP107, encoded by the npp-5 gene (standard C. elegans nomenclature is used hereafter; see Table 1). Allele npp-5(tm3039) is a 524–base pair deletion from the first intron to the fourth exon, whereas 1291 base pairs from the fourth exon to the sixth exon are deleted in allele npp-5(ok1966) (Figure 1A). Reverse transcription (RT)–PCR revealed the activation of a cryptic 3′ splice site in npp-5(tm3039), which creates a premature termination codon (PTC) after 35 amino acid residues (Supplemental Figure S1), implying that npp-5(tm3039) is a null allele. The deletion in npp-5(ok1966) similarly induces a PTC downstream of the mutation, but in this case approximately one-third of the open reading frame is intact (Supplemental Figure S1). We raised an antibody against a peptide from the N-terminus of NPP-5. As expected from the molecular lesion, neither Western blotting (Figure 1B) nor immunofluorescence analysis (Figure 1C) revealed a specific signal in npp-5(tm3039) mutants. The absence of signal in npp-5(ok1966) mutants suggested that the PTC renders the mRNA unstable. Both mutants can be propagated as homozygous strains; however, we introduced a balancer chromosome into each strain to avoid selection for suppressor mutations or epigenetic changes. Adult offspring from heterozygous animals consisted of 26.2–26.7% homozygous mutants (npp-5(ok1966), n = 806; tm3039, n = 757), indicating that maternal contribution of NPP-5 enables mutants to complete embryonic and larval development. Analyzing brood size of F1 homozygous mutants revealed a decrease of 21.9% in npp-5(ok1966) and 33.4% in npp-5(tm3039) (Table 2; p < 0.005). For both alleles we observed a low but statistically significant increase in the frequency of lethality among F2 embryos produced by F1 homozygous mutants (Table 2; 5.3–7.3%). F2 larval development was severely compromised in both mutants, with only 8.9–12.0% of the offspring developing into adults at 20°C (Figure 1D). At 25°C no offspring developed into fertile adults, suggesting a higher requirement for NPP-5 when developmental pace is increased (Table 2). Of importance, ectopic expression of green fluorescent protein (GFP)–NPP-5 fully restored embryonic and larval development of npp-5(ok1966) and npp-5(tm3039) mutants (Table 2 and Figure 1D). This observation, together with the identical behavior of the two mutant alleles, confirmed that the observed phenotypes could be attributed to the npp-5 gene. We therefore conclude that NPP-5 plays a critical role in C. elegans development.
FIGURE 1:
npp-5(tm3039) and npp-5(ok1966) are null mutations of NUP107/npp-5. (A) Schematic representation of the C. elegans npp-5 gene and deletion alleles npp-5(ok1966) and npp-5(tm3039). Exons and introns are indicated by boxes and lines, respectively. (B) Western blot analysis of ∼1500 wild-type (WT), npp-5(ok1966), and npp-5(tm3039) embryos probed with anti-NPP-5 and anti–α-tubulin antibodies. NPP-5 appeared with a molecular weight of ∼92 kDa in wild-type embryos only. Asterisk indicates nonspecific cross-reactivity. (C) Wild type and npp-5(tm3039) embryos were fixed and stained with anti–NPP-5 antiserum (red) and monoclonal antibody mAb414 (green). Chromatin was detected using Hoechst 33258 (blue). Boxed regions in the merged panels are shown at higher magnification to the left. Scale bar, 10 μm. (D) Percentage of wild-type, npp-5(ok1966) and npp-5(tm3039) offspring dying during embryogenesis (Emb) or larval development (Lvl) or reaching adulthood (Adult) at 20°C. Error bars, SE of the mean.
TABLE 2:
Development of npp-5(ok1966) and npp-5(tm3039) mutants.
| Genotype | na | Brood size | Embryonic lethality (%) | Larval lethality (%) | Adults (%) |
|---|---|---|---|---|---|
| Wild type, 20ºC | 6 | 319 ± 6 | 0.7 ± 0.2 | 0.3 ± 0.1 | 99.0 ± 0.1 |
| Wild type, 25ºC | 3 | 217 ± 25 | 1.8 ± 1.0 | 0.2 ± 0.2 | 98.0 ± 1.1 |
| ok1966/+, 20ºC | 5 | 273 ± 10† | 2.2 ± 0.5 | 1.0 ± 0.3 | 96.8 ± 0.6 |
| ok1966, 20ºC | 6 | 249 ± 15† | 5.3 ± 1.0* | 82.7 ± 1.8* | 12.0 ± 2.1* |
| tm3039/+, 20ºC | 5 | 257 ± 11† | 1.8 ± 0.7 | 0.1 ± 0.1 | 98.1 ± 0.7 |
| tm3039, 20º | 10 | 212 ± 14† | 7.3 ± 1.4* | 83.8 ± 1.4* | 8.9 ± 1.3* |
| tm3039; GFP-NPP-5, 20ºC | 9 | 259 ± 19 | 1.1 ± 0.7 | 0.8 ± 0.2 | 98.1 ± 0.7 |
| tm3039, 25ºC | 4 | 145 ± 14† | 18.2 ± 4.1* | 81.8 ± 4.1* | 0.0 ± 0.0* |
Heterozygous or first-generation homozygous mutant L4 hermaphrodites were incubated on nematode growth medium plates at indicated temperatures and moved to fresh plates every 8–16 h. Wild-type N2 strain was used as control.
aNumber of founders. For each founder, brood sizes and the percentages of embryonic lethality, arrested or dead larvae, and adults were determined after 0, 24, and 96 h (average ± SE of the means).
†Significant differences by two-tailed t test: different from the wild type at same temperature (p < 0.005).
*Significant differences by chi-square test: different from the wild type at same temperature (p < 0.001).
NPP-5 is dispensable for nuclear protein import
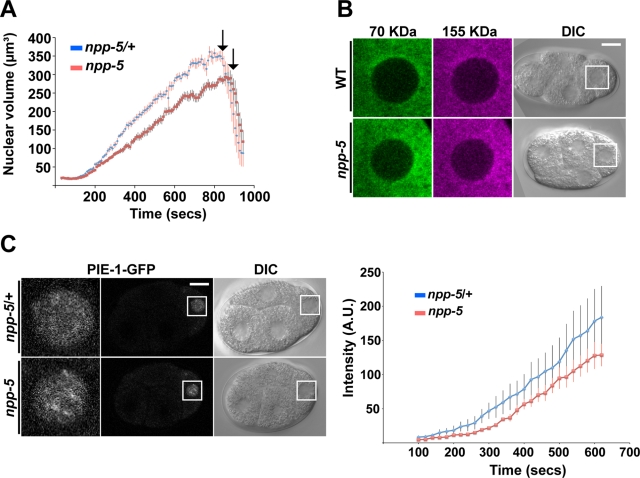
The developmental arrest could potentially reflect defects in NE function, including nucleocytoplasmic transport. To test this, we monitored the growth rate of P1 cell nuclei following the first mitosis in embryos expressing GFP fused to histone H2B/HIS-58 (Figure 2A). Whereas nuclei in embryos produced by heterozygous siblings (which are referred to as control embryos hereafter) grew by 0.56 ± 0.02 µm3/s (time interval 152–696 s after anaphase onset; n = 12), the rate was reduced to 0.42 ± 0.02 µm3/s in F2 mutant embryos (n = 13; 25.6% decrease, p < 2 × 10−5). The final size of P1 nuclei was reduced by 12.8% (p < 0.05). In both control and npp-5(tm3039) embryos P1 nuclei kept growing until entry into mitosis, which occurred after ∼832 s in control and ∼880 s in mutant embryos. Thus absence of NPP-5 reduced nuclear growth and delayed entry into mitosis.
FIGURE 2:
Nuclear protein import occurs in the absence of NPP-5. (A) Size of P1 nuclei was determined by time-lapse microscopy, revealing a significantly slower nuclear growth rate in npp-5(tm3039) embryos (n = 13) compared with npp-5(tm3039)/+ embryos (n = 12). Time is relative to P0 anaphase onset. (B) Gonads of wild-type (n = 14) and npp-5(tm3039) (n = 16) animals were injected with a mixture of 70- (green) and 155-kDa (magenta) dextrans. Exclusion of the dextrans from embryonic nuclei was observed by live confocal microscopy. (C) Nuclear import of PIE-1-GFP into P2 nuclei was observed by time-lapse microscopy (left) and quantified (right). Import in npp-5(tm3039) embryos (n = 10) was comparable to that in control embryos (n = 10). Time is relative to P1 anaphase onset. Error bars, SE of the mean. Scale bars, 10 μm.
Structural changes in NPCs can affect NE permeability. Inert molecules larger that ∼45 kDa are normally unable to cross the NE, but depletion of Nups can increase this permeability barrier. We investigated whether this was also the case in npp-5(tm3039) embryos by injecting the gonad arms of hermaphrodites with a mixture of fluorescent dextrans (Galy et al., 2003). We observed that 70- and 155-kDa dextrans were effectively excluded from nuclei from both control and npp-5(tm3039) embryos, indicating that the permeability barrier was not grossly affected in the absence of NPP-5 (Figure 2B). Next we monitored nuclear accumulation of several protein substrates fused to GFP. PIE-1 is a transcriptional regulator found specifically in germline blastomeres, where it is imported into the nucleus by an unknown transport pathway (Reese et al., 2000). In both control and npp-5(tm3039) four-cell-stage embryos robust nuclear accumulation of PIE-1–GFP was observed in the P2 cell, although time-course analysis indicated a trend for slower import in the absence of NUP107 (Figure 2C and Supplemental Movie S1; p = 0.40). Similarly, stress-induced nuclear import of the transcription factor DAF-16, as well as constitutive nuclear import of lacZ-GFP coupled to the SV40 T-antigen nuclear localization signal in intestine and vulva cells, was observed in npp-5 mutants (Supplemental Figure S2, A and B). We therefore concluded that NPP-5 is dispensable for importin α/β–mediated nuclear protein import, but we cannot rule out that nucleocytoplasmic transport of other substrates may be NPP-5 dependent.
NPC assembly can occur in the absence of NPP-5
The NUP107 complex is essential for postmitotic and interphase NPC assembly (Harel et al., 2003; Walther et al., 2003; D’Angelo et al., 2006), but the relative contribution of each member of the subcomplex has remained largely unknown, partly because small interfering RNA (siRNA) approaches in vertebrate cells lead to a general down-regulation of most or all components. To address the issue of coregulation, embryonic extracts from wild-type and npp-5(tm3039) animals were analyzed by Western blotting using antibodies against NUP96/NPP-10C and NUP133/NPP-15 from the NUP107 complex, as well as NUP35/NPP-19, NUP98/NPP-10N, and NUP153/NPP-7. Except for NPP-15, all these Nups were present at normal levels in npp-5(tm3039) embryos (Figure 3A and Supplemental Figure S3). Moreover, we previously showed that knockdown of NPP-5 does not affect expression of ELYS/MEL-28 in C. elegans (Galy et al., 2006). Conversely, RNAi against npp-19 (Rodenas et al., 2009), NUP155/npp-8 (Franz et al., 2005), or mel-28 (Galy et al., 2006) was shown not to affect expression of NPP-5. The independent expression of individual Nups tested so far underlines the usefulness of C. elegans as genetic system to dissect the function of NPC components (Gorjanacz et al., 2007). Affinity-purified antibodies against NPP-15 gave rise to two bands of the expected size (∼128 kDa) in wild-type extracts but, intriguingly, only a single band in npp-5(tm3039) (Figure 3A). Comparing extracts from wild-type embryos to extracts from embryos produced by animals heterozygous for a npp-15 deletion allele (ok1954) showed that both bands correspond to NPP-15 protein (Supplemental Figure S3F).
FIGURE 3:
Expression and localization of most Nups are NPP-5 independent. (A) Western blot analysis of embryonic extracts showed similar expression levels of NPP-7, NPP-10N, NPP-10C, and NPP-19 in npp-5(tm3039) embryos compared with the wild type. In contrast, NPP-15 appeared as a duplet in the wild type but not in the mutant. (B–D) Wild type, npp-5(tm3039) (B, D), and npp-5(ok1966) (C) embryos were fixed and stained with serum against NPP-10C (B), MEL-28 (C), or NPP-15 (D) (red) and mAb414 (green). Chromatin was detected using Hoechst 33258 (blue). (E, F) Expression of GFP-NPP-23 and GFP-NPP-2/mCherry-HIS-58 was analyzed by live microscopy. Whereas depletion of NPP-5 inhibited recruitment of GFP-NPP-23 (n ≥ 24 for each treatment; E), GFP-NPP-2 was unaffected (F; n ≥ 8 for each treatment). Boxed regions in the merged panels are shown at higher magnification to the left. Scale bars, 10 μm.
We next analyzed whether the lack of NPP-5 prevented proper localization of components of the NUP107 complex. MEL-28 and NPP-10C localized at the nuclear periphery in interphase and to kinetochores during mitosis in both wild-type and npp-5(tm3039) embryos (Figure 3, B and C). Moreover, the monoclonal antibody mAb414, a general NPC marker, showed normal staining in the absence of NPP-5. Nuclear rim accumulation of NPP-15 was similarly unaffected by NPP-5 depletion (Figure 3D). However, recruitment to kinetochores in mitosis was detected only in wild-type embryos but not in npp-5(tm3039) embryos. To investigate additional members of the NUP107 complex, we made transgenic strains expressing GFP fused to either NUP43/NPP-23 (D’Angelo et al., 2009) or NUP85/NPP-2, neither of which had been visualized previously in C. elegans. As expected, both Nups localized to NPCs and kinetochores in the wild type (Figure 3, E and F; and data not shown). Whereas nuclear rim and kinetochore recruitment of GFP-NPP-23 was diminished in embryos depleted for NPP-5 by RNAi (Figure 3E), GFP-NPP-2 was present at normal levels at the nuclear periphery in npp-5(tm3039) animals (Figure 3F). Our GFP-NPP-2 reporter was only expressed in postembryonic cells, which precluded a detailed analysis of mitosis in second-generation mutants since divisions have largely ceased in these animals. In first-generation mutants at the larval L2–L3 stage GFP-NPP-2 localized to kinetochores during division of seam cells (data not shown), but we cannot exclude the presence of low amounts of maternally contributed NPP-5 protein in these animals. We conclude that in the absence of NPP-5 only localization of NPP-23 and NPP-15 is affected, suggesting that NPC assembly and stability of the NUP107 complex are largely NPP-5 independent.
Localization of NPP-5 depends on most other subcomplex members
Having observed that most Nups behave normally in NPP-5-depleted animals, we asked the reciprocal question: which members of the NUP107 complex are required to properly localize NPP-5? Using the rescuing GFP-NPP-5 strain (Table 2), we analyzed NPP-5 dynamics in control and RNAi-treated embryos. On depletion of NPP-2, the signal of GFP-NPP-5 was reduced both at the nuclear periphery and at kinetochores, whereas RNAi against npp-10C and NUP160/npp-6 completely abolished NPP-5 recruitment (Figure 4 and Supplemental Movies S2–S5). Targeting npp-10C caused, moreover, severe chromosome segregation defects in mitosis. Because NPP-10N and NPP-10C are translated as a precursor polypeptide from a common mRNA, RNAi efficiently knocks down expression of both Nups (Galy et al., 2003). Thus these phenotypes might contribute to the lack of NPP-10N, NPP-10C, or both. Analyzing endogenous NPP-5 by immunofluorescence confirmed the dependence on NPP-2, NPP-10N/C, NPP-6, and MEL-28 for correct localization of NPP-5 (Supplemental Figure S4). In contrast, we found that RNAi against SEH1/npp-18 did not impede recruitment of NPP-5. Together these results suggest that whereas the NUP107 complex is stable in the absence of NPP-5, NUP107/NPP-5 itself is very sensitive to perturbations of the NPC subcomplex.
FIGURE 4:
NPP-5 localization is sensitive to perturbations of other NUP107 complex members. Still images from time-lapse confocal microscopy of embryos expressing GFP-NPP-5 (green) and mCherry-HIS-58 (magenta). RNAi against npp-2, npp-10, or npp-6 inhibited proper localization of GFP-NPP-5 during interphase (top rows) and metaphase (bottom rows; n ≥ 8 for each treatment). Boxed regions on the right are shown at higher magnification to the left. Scale bar, 10 μm.
NPP-5 is required for proper kinetochore assembly and Aurora B/AIR-2 recruitment
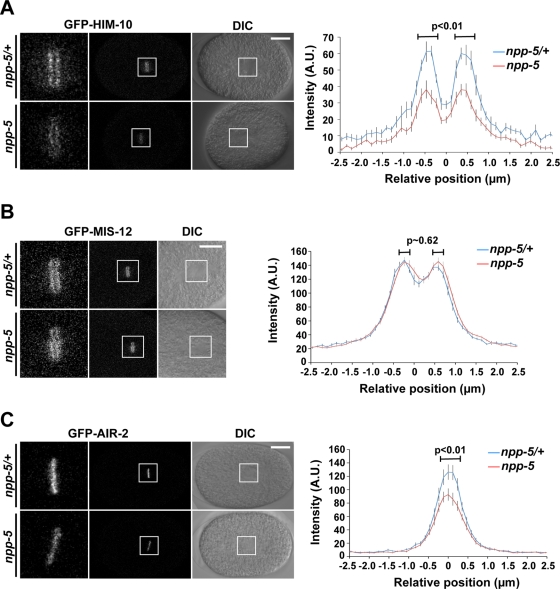
The mitotic localization of the NUP107 complex to kinetochores led to the hypothesis that Nups could be directly involved in chromosome segregation (Belgareh et al., 2001). In support of this, HeLa cells with reduced NUP107 and SEH1 loading onto kinetochores showed increase frequency of misaligned chromosomes during metaphase (Zuccolo et al., 2007). To test whether depletion of NPP-5 affects kinetochore assembly in C. elegans, we monitored the behavior of outer kinetochore proteins NUF2/HIM-10 and MIS12/MIS-12, which are members of the NDC80 and MIS12 complexes, respectively. Quantification of GFP-HIM-10 at kinetochores of metaphase chromosomes revealed a 39% reduction in npp-5(tm3039) embryos (Figure 5A and Supplemental Movie S6; p < 0.01), whereas GFP-MIS-12 localization was NPP-5 independent (Figure 5B and Supplemental Movie S7; p ≈ 0.62). These results suggest that NPP-5 is required for specific aspects of outer kinetochore assembly.
FIGURE 5:
NPP-5 is required for efficient localization of HIM-10 and AIR-2. Still images from time-lapse confocal microscopy of control (npp-5(tm3039)/+) and npp-5(tm3039) embryos expressing GFP-HIM-10 (A), GFP-MIS-12 (B), or GFP-AIR-2 (C). Boxed regions (middle) are shown at higher magnification to the left. Scale bars, 10 μm. Graphs on the right represent mean fluorescence intensities measured in 5 × 2.3 μm2 rectangles perpendicular to the metaphase plates. Position is relative to the center of metaphase chromosomes. Probability values (p) from two-tailed t tests are shown. Error bars, SE of the mean. n ≥ 15 in all experiments.
The fidelity of bipolar microtubule attachment to kinetochores is monitored by the CPC, which includes INCENP, Survivin, Borealin, and the protein kinase Aurora B. We tested whether CPC recruitment is NPP-5 dependent by measuring GFP-Aurora B/AIR-2 levels on mitotic chromatin. This revealed a 34% reduction of chromatin-associated GFP-AIR-2 during metaphase in npp-5(tm3039) embryos as compared with control embryos (Figure 5C and Supplemental Movie S8; p < 0.01). Thus we concluded that NPP-5 is required for proper localization of specific kinetochore and CPC components.
Spindle assembly checkpoint protein MAD1/MDF-1 accumulates at kinetochores in the absence of NPP-5
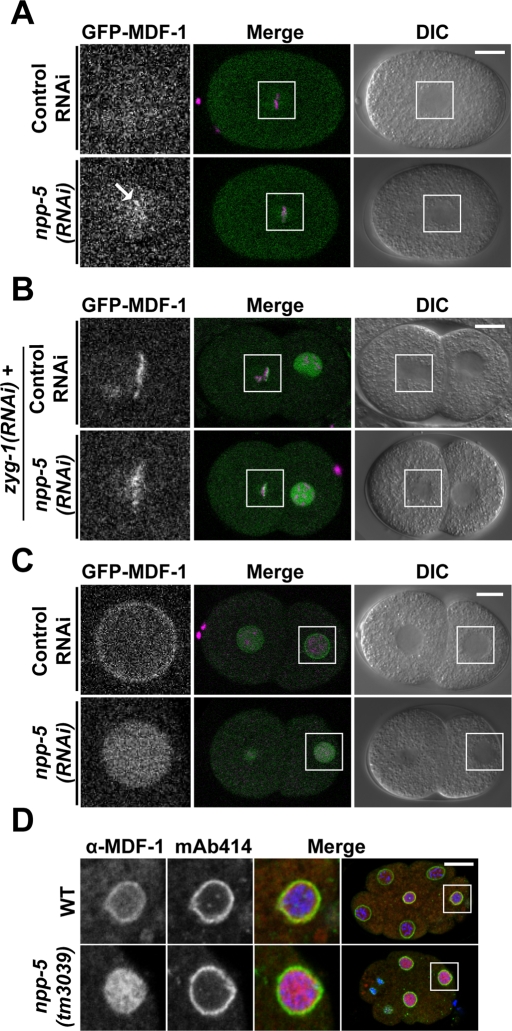
Unattached kinetochores due either to defective kinetochore assembly or Aurora B–mediated correction of syntelic and merotelic microtubule (MT)–kinetochore attachments are detected by the SAC (Tanaka, 2010; Lampson and Cheeseman, 2011). On the basis of our observation that HIM-10 and AIR-2 localization is partially compromised in npp-5(tm3039) embryos, we addressed two questions: Does the absence of NPP-5 from kinetochores trigger a signal to the SAC? Is the SAC functional in cells lacking NPP-5? To answer the first question, we investigated whether SAC proteins MAD1/MDF-1 and MAD2/MDF-2 localize to mitotic chromatin in npp-5(tm3039) embryos. In contrast to the situation in most vertebrate cells, C. elegans MDF-1 and MDF-2 accumulate on chromatin only if cells are stressed during mitosis (Yamamoto et al., 2008; Essex et al., 2009). Live recordings of npp-5(RNAi) embryos revealed faint but reproducible accumulation of GFP-MDF-1 on chromatin during metaphase, suggesting that absence of NPP-5 is detected by the SAC (Figure 6A and Supplemental Movie S9). GFP-MDF-2 recruitment was not observed, however, possibly due to a strong fluorescence signal from soluble protein (Supplemental Figure S5A). We next induced a situation of massive monotelic and syntelic MT–kinetochore attachment by preventing centrosome duplication through RNAi-mediated loss of ZYG-1 protein (Yamamoto et al., 2008; Essex et al., 2009). As previously shown, SAC proteins strongly accumulated on mitotic chromatin of two-cell-stage zyg-1(RNAi) embryos (Figure 6B and Supplemental Figure S5B). Because depletion of NPP-5 did not interfere with localization of GFP-MDF-1 or GFP-MDF-2 in zyg-1(RNAi) embryos (Figure 6B and Supplemental Figure S5B), we conclude that NPP-5 is not required for robust SAC signaling upon strong stimuli, such as monopolar spindle formation. Similarly, the NUP107 complex is not required for SAC function in cell-free Xenopus extracts (Orjalo et al., 2006).
FIGURE 6:
NPC localization of spindle assembly checkpoint protein MDF-1 depends on NPP-5. (A–C) Still images from time-lapse confocal microscopy of embryos expressing GFP-MDF-1 (green) and mCherry-HIS-58 (magenta). (A) Depletion of NPP-5 induces recruitment of GFP-MDF-1 to metaphase chromosomes (arrow; n = 10). (B) GFP-MDF-1 accumulation on chromosomes attached to monopolar spindles in zyg-1(RNAi) embryos is unaffected by depletion of NPP-5 (n = 3). (C) Localization of GFP-MDF-1 to NPCs in interphase is abolished in the absence of NPP-5 (n = 14). (D) Wild-type and npp-5(tm3039) embryos were fixed and stained with anti–MDF-1 antiserum (red) and mAb414 (green). Chromatin was detected using Hoechst 33258 (blue). Boxed regions in the merged panels are shown at higher magnification to the left. Scale bars, 10 μm.
MDF-1 localizes to NPCs through NPP-5
Initially described in budding yeast, Mad1p and to some degree also Mad2p accumulate at NPCs in several cell types during interphase (Campbell et al., 2001; Iouk et al., 2002). In interphase cells of C. elegans embryos, GFP-MDF-1 also showed enhanced signal at the NE in addition to diffuse nucleoplasmic staining (Figure 6C). It is striking that the NE accumulation of GFP-MDF-1 was absent in NPP-5–depleted embryos, whereas the intranuclear signal was normal (Figure 6C and Supplemental Movie S9). Immunofluorescence analysis of endogenous MDF-1 in control and npp-5(tm3039) embryos confirmed these observations (Figure 6D), demonstrating that NPP-5 is strictly required to localize MDF-1 to the NE.
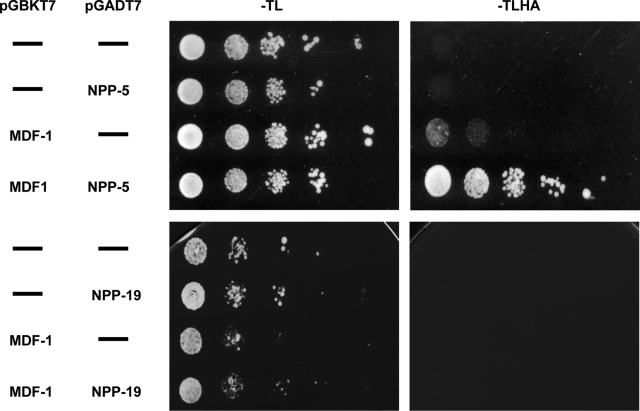
To investigate whether MDF-1 accumulates at NPCs via a physical interaction with NPP-5, we expressed the two proteins in budding yeast, fused to either the Gal4 activation domain or the Gal4 DNA-binding domain. Only when MDF-1 and NPP-5 fusion proteins were expressed simultaneously did the yeast grow on media lacking the selectable markers histidine and adenine (Figure 7, top), indicating that the two proteins interact directly. Because budding yeast Mad1p and Nup53p have been demonstrated to interact physically (Scott et al., 2005), we also tested whether this was the case for the C. elegans orthologues MDF-1 and NPP-19. However, we did not observe any interaction in the yeast-two-hybrid assay (Figure 7, bottom), which suggests that MAD1 is anchored at the NPC in a species-specific manner.
FIGURE 7:
NPP-5 physically interacts with MDF-1 in yeast-two-hybrid assays. Full-length npp-5 and mdf-1 cDNAs were cloned into prey and bait vectors, respectively, and used to transform yeast cells. Growth on selective (–Trp-Leu-His-Ade) medium was only supported when the two genes were present together. No interaction was observed between NPP-19 and MDF-1.
NPP-5–deficient embryos are hypersensitive to MDF-1 perturbation
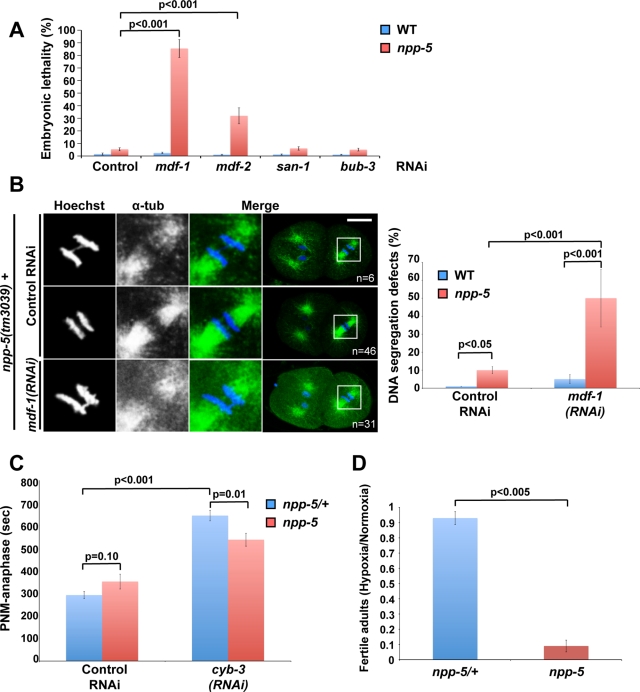
Our observation that NPP-5 is partially responsible for MDF-1 localization prompted us to investigate whether they interact genetically. Indeed, we found that whereas depletion of the proteins singly caused only 3–5% embryonic lethality, RNAi against mdf-1 in npp-5(tm3039) animals led to 85 ± 7% embryonic lethality (Figure 8A; p < 0.001). MDF-2, which forms a complex with MDF-1, showed an intermediate degree of synthetic embryonic lethality with NPP-5 (32 ± 6%; p < 0.001), whereas depletion of SAC protein BUBR1/SAN-1 or BUB3/BUB-3 did not enhance lethality in npp-5(tm3039) embryos (p > 0.10). Thus NPP-5 displays a synthetic lethality phenotype specifically with the MDF-1/MDF-2 branch of the SAC.
FIGURE 8:
NPP-5 mutants are hypersensitive to MDF-1 depletion and anoxic stress. (A) Embryonic lethality was measured following RNAi against different members of the SAC in wild-type and npp-5(tm3039) animals (n > 600 embryos in all experiments). (B) Wild-type and npp-5(tm3039) animals treated with control or MDF-1 RNAi were fixed and stained with anti–α-tubulin (green). Chromatin was detected using Hoechst 33258 (blue). Boxed regions in the merged panels are shown at higher magnification to the left. Percentages of embryos presenting DNA segregation defects are indicated in the graph, demonstrating a synthetic phenotype in embryos simultaneously depleted for NPP-5 and MDF-1. (C) Time from pronuclear meeting (PNM) to anaphase was measured following incubation on control or cyb-3 RNAi plates for 30 h (n ≥ 5 for all treatments). (D) Embryos from npp-5(tm3039)/+ and npp-5(tm3039) animals were exposed to hypoxia for 21 h. After recovery under normoxic conditions development into fertile adults was determined, revealing a severe effect in npp-5(tm3039) mutants (n > 600 embryos in all experiments). Probability values (p) are shown from chi-square tests (A, B), two-tailed t test (C), and Wilcoxon rank sum test (D). Error bars, SE of the mean.
The implication of NPP-5 in kinetochore assembly and AIR-2 localization suggested that chromatin segregation might be impaired in the absence of NPP-5. Moreover, the synthetic interaction of NPP-5 with MDF-1 and MDF-2 could reflect that these SAC proteins are required to prevent chromatin segregation defects in npp-5(tm3039) embryos by inducing an anaphase delay. Indeed, immunofluorescence analysis of npp-5(tm3039) mutants revealed chromatin bridges in 10% of the embryos (p < 0.05), a phenotype never observed in control embryos (Figure 8B). RNAi against mdf-1 caused 5% chromatin bridges in control embryos, whereas a dramatic increase to 49% (p < 0.001) was observed upon depletion of MDF-1 from npp-5(tm3039) embryos. Thus MDF-1 is required to prevent chromosome missegregation in the absence of NPP-5.
It was recently found that loss of Cyclin B3/CYB-3 induces a strong SAC-dependent mitotic arrest (Deyter et al., 2010), which we hypothesized might also require NPP-5. Because chromatin segregation is virtually abolished upon depletion of CYB-3 (Deyter et al., 2010; our data not shown), we used other events to monitor mitotic progression: we defined “anaphase” on the basis of centrosome behavior and cleavage furrow ingression (∼80 s after anaphase onset in control embryos). Compared to control embryos, time from pronuclear meeting (prophase) to anaphase was increased by 115% in embryos depleted for CYB-3 (Figure 8C; p < 0.001). Timing was unaffected in npp-5 mutants incubated on control RNAi plates (p = 0.10), but the cyb-3(RNAi)–induced mitotic delay was partially alleviated by removal of NPP-5 (17%; p = 0.01).
As an alternative way to test whether NPP-5 is required for proper SAC function, we examined response to oxygen deprivation. C. elegans embryos have a remarkable capacity to withstand anoxic stress. Under conditions of low oxygen, embryos arrest development and enter a stage known as suspended animation. This behavior is SAC dependent, implying a metaphase arrest, although cells may also be blocked in prophase (Nystul et al., 2003; Hajeri et al., 2010). To investigate whether embryos lacking NPP-5 are fully competent to enter suspended animation, we placed control and npp-5(tm3039) embryos under hypoxic conditions for 21 h before returning them to normal atmosphere. It is striking that, whereas the hypoxia stress only modestly decreased the frequency of control embryos completing embryogenesis and developing into fertile adults (2.4 and 7.5% decrease, respectively), the frequency of viable npp-5(tm3039) embryos decreased 13.3%, and only 1.1% of npp-5(tm3039) embryos developed into fertile adults following the hypoxia treatment (86.9% decrease; Figure 8D and Supplemental Table S1). This suggests that loss of NPP-5 sensitizes cells so they cannot withstand stress that otherwise would have been tolerated via SAC-induced mitotic arrest. From these results we conclude that NPP-5 is required for efficient cell cycle arrest under stress induced by either perturbation of cell cycle regulators or hypoxia.
DISCUSSION
Several studies determined that the NUP107 complex plays a crucial role in postmitotic and interphase NPC formation (Harel et al., 2003; Walther et al., 2003; D’Angelo et al., 2006). Deeper understanding of the initial steps of NPC assembly has, however, been hindered by technical difficulties preventing the functional dissection of the NUP107 complex.
We provide here the first in vivo characterization of a metazoan NUP107 mutant. Moreover, we demonstrate that C. elegans NUP43/NPP-23, NUP85/NPP-2, and NUP133/NPP-15 localize at the nuclear periphery in interphase and at kinetochores during mitosis, thus behaving like bona fide NUP107 complex components. Previous depletion of NUP107 with siRNAs in HeLa cells or by immunoprecipitation from Xenopus extracts caused a codepletion of several NUP107 complex members, hampering a detailed understanding of how this ∼720-kDa complex functions (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003). However, we show that the majority of Nups are expressed at normal levels in a C. elegans NUP107-null mutant, npp-5(tm3039), paving the way to study the role of individual NUP107 complex members.
It is striking that, although most npp-5 mutant animals do not complete development to adulthood, NPCs are formed, and neither nuclear protein import nor nuclear exclusion of soluble cytoplasmic content is grossly affected by the absence of NPP-5. The high lethality, in particular at 25°C, led us to conclude that npp-5 is an essential gene in C. elegans, as also reported for Schizosaccharomyces pombe (Bai et al., 2004) and Drosophila melanogaster (Katsani et al., 2008). In contrast, deletion of the orthologous Nup84 gene from S. cerevisiae or Aspergillus nidulans causes milder, temperature-sensitive growth defects (Siniossoglou et al., 1996; Osmani et al., 2006).
Dissection of the Y-shaped NUP107 complex
Among Nups that belong to the NUP107 complex, we found that only NPP-23 and NPP-15 depend on NPP-5 for their correct localization. Recruitment of NPP-23 is diminished both at kinetochores during mitosis and at the NPC during interphase in the absence of NPP-5. The position of NUP43/NPP-23 within the Y-shaped NUP107 complex is unknown (Lutzmann et al., 2002), but on the basis of our observation that other Nups are still at the NPC in the absence of NPP-5, we suggest that NPP-23 and NPP-5 may interact directly. On the other hand, NPP-15 localizes normally at NPCs during interphase in npp-5(tm3039) embryos but does not localize at kinetochores in mitosis. The observation that NPC localization of endogenous NPP-15 is NPP-5 independent was unexpected since GFP-tagged human NUP133 mutated for the residues that mediate its interaction with NUP107 in vitro does not accumulate at the nuclear periphery in HeLa cells (Boehmer et al., 2008). We propose that the addition of a GFP tag to NUP133 renders it more dependable on NUP107 for its NPC incorporation. Indeed, we observe that also C. elegans GFP-NPP-15 is dependent on NPP-5 for its localization to the NPC (Supplemental Figure S6). The position of NUP133 at the base of the Y-shaped complex “below” NUP107 (Lutzmann et al., 2002) implies that removal of NUP107 should release NUP133 from the rest of the NUP107 complex. Thus, how does NPP-15 localize to the NPC in the absence of NPP-5? One possibility is that NPP-15 may interact physically with other Nups. In fact, it was recently suggested that the NUP107 complex is arranged in a head-to-tail manner within the NPC, through direct contacts between the NUP133 and Nup120p/NUP160, forming closed rings of eight NUP107 complexes (Seo et al., 2009). Moreover, NUP133 contains an ALPS motif capable of sensing membrane curvature (Drin et al., 2007), which plays a crucial role for NUP133 recruitment during de novo interphase NPC assembly (Doucet et al., 2010). On the basis of these observations we propose that NUP133/NPP-15 may use several mechanisms for its NPC incorporation and that binding to NUP107/NPP-5 is only required for kinetochore targeting. It is interesting to note that Western blot analysis of npp-5(tm3039) extracts reveals only a single NPP-15 band, whereas two protein species are detected in wild-type extracts. We do not have information on the nature of the doublet in the wild type, but the differential behavior of NPP-15 in terms of NPC and kinetochore localization in the absence of NPP-5 leads us to speculate that the two NPP-15 bands represents different isoforms of NPP-15—one localizing to kinetochores during mitosis and another to NPCs during interphase in C. elegans. Without NPP-5 the former NPP-15 isoform becomes unstable, whereas the latter is stabilized through interactions with other Nups and/or pore membrane components. We found that depletion of other NUP107 complex proteins, such as NPP-2, NPP-10C, and NPP-6, efficiently abrogated NPP-5 recruitment and NPC assembly, suggesting that these proteins are critical for NUP107 complex stability and postmitotic NE reformation. An important conclusion from our work is therefore that the requirement for the NUP107 complex during NPC assembly is independent of NUP107/NPP-5 itself.
NPP-5 acts at multiple steps of mitosis
Although it is well established that NUP107/NPP-5 localizes to kinetochores in vertebrate and nematode cells (Belgareh et al., 2001; Loiodice et al., 2004; Franz et al., 2005) but not in Drosophila (Katsani et al., 2008), little is known about the functional implications. By depleting NUF2 from HeLa cells, Zuccolo and colleagues concluded that the NDC80 complex plays a major role in the recruitment of the NUP107 complex to kinetochores (Zuccolo et al., 2007). Our observation that kinetochore accumulation of NUF2/HIM-10 is reduced in embryos lacking NPP-5 suggests that proper localization of the two protein complexes is interdependent. This is in contrast to the experiments of Zuccolo and colleagues, in which inhibition of NUP107 complex recruitment to kinetochores through SEH1 siRNA treatment did not affect localization of HEC1, which forms heterodimers with NUF2 (Zuccolo et al., 2007). However, since detectable levels of SEH1 were still present upon siRNA treatment, we propose that residual NUP107 complex activity at kinetochores was sufficient for the HEC1 accumulation observed by Zuccolo and coworkers, whereas complete absence of NPP-5 causes a ∼40% decrease in HIM-10 as reported here. Of importance, MIS-12 localized normally in npp-5(tm3039) embryos, which suggest that NPP-5 acts at specific steps of kinetochore assembly.
Our observation that 10% of dividing npp-5(tm3039) embryos contained lagging chromosomes during mitosis is compatible with reduced correction of syntelic microtubule–kinetochore attachments by Aurora B/AIR-2 (Lampson and Cheeseman, 2011), as also proposed for SEH1-depleted HeLa cells (Platani et al., 2009). Moreover, alterations in kinetochore structure upon NPP-5 depletion, including reduced HIM-10 accumulation, are likely to be detected by the SAC. Indeed, we observed accumulation of GFP-MAD1/MDF-1 on mitotic chromosomes in embryos depleted for NPP-5, as also reported upon interfering with kinetochore localization of the entire NUP107 complex though depletion of SEH1 in HeLa cells (Zuccolo et al., 2007). Finally, npp-5(tm3039)-mutant embryos were hypersensitive to depletion of MDF-1, resulting in 85% embryonic lethality and lagging chromosomes in 49% of dividing cells, which further supports the possibility that the SAC is responsible for detection and correction of mitotic spindle defects caused by the absence of NPP-5.
Our experiments unexpectedly revealed that MDF-1 depends strictly on NPP-5 in order to accumulate at the nuclear periphery during interphase. MAD1 localization to NPCs was previously described in different organisms, but a clear functional implication of this behavior is lacking. In S. cerevisiae the interaction with the NPC is mediated via Nup53p and Mlp1p/Mlp2p (Scott et al., 2005), whereas in HeLa cells localization of MAD1 to the NPC is dependent on the Mlp1p/Mlp2p homologue TPR (Lee et al., 2008) and NUP153 (Lussi et al., 2010). Yeast-two-hybrid experiments presented here demonstrate that C. elegans MDF-1 interacts directly with NPP-5 but not with NUP35/NPP-19. Thus our data show that accumulation of MAD1 at NPCs is conserved and that different MAD1–Nup interactions may have arisen during evolution.
Cells are more dependent on the SAC in situations of stress. Our observation that suspended animation and delay in mitotic progression were compromised in the absence of NPP-5, combined with the genetic and physical interaction between NPP-5 and MDF-1, suggests that NPP-5 may play an active role in control of chromosome segregation and cell cycle progression. We propose that NUP107/NPP-5 may serve as binding site for MAD1/MDF-1 both at the NPC and at kinetochores and that the two proteins may even translocate together at the entry into mitosis. However, future experiments are required to analyze this interaction in further detail.
MATERIALS AND METHODS
Nematode strains and transgenesis
The wild-type strain used was the C. elegans Bristol strain N2. BN51 bqIs51[Ppie-1::gfp::npp-5] and XA3545 qaIs3545[Ppie-1::gfp::him-10] were obtained by microparticle bombardment (Praitis et al., 2001) of DP38 using either plasmid pUP1 or plasmids pPAG27 and pDP#MM051, respectively. BN128 bqEx128[Pnpp-2::gfp::npp-2; Plmn-1::mCherry::his-58] was obtained by microinjection of plasmids pBN1 and pBN29 into N2. BN150 bqSi150[Phsp-16.41::gfp::C09G9.2a] and BN168 bqSi168[Phsp-16.41::gfp::npp-15] were obtained by MosSCI transformation (Frokjaer-Jensen et al., 2008) of EG4322 with plasmids pBN27 and pBN23, respectively. BN68 bqIs51[Ppie-1::gfp::npp-5]; ltIs37[Ppie-1::mCherry::his-58] was made by crossing BN51 and OD57 (McNally et al., 2006). npp-5(tm3039) was provided by Shohei Mitani of the Japanese National Bioresource Project (Tokyo, Japan) and backcrossed to the wild type six times before balancing to obtain strain BN40. npp-5(ok1966) was provided by the International C. elegans Gene Knockout Consortium (http://celeganskoconsortium.omrf.org/) and backcrossed to the wild type six times to obtain the homozygous strain BN28 and the balanced strain BN85. Subsequently, BN40 was crossed with the following strains: TJ356 (Henderson and Johnson, 2001) to generate BN22 npp-5(tm3039)/mIn1; zIs356[daf-16::gfp]; BN68 to generate BN69 npp-5(tm3039)/mIn1; bqIs51[Ppie-1::gfp::npp-5]; ltIs37[Ppie-1::mCherry::his-58]; JH1327 (Reese et al., 2000) to generate BN73 npp-5(tm3039)/mIn1; axIs[Ppie-1::pie-1::gfp]; XA3501 (Askjaer et al., 2002) to generate BN106 npp-5(tm3039)/mIn1; ruIs32[Ppie-1::gfp::his-58]; XA3545 to generate BN114 npp-5(tm3039)/mIn1; qaIs3545[Ppie-1::gfp::him-10]; BN128 to generate BN133 npp-5(tm3039)/mIn1; bqEx128[Pnpp-2::gfp::npp-2; Plmn-1::mCherry::his-58]; OD27 (CGC) to generate BN153 npp-5(tm3039)/mIn1; ltIs14[Ppie-1::gfp::air-2]; and OD8 (Cheeseman et al., 2004) to generate BN159 npp-5(tm3039)/mIn1; ltIs4[Ppie-1::gfp::mis-12]. Similarly, BN28 was crossed with PS3808 (Gupta and Sternberg, 2002) to generate BN59 npp-5(ok1966); syIs80[Plin-11::nls::gfp::lacZ]. npp-15(ok1954) was provided by the International C. elegans Gene Knockout Consortium and backcrossed to the wild type six times before balancing to obtain strain BN126. RQ244 mdf-1(gk2) V; jzIs1[Ppie-1::gfp:: mdf-1]; ltIs37[Ppie-1::mCherry::his-58] and OD110 ltIs52[Ppie-1::gfp::mdf-2] ltIs37[Ppie-1::mCherry::his-58] were as described (Yamamoto et al., 2008; Essex et al., 2009). For further details on strains refer to Supplemental Table S2. All strains were cultured using standard C. elegans methods (Stiernagle, 2006).
Plasmids and RNAi
Plasmid pQE30-NPP-15 (amino acids 739–1024) for expression of hexahistidine-tagged NPP-15 antigen was generated by PCR amplification of C. elegans genomic DNA (primers B153 + B154). Plasmid pUP1 for expression of GFP-NPP-5 was made by insertion of unc-119 derived from plasmid pDP#MM051 (Maduro and Pilgrim, 1995) into pPAG1 (Franz et al., 2005). Plasmid pPAG27 for expression of GFP-HIM-10 was constructed by replacing the lmn-1 gene of plasmid pPAG4 (Galy et al., 2003) with a PCR-amplified him-10 sequence (primers H556 + H557). Plasmid pBN29 for expression of GFP-NPP-2 was generated by cloning the npp-2 gene (primers B069 + B200 + B071 + B072) into plasmid pBN8, a derivative of pCFJ151 (Frokjaer-Jensen et al., 2008) containing a longer polylinker. pBN29 includes 1993 base pairs upstream of the start codon and 560 base pairs downstream of the stop codon, as well as GFP inserted into a PCR-engineered BsrGI site immediately after the start codon. Plasmids pBN23 and pBN27 for expression of GFP-NPP-15 and GFP-NPP-23 were generated by cloning npp-15 (primers B236 + B237) and C09G9.2a (primers B248 + B249) into plasmid pBN16, respectively. Plasmid pBN16 for single-copy integration of heat shock–inducible transgenes into the C. elegans genome was made by insertion of the hsp-16.41 promoter (primers B216 + B232), a polylinker, and the unc-54 3′ untranslated region (UTR; B233 + B217) into pBN8. Plasmid pBN1 for expression of mCherry-HisH2B in somatic cells contains the lmn-1 promoter (5088 base pairs) in front of a mCherry-his-58 fusion gene and the pie-1 3′UTR (1822 base pairs).
Plasmids pGADT7-npp-5 and pAD-npp-19 encoding the yeast Gal4 activation domain fused to either C. elegans NPP-5 or NPP-19 were constructed by RT-PCR amplification with primers B364 + B366 and primers H624 + H633, respectively. Plasmid pGBKT7-mdf-1 encoding the yeast Gal4 DNA–binding domain fused to C. elegans MDF-1 was constructed by PCR amplification of mdf-1 cDNA from plasmid pACT-Ptac-GST (Watanabe et al., 2008), using primers B367 + B368 into pGBKT7 (Clontech, Mountain View, CA).
For primer sequences refer to Supplemental Table S3. All constructions were verified by sequencing.
RNAi constructs were described previously (Galy et al., 2003; Kamath et al., 2003). The empty pPD129.36 vector was used as negative control. RNAi experiments were performed by feeding at 20°C for ∼36 h unless otherwise specified (Galy et al., 2003).
Production and purification of antibodies
NPP-5 and NPP-15 antibodies were raised in rabbits against a synthetic NPP-5 peptide (amino acids 11–26) and recombinant NPP-15 (amino acids 738–1023), respectively. Antibodies were affinity purified from sera using either an Affi-Gel 15 (Bio-Rad, Hercules, CA) column coupled with the synthetic NPP-5 peptide or Immobilon-P (Millipore, Billerica, MA) membrane coated with the NPP-15 antigen. Bound antibodies were eluted with 0.1 M glycine (pH 2.5) and immediately neutralized with 1 M Tris (pH 8.0).
Dextran microinjection and live embryo imaging
Fluorescein isothiocyanate–labeled, 70-kDa dextran (FD70S; Sigma-Aldrich, St. Louis, MO) and tetramethylrhodamine isothiocyanate–labeled, 155-kDa dextran (T1287; Sigma-Aldrich) were purified using a Nanosep 10K centrifugal device (OD010C33; Nanosep, Lund, Sweden) until a final concentration of 2 mg/ml in phosphate-buffered saline (PBS). A 1:1 mixture of the two dextrans was injected into the gonads of N2 and BN40 animals, followed by incubation at 20°C for 5 h before dissection.
Fluorescent reporters driven by the hsp-16.41 promoter were induced by a 32.7°C heat shock for 15 min, followed by 5-h recovery at 20°C prior to observation.
Embryos were mounted in M9 buffer (86 mM NaCl, 22 mM KH2PO4, 34 mM Na2HPO4, 1 mM MgSO4) between a coverslip and a 2% agarose pad. Epifluorescence and transmitted light were recorded at 22–24°C with a Leica confocal TCS SP2 microscope through an HCX PL APO 63×/1.4 objective (Leica, Wetzlar, Germany). Images were captured using integrated Leica software and processed with ImageJ (National Institutes of Health, Bethesda, MD) and Adobe Photoshop (Adobe, San Jose, CA). The laser intensity was adjusted so that no effect on development was observed. Images were collected at 20-s intervals for a total of 20–40 min, except for Figure 5B, where images were collected at 5-s intervals.
Immunofluorescence
Gravid hermaphrodites were dissected in 3 μl of M9 buffer directly on poly-l-lysine–coated glass slides and covered with 12 × 12 mm coverslips. To crack the eggshells, slides were transferred immediately to metal plates on top of dry ice. After 15 min, coverslips were flicked off and slides were placed in methanol for 15–18 min at −18°C. After rehydration for 30 min in PBS with 0.1% Tween 20 (PBST) and blocking for 30 min with 10% fetal calf serum in PBST (PBST-F) the embryos were incubated for 2 h with primary antibodies diluted in PBST-F as indicated in Supplemental Table S4. Embryos were washed for 1 h in PBST, followed by incubation for 2 h with secondary antibodies diluted in PBST-F. Secondary antibodies were Alexa Fluor 546–conjugated goat anti-mouse antibodies (1:1000; Invitrogen, Carlsbad, CA) and Alexa Fluor 633–conjugated goat anti-rabbit antibodies (1:1000; Invitrogen). Embryos were washed again for 1 h in PBST and finally mounted with Mowiol containing 5 μg/ml Hoechst 33258. All steps were at room temperature. Confocal images were obtained with a Leica confocal SPE microscope equipped with a ACS APO 63×/1.3 objective and processed with ImageJ and Adobe Photoshop.
Western Blot
Embryos were obtained by hypochlorite treatment (1 N NaOH, 30% bleach solution) and disrupted by boiling and vortexing in SDS sample buffer together with 0.5-μm-diameter glass beads and separated by 10% SDS–PAGE. Proteins were transferred to Immobilon P membranes (Millipore), which were blocked with PBS containing 0.05% Tween-20 and 3% low-fat milk (PBST-M) and probed for 2 h at room temperature with antibodies diluted in PBST-M as described in Supplemental Table S4. Next membranes were washed with PBST for 1 h, incubated with peroxidase-conjugated secondary antibodies (1:5000–1:10,000; Sigma-Aldrich) for 2 h at room temperature, and developed with ECL Plus (GE Amersham, Piscataway, NJ).
Anoxia experiments
Ten gravid young hermaphrodites per condition were placed on a nematode growth medium plate for 1 h to lay embryos. The hermaphrodites were removed, and plates were placed in an anaerobic jar (Schütt Labortechnik, Göttingen, Germany) that was flushed with nitrogen gas (O2 < 0.1%). After 21 h at 20°C, plates were removed from the jar, and development was monitored for 4 d.
Yeast-two-hybrid assay
Yeast strain AH109 (Clontech; MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4?, gal80?, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1TATA-lacZ) was transformed using the LiAc method and selected in the appropriate synthetic complete (SC) minimal medium. Transformants containing pGBKT7-mdf-1 and either pGADT7-npp-5 or pAD-npp-19 were grown at 30°C to OD600 0.5 in SC-Leu-Trp medium and spotted as 10-fold serial dilutions to detect the ability to grow on minimal-medium plates lacking adenine and histidine. Growth was assayed after 3 d at 30°C. Combinations of empty pGADT7 and pGBKT7 vectors were also transformed into AH109 with each construct to assess self-activation.
Supplementary Material
Acknowledgments
We thank A. Arroyo, A. Desai, K. Kemphues, R. Kitagawa, and C. Luque for reagents and P. Alarcón and M. Rodríguez for technical assistance. V. Galy, A. Miranda-Vizuete, J. R. Martínez-Morales, and members of the Askjaer lab are acknowledged for discussion on the manuscript. This work was funded by the Spanish Ministry of Science and Innovation (BFU-2007-60116, BFU-2010-15478), the Spanish National Research Council (200820I028), the European Regional Development Fund, and the Regional Government of Andalusia (P07-CVI-02697). In addition, we acknowledge the Fundación Ramón Areces for a fellowship to E.R. and the Junta de Andalucía for institutional support. Some nematode strains used in this work were provided by the National Bioresource Project for the Nematode C. elegans (directed by Shohei Mitani), the C. elegans Gene Knockout Consortium, and the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN), which is funded by the National Institutes of Health, National Center for Research Resources.
Abbreviations used:
- CPC
chromosomal passenger complex
- NE
nuclear envelope
- NPC
nuclear pore complex
- Nup
nucleoporin
- PTC
premature termination codon
- SAC
spindle assembly checkpoint
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0927) on January 11, 2012.
REFERENCES
- Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Askjaer P, Galy V, Hannak E, Mattaj IW. Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell. 2002;13:4355–4370. doi: 10.1091/mbc.E02-06-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai SW, Rouquette J, Umeda M, Faigle W, Loew D, Sazer S, Doye V. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol Cell Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci USA. 2003;100:981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter GM, Furuta T, Kurasawa Y, Schumacher JM. Caenorhabditis elegans cyclin B3 is required for multiple mitotic processes including alleviation of a spindle checkpoint-dependent block in anaphase chromosome segregation. PLoS Genet. 2010;6:e1001218. doi: 10.1371/journal.pgen.1001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Essex A, Dammermann A, Lewellyn L, Oegema K, Desai A. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol Biol Cell. 2009;20:1252–1267. doi: 10.1091/mbc.E08-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–3531. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M, Jaedicke A, Mattaj IW. What can Caenorhabditis elegans tell us about the nuclear envelope? FEBS Lett. 2007;581:2794–2801. doi: 10.1016/j.febslet.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta BP, Sternberg PW. Tissue-specific regulation of the LIM homeobox gene lin-11 during development of the Caenorhabditis elegans egg-laying system. Dev Biol. 2002;247:102–115. doi: 10.1006/dbio.2002.0688. [DOI] [PubMed] [Google Scholar]
- Hajeri VA, Little BA, Ladage ML, Padilla PA. NPP-16/Nup50 function and CDK-1 inactivation are associated with anoxia-induced prophase arrest in Caenorhabditis elegans. Mol Biol Cell. 2010;21:712–724. doi: 10.1091/mbc.E09-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Katsani KR, Karess RE, Dostatni N, Doye V. In vivo dynamics of Drosophila nuclear envelope components. Mol Biol Cell. 2008;19:3652–3666. doi: 10.1091/mbc.E07-11-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussi YC, Shumaker DK, Shimi T, Fahrenkrog B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus. 2010;1:71–84. doi: 10.4161/nucl.1.1.10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystul TG, Goldmark JP, Padilla PA, Roth MB. Suspended animation in C. elegans requires the spindle checkpoint. Science. 2003;302:1038–1041. doi: 10.1126/science.1089705. [DOI] [PubMed] [Google Scholar]
- Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M, Santarella-Mellwig R, Posch M, Walczak R, Swedlow JR, Mattaj IW. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol Biol Cell. 2009;20:5260–5275. doi: 10.1091/mbc.E09-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Rodenas E, Klerkx EP, Ayuso C, Audhya A, Askjaer P. Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev Biol. 2009;327:399–409. doi: 10.1016/j.ydbio.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4362–4374. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci USA. 2009;106:14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. In WormBook. The Online Review of C. elegans Biology. http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html.
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Tanaka TU. Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J. 2010;29:4070–4082. doi: 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Yamamoto TG, Kitagawa R. Spindle assembly checkpoint gene mdf-1 regulates germ cell proliferation in response to nutrition signals in C. elegans. EMBO J. 2008;27:1085–1096. doi: 10.1038/emboj.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto TG, Watanabe S, Essex A, Kitagawa R. SPDL-1 functions as a kinetochore receptor for MDF-1 in Caenorhabditis elegans. J Cell Biol. 2008;183:187–194. doi: 10.1083/jcb.200805185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.